Abstract
Background
Hepatocellular carcinoma (HCC) is the most common type of liver malignancy. Despite significant progress in HCC treatment, resistance to chemotherapy and tumor metastasis are the main reasons for the unsatisfactory prognosis of HCC. Circular RNAs (circRNAs) have been extensively documented to play a role in the development of various types of cancer.
Aims
Here, we investigated the role of DEAD-box helicase 17 circRNA (circDDX17) in HCC and its underlying molecular mechanisms.
Methods
Our research employed various techniques including reverse transcription-quantitative polymerase chain reaction (RT-qPCR), cell counting kit-8 (CCK-8), flow cytometry, dual luciferase reporter assay, RNA immunoprecipitation (RIP), and western blot analysis. Additionally, we conducted a tumor xenograft assay to investigate the in vivo function of circDDX17.
Results
Firstly, the expression of circDDX17 was downregulated in HCC tissues and cells. Through functional experiments, it was observed that the overexpression of circDDX17 enhanced the sensitivity of sorafenib, promoted apoptosis, and inhibited the process of epithelial-mesenchymal transition (EMT) in vitro. Additionally, in vivo studies revealed that circDDX17 reduced tumor growth and increased sorafenib sensitivity. Mechanically, circDDX17 competitively combined miR-21-5p to suppress PTEN expression and activate the PI3K/AKT pathway. Furthermore, our rescue assays demonstrated that circDDX17 act as a tumor suppressor by blocking sorafenib resistance and tumorigenesis, while the inhibitory effect caused by circDDX17 upregulation was neutralized when miR-21-5p was overexpressed, PTEN was silenced, or the PI3K/AKT pathway was activated.
Conclusion
Our findings firstly confirmed that circDDX17 suppressed sorafenib resistance and HCC progression by regulating miR-21-5p/PTEN/PI3K/AKT pathway, which may provide novel biomarkers for the diagnosis, treatment and prognosis of HCC.
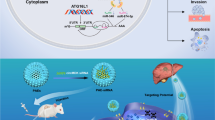
Graphical Abstract
Schematic illustration showed that the molecular mechanism by which circDDX17 regulates sorafenib resistance and tumor progression of HCC via the miR-21-5p/PTEN/PI3K/AKT pathway. In HCC cells, the expression of circDDX17 was decreased, and overexpression of circDDX17 promoted cell apoptosis, and enhanced sorafenib sensitivity and reduced EMT process. Besides, circDDX17 competitively combined miR-21-5p and miR-21-5p targeted suppression PTEN to regulate the activation of PI3K/AKT pathway, thus promoting the tumorigenesis of HCC.

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Hepatocellular carcinoma (HCC) is a prevalent primary liver cancer globally [1]. Major risk factors for the development of HCC include hepatitis B virus (HBV) and hepatitis C virus (HCV) infection [2]. Treatment options for HCC include surgical resection, liver transplantation, liver directed therapy and combination therapy [3]. Despite advancements in treatment, only a small percentage of patients achieve complete cure due to late-stage diagnosis [4]. Sorafenib is an oral multi-kinase inhibitor used to treat advanced HCC [5]. However, sorafenib resistance severely limits its therapeutic effect. Therefore, elucidation of the underlying mechanism of HCC onset and prognosis is essential for the treatment of HCC.
Circular RNAs (circRNAs) are highly conserved non-coding RNAs that are covalently closed structure produced by the back-splicing mechanism of precursor mRNAs during transcription [6,7,8]. Recently, more and more circRNAs have been identified, indicating that abnormally expressed circRNAs have a significant impact on HCC tumorigenesis and chemoresistance [9]. For example, circMRPS35 exerts a carcinogenic effect and resistance to cisplatin in HCC [7]. On the other hand, Circ_0003418 is downregulated in HCC, and silencing circ_003418 promotes cell proliferation, invasion, migration, cisplatin resistance and reduces cell apoptosis of HCC cells [10]. Strikingly, DEAD-box helicase 17 circRNA (circDDX17) is produced through reverse splicing from a linear transcript of exons 2–5 of the DDX17 gene [11]. Besides, circDDX17 has been reported as a tumor suppressor in many diseases, including colorectal cancer [12], prostate cancer [13], breast cancer [14] and myocarditis [11]. However, the role of circDDX17 in HCC has not been investigated.
MicroRNAs (miRNAs) are a class of small RNAs that are approximately 20–24 nt in length [15]. Accumulating evidences showed that miRNA also play an essential role in HCC tumorigenesis and chemoresistance. For instance, miR-541 acts as a tumor suppressor in HCC, and its overexpression inhibits HCC cells proliferation, metastasis and autophagy [16]. miR-182 is upregulated in cisplatin-resistant HCC cells, and its increases HCC growth and cisplatin resistance [17]. Notably, it is reported that miR-21-5p involved in the development of human cancers, including lung cancer [18], oropharyngeal cancer [19] and breast cancer [20]. In addition, increasing research focuses on the interaction between circRNAs and miRNAs. For example, circ_104348 has been found to promote cell growth, migration, invasion and suppress apoptosis in HCC by acting as a sponge for miR-187-3p [21]. However, the relationship between miR-21-5p and circDDX17 in HCC remains unclear.
As a classical pathway, the PTEN/PI3K/AKT signaling pathway has been reported to be involved in multiple biological processes in human diseases [22]. These diseases include prostate cancer [23], non-small-cell lung cancer [24], colon cancer [25] and HCC [26]. For example, brusatol suppresses cell growth and induces cell apoptosis of renal cancer cells through enhancing PTEN expression and blocking the expression levels of p-PI3K and p-AKT [27]. The combination of curcumin and glycyrrhetinic acid has been found to decrease cell proliferation and increase cell apoptosis and cell cycle arrest in HCC cells [28]. Moreover, in the non-tumor field, miR-26a-5p has been reported to activate the PI3K/AKT signaling pathway by silencing PTEN expression, thereby improving cardiomyocyte survival during myocardial ischemia/reperfusion (I/R) injury by inhibiting the PTEN/PI3K/AKT pathway [29]. Besides, miRNAs have been shown to regulate the PTEN/PI3K/AKT pathway in human cancer. For instance, miR-301a as an oncogene promotes ovarian cancer growth and resistance through inhibiting PTEN expression and increasing the expression of p-PI3K and p-AKT [30]. However, the relationship between PTEN/PI3K/AKT and miR-21-5p remains unknown.
Overall, our research focused on exploring the regulatory role and underlying mechanism of circDDX17 in HCC progression. We firstly indicated that circDDX17 was downregulated in HCC tissues and cells, and overexpression of circDDX17 promoted cell apoptosis, and enhanced sorafenib sensitivity and reduced EMT process. Furthermore, we identified that circDDX17 competitively bound to miR-21-5p and suppressed its activity, thereby targeting, a tumor suppressor gene. This interaction ultimately regulated the activation of the PI3K/AKT pathway. Further investigations suggested that overexpression of miR-21-5p, suppression of PTEN and activation of PI3K/AKT pathway by 740Y-P counteracted the anti-cancer effects induced by overexpression of circDDX17. These data hinted that circDDX17, miR-21-5p and the PTEN/PI3K/AKT pathway may hold significant potential for the development of effective treatment strategies for HCC.
Materials and Methods
Patient Sample
Tumor and adjacent normal tissues were collected from HCC patients from May 2021 to December 2022. Patients who underwent initial hepatectomy and were pathologically diagnosed with HCC were included in the study; however, patients with hepatocellular carcinoma who had received any treatment or experienced recurrence or other systemic diseases and subjects who refused to give informed consent were not included in the study. This study was approved by the Ethics Committee of The First Affiliated Hospital of Harbin Medical University, and was conducted in accordance with the Declaration of Helsinki. All patients provided informed written consent for the use of their tissues in our study.
Cell Culture and Vectors Transfection
Human HCCLM3, Huh7 cells and Transformed Human Liver Epithelial-2 (THEL-2) cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM) (Gibco BRL, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (FBS) with 5% CO2. oe-circDDX17, kd-PTEN, miR-21-5p mimic and their controls were transfected into HCC cells using Lipofectamine 2000 transfection reagent (Invitrogen, Madison, CA, USA) according to the manufacturer's instructions.
Reverse Transcription-Quantitative Polymerase Chain Reaction (RT-qPCR)
RNA was extracted from tissues and cells using the TRIzol reagent (Invitrogen), and reverse transcription was performed using the First-Strand cDNA Synthesis Kit (Fermentas, Lithuania). RT-qPCR was performed using SYBR Premix Ex Taq II (TaKaRa) with 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). The relative expression of target genes was calculated by the 2−ΔΔCt method and normalized by GAPDH or U6. For RNase R treatment, total RNA (2 μg) was incubated with or without 3 U/μg of RNase R (Epicentre Technologies, Madison, WI, USA) for 20 min at 37 °C. After treatment with RNase R, RT-qPCR was performed to determine the expression levels of DDX17 and circDDX17 mRNA. Primer sequences are shown in Table 1.
Western Blot
The total protein from HCCLM3 and Huh7 cells was isolated and quantified. Protein samples were then separated on SDS-PAGE gel and transferred to a nitrocellulose membrane with primary antibodies against PTEN, p-PI3K, PI3K, p-AKT (Ser473) and AKT (Cell Signaling Technology, Boston, MA, USA) at 4 ℃ overnight, then followed by adding the corresponding secondary antibody. The protein bands were visualized with ECL kit (Promoter, Wuhan, China) and quantified by Image J software (Image J Software, Inc., USA).
Dual Luciferase Reporter Assay
The sequence of circDDX17 containing miR-21-5p targeting sites was constructed into pmirGLO vectors to form WT-cricDDX17 and MUT-circDDX17 vectors. These vectors, along with control mimic or miR-21-5p mimic, were co-transfected into HCCLM3 and Huh7 cells. The luciferase activity was measured by a luciferase assay kit (Promega, Madison, WI).
RNA Immunoprecipitation (RIP)
The RIP assay was performed using the immunoprecipitation kit (Millipore, Bedford, MA, USA). To analyze the interaction of circDDX17 and miR-21-5p, HCCLM3 and Huh7 cells were lysed and incubated with magnetic heads containing anti-Ago2 or anti-IgG. Finally, RNA was extracted and analyzed by RT-qPCR.
Xenografted Tumor Model
Male BALB/c nude mice (6–8 weeks) were obtained from SLAC Laboratory Animal Co., Ltd. (Shanghai, China). Briefly, mice were randomly assigned to four groups: Control, sorafenib (SF), oe-circDDX17 and SF + oe-circDDX17. Treated HCCLM3 cells (5 × 106) from the above four groups were subcutaneously injected into mice. The tumor volume was measured every week. Finally, the mice were sacrificed and tumors were collected and weighted. All animal experiments were performed in accordance with ARRIVE guidelines, and were approved by the Ethics Committee of the First Affiliated Hospital of Harbin Medical University.
Cell Counting Kit-8 (CCK-8)
HCC cells were seeded into 96-well plates and then treated with different doses of sorafenib for 48 h. Then, the cells were cultured with 10 μL of CCK-8 solution for 2 h. Then, the absorbance was measured at 450 nm suing a Multi-Mode Reader (BioTek, Burlington, VT, USA). The half inhibition concentration (IC50) was assessed by the cell viability curve.
Flow Cytometry
Cell apoptosis was determined by using the Annexin V Apoptosis Detection Kit (Trevigen, Gaithersburg, MD, USA). HCCLM3 and Huh7 cells were washed and collected. Annexin V-fluorescein isothiocyanate (FITC) and propidium iodide (PI) (10 μL) were added to of the cell and incubated for 20 min in the dark at room temperature. Samples were analyzed by using a FACScan flow cytometer (Becton Dickinson, San Jose, CA, USA). The extent of cells in each group was circled by forward (FSC) and sideways (SSC), respectively. Since our sample was single cells, we chose the cross gate for circling; single-positive cells labeled with Annexin V were early apoptotic cells; double-positive cells labeled with both Annexin V and PI were late apoptotic cells; single-positive cells labeled with PI were necrotic cells; double-negative cells not labeled by either were living cells.
Statistical Analysis
Statistical analysis in our study was assessed by GraphPad Prism 8 software (GraphPad, San Diego, CA, USA), and all data were presented as the mean ± standard deviation (SD). Unpaired two-tailed Student’s t test was used to compare the difference between two groups, and the one-way ANOVA analysis was conducted to compare multiple groups. P < 0.05 was considered as statistically significant.
Results
CircDDX17 Expression Was Decreased in HCC Tissues and Cells
Firstly, we collected the HCC tissues and adjacent normal tissues, and we measured the expression levels of circDDX17 in 34 pairs of HCC tissues by RT-qPCR. As shown in Fig. 1A, circDDX17 was significantly reduced in HCC tissues compared with the adjacent normal tissues. Then, we detected the expression levels of circDDX17 in Transformed Human Liver Epithelial-2 (THEL-2) cells and human HCC cell lines (HCCLM3 and Huh7 cells), and we found that HCCLM3 and Huh7 cells show low expression compared to THEL-2 cells (Fig. 1B). Furthermore, in sorafenib-treated cells, the expression of circDDX17 was decreased in HCCLM3 and Huh7 cells compared with the THEL-2 cells (Fig. 1C). Besides, 35 HCC patient samples were divided into circDDX17 high expression and low expression groups based on mean values. The relationship between circDDX17 and the clinicopathological characteristics of HCC was explored, revealing a positive association between circDDX17 expression and HBV infection (Table 2). In addition, linear RNA expression was significantly reduced by RNase R treatment, while circDDX17 expression remained relatively stable, demonstrating that the circDDX17 that we used was a circular RNA (Fig. 1D). The above results indicated that circDDX17 was repressed in HCC.
CircDDX17 expression was decreased in HCC tissues and cells. A The expression of CircDDX17 in HCC tissues and adjacent normal tissues was detected by RT-qPCR. B The expression of CircDDX17 was measured by RT-qPCR in THEL-2 and HCC cells (HCCLM3 and Huh7). C The expression of CircDDX17 was measured in sorafenib (SF)-treated THEL-2, HCCLM3 and Huh7 cells by RT-qPCR. D The expression of linear and circular RNA with or without RNase R treatment. Data are expressed as the mean ± SD. Statistical analyses used Student’s t-test (A, D) and one-way ANOVA analysis (B, C). **P < 0.01; ***P < 0.001
Overexpression of circDDX17 Elevated Sorafenib Sensitivity, Apoptosis and Hampered EMT in HCC Cells
To investigated the impact of circDDX17 on HCC progression, we constructed and transfected circDDX17 overexpressing vector (oe-circDDX17) into HCCLM3 and Huh7 cells. First of all, RT-qPCR analysis illustrated a remarkably elevation of circDDX17 levels was observed in HCCLM3 and Huh7 cells after transfection of oe-circDDX17 vector compared with oe-NC vector (Fig. 2A). Then CCK-8 assay showed that IC50 of sorafenib was visibly reduced in HCCLM3 and Huh7 cells overexpressing circDDX17 compared to the oe-NC group (Fig. 2B). Cell apoptosis analysis suggested that overexpression of circDDX17 caused increased apoptosis levels of HCCLM3 and Huh7 cells under both control and sorafenib treatment conditions (Fig. 2C, D). Meanwhile, the expression of the anti-apoptotic gene Bcl-2 decreased while the expression of the pro-apoptotic gene Bax increased in both HCCLM3 and Huh7 cells after transfection with the oe-cricDDX17 vector, whether in the control or sorafenib-treated group (Fig. 2E, F). Furthermore, in HCCLM3 and Huh7 cells treated with either control or sorafenib, upregulation of circDDX17 induced the expression of E-cadherin and inhibited the expression of N-cadherin and Vimentin, suggesting overexpression of circDDX17 suppressed the EMT process (Fig. 2G–I). Taken together, circDDX17 induced sorafenib sensitivity and blocked HCC cell growth in vitro.
Overexpression of circDDX17 elevated sorafenib sensitivity, apoptosis and hampered EMT in HCC cells. A The expression of circDDX17 after transfection of overexpressing circDDX17 vectors in HCCLM3 and Huh7 cells. B IC50 of sorafenib was evaluated by CCK-8 assay after treatment with sorafenib. C, D Percentage of apoptotic cells was detected by flow cytometry in HCCLM3 and Huh7 cells treated with or without sorafenib after overexpression of circDDX17. E, F The apoptosis-related genes (Bcl-2 and Bax) was detected by RT-qPCR assay in HCCLM3 and Huh7 cells. G–I EMT-related markers (E-cadherin, N-cadherin and Vimentin) were measured by RT-qPCR analysis in HCCLM3 and Huh7 cells. Data are expressed as the mean ± SD. Statistical analyses used Student’s t-test (A–I). *P < 0.05; **P < 0.01; ***P < 0.001
Upregulation of circDDX17 Blocked Tumor Growth and Increased Sorafenib Sensitivity In Vivo
To further evaluate the role of circDDX17 in tumor growth and sorafenib sensitivity in vivo, the xenograft model was established. The results indicated that sorafenib injection and circDDX17 overexpression hampered tumor size, while the upregulation of circDDX17 combined with sorafenib inhibited tumor growth more effectively (Fig. 3A). Coherently, sorafenib and circDDX17 overexpression reduced the tumor volume and tumor weight, and the combination of the two exacerbated this inhibitory effect (Fig. 3B, C). These data disclosed that upregulation of circDDX17 enhanced sorafenib sensitivity and inhibited the growth of HCC xenografts.
Upregulation of circDDX17 blocked tumor growth and increased sorafenib sensitivity in vivo. A Tumors formed in subcutaneous xenograft tumors from Control, oe-DDX17, SF and SF + oe-DDX17 group. B Tumor volume in four groups was measured every week after inoculation. C Tumor weight was examined after mice were sacrificed. Data are expressed as the mean ± SD. Statistical analyses used one-way ANOVA analysis (B, C). *P < 0.05; **P < 0.01; ***P < 0.001
miR-21-5p Was the Downstream Target of circDDX17
According to a report, circRNAs have the ability to act as sponges for miRNAs [31]. Through the use of bioinformatics databases, we identified potential miRNAs that could directly bind to circDDX17. We utilized CircBank, miRanda, and StarBase to predict miRNAs that target CircDDX17. As shown in Fig. 4A, we screened six possible candidate miRNAs, among the six miRNAs, miR-371a-5p, miR-346, miR-21-5p and miR-191-5p were highly expressed in HCC according to previous reports (Fig. 4A). Next, the expression levels of miR-371a-5p, miR-346, miR-21-5p and miR-191-5p in HCC cells were measured by RT-qPCR, and we found that these miRNAs were all increased in HCC cells compared to THLE-2 cells (Fig. 4B). Among them, miR-21-5p exhibited a higher expression level and was selected for subsequent studies. The binding site between circDDX17 and miR-21-5p was shown in Fig. 4C. The dual luciferase reporter assay showed that the luciferase activity of circDDX17-WT was visibly reduced and circDDX17-MUT was not changed after treatment with miR-21-5p mimic in both HCCLM3 and Huh7 cells (Fig. 4D). Simultaneously, RIP assays revealed that Ago2 could accumulate circDDX17 and miR-21-5p (Fig. 4E). Additionally, RT-qPCR analysis highlighted that overexpression of circDDX17 notably depleted the mRNA expression of miR-21-5p in HCCLM3 and Huh7 cells (Fig. 4F). These data illustrated that miR-21-5p could bind to circDDX17.
miR-21-5p was the downstream target of circDDX17. A The online tools CircBank, miRanda and StarBase were used to predict miRNAs that might target circDDX17. B The expressions of miR-371a-5p, miR-346, miR-21-5p, and miR-191-5p were examined in pancreatic cancer and noncancerous tissues by RT-qPCR. C The binding sites between circDDX17 and miR-21-5p were predicted by bioinformatics websites. D The relationship between circDDX17 and miR-21-5p confirmed by dual luciferase reporter assay. E CircDDX17 and miR-21-5p co-immunoprecipitated with Ago2 revealed by RIP assay. F The level of miR-21-5p was measured in HCCLM3 and Huh7 cells after transfection with oe-circDDX17 and oe-NC. Data are expressed as the mean ± SD. Statistical analyses used Student’s t-test (D–F) and one-way ANOVA analysis (B). **P < 0.01; ***P < 0.001
CircDDX17 Regulated Chemosensitivity and Tumorigenesis of HCC by Sponging miR-21-5p
We have confirmed that circDDX17 can bind and negatively regulate miR-21-5p. Subsequently, we examined whether miR-21-5p impacts the regulatory role of circDDX17 in HCC. We upregulated both circDDX17 and miR-21-5p, and functional experiments was performed. The CCK-8 assay indicated that overexpression of circDDX17 greatly hampered IC50 of sorafenib in HCCLM3 and Huh7 cells, whereas miR-21-5p mimic diminished oe-circDDX17-induced inhibitory effects (Fig. 5A). Moreover, flow cytometry analysis suggested that the facilitation of apoptosis by upregulating circDDX17 could be ameliorated by miR-21-5p mimic in control or sorafenib-treated HCC cells (Fig. 5B, C). Consistent with the results above, in both control and sorafenib-treated HCCLM3 and Huh7 cells, the depletion of Bcl-2 and the upregulation of Bax induced by overexpressing circDDX17 was abolished by miR-21-5p mimic (Fig. 5D, E). Furthermore, RT-qPCR analysis showed that the upregulation of E-cadherin and the suppression of N-cadherin and Vimentin was observed in normal control or sorafenib-treated HCCLM3 and Huh7 cells overexpressing circDDX17 compared to their oe-NC counterparts, while this effect can be restored by overexpressing miR-21-5p, suggesting that miR-21-5p alleviated the inhibition of EMT caused by the upregulation of circDDX17 (Fig. 5F–H). Based on these data, we demonstrated that overexpression of miR-21-5p counteracted the inhibitory effect of circDDX17 on chemoresistance and tumorigenesis.
CircDDX17 regulated chemosensitivity and tumorigenesis of HCC by sponging miR-21-5p. A IC50 of sorafenib was evaluated by CCK-8 assay in HCCLM3 and Huh7 cells after transfection with control, oe-DDX17, oe-DDX17 + miR-NC, and oe-DDX17 + miR-mimic. B, C Percentage of apoptotic cells was detected by flow cytometry in HCCLM3 and Huh7 cells treated with or without sorafenib after transfection with control, oe-DDX17, oe-DDX17 + miR-NC, and oe-DDX17 + miR-mimic. D, E The expression of Bcl-2 and Bax was detected by RT-qPCR assay in HCCLM3 and Huh7 cells. F–H The expression of E-cadherin, N-cadherin and Vimentin was detected by RT-qPCR analysis in HCCLM3 and Huh7 cells. Data are expressed as the mean ± SD. Statistical analyses used one-way ANOVA analysis (A–H). *P < 0.05; **P < 0.01; ***P < 0.001
miR-21-5p Suppressed PTEN Activity and Enhanced the PI3K/AKT Signaling Pathway
To investigate the effect of miR-21-5p on PTEN/PI3K/AKT pathway, Western blot analysis was used to assessed the protein expression levels in HCCLM3 and Huh7 cells. The results revealed a decrease in PTEN levels and an increase in the expression of phosphorylated PI3K and AKT, suggesting that PTEN expression was inhibited and PI3K/AKT signal was activated after miR-21-5p mimic treatment inhibited PTEN expression and activated the PI3K/AKT signal, as compared to the NC mimic group (Fig. 6A–C). Additionally, we detected PTEN expression in both control and sorafenib treated HCC cells, we found that the levels of PTEN were greatly decreased in sorafenib-treated HCCLM3 and Huh7 cells compared with the sorafenib-untreated HCC cells (Fig. 6C). The above findings uncovered that miR-21-5p negatively regulated PTEN and activated PI3K/AKT pathway.
miR-21-5p suppressed PTEN activity and enhanced the PI3K/AKT signaling pathway. A, B The levels of PTEN, p-PI3K, PI3K, p-AKT, AKT were measured by Western blot analysis in NC mimic and miR-21-5p mimic treated HCCLM3 and Huh7 cells. C, D The protein expression of PTEN in HCC cells treated with or without sorafenib. Data are expressed as the mean ± SD. Statistical analyses used Student’s t-test (A–D). **P < 0.01; ***P < 0.001
CircDDX17 Regulated HCC Progression by Regulating PTEN/PI3K/AKT Pathway
Finally, we tested whether circDDX7 regulated HCC progression through the PTEN/PI3K/AKT pathway. We overexpressed circDDX17 and either knocked down of PTEN or activated the PI3K/AKT pathway by PI3K activator 740Y-P in control or sorafenib-treated HCCLM3 and Huh7 cells. The CCK-8 assay showed that the overexpression of circDDX17 effectively counteracted the inhibition of sorafenib's IC50, while this effect was reversed by PTEN knockdown or 740Y-P treatment (Fig. 7A). In addition, in control or sorafenib-treated HCC cells, cell apoptosis was measured by flow cytometry analysis, and we found that circDDX17-induced apoptotic cell death could be alleviated by either kd-PTEN or 740Y-P (Fig. 7B, C). Likewise, in both control and sorafenib-treated HCCLM3 and Huh7 cells overexpressing circDDX17, Bcl-2 was repressed and Bax was upregulated, but this pro-apoptotic effect of upregulation of circDDX17 was abrogated by inhibition of PTEN or 740Y-P co-treatment (Fig. 7D, E). Furthermore, the overexpression of circDDX17 in control or sorafenib-treated cells exhibited an anti-EMT effect, as shown in Fig. 7F–H. However, this effect was reversed by silencing PTEN or activating the PI3K/AKT pathway using 740Y-P. In conclusion, overexpressing circDDX17 facilitated sorafenib sensitivity, apoptosis and hampered EMT, while inhibition of PTEN or activation of the PI3K/AKT pathway offset the suppression of tumorigenesis by circDDX17.
CircDDX17 regulated HCC progression by regulating PTEN/PI3K/AKT pathway. A IC50 of sorafenib was evaluated by CCK-8 assay in control or sorafenib-treated HCCLM3 and Huh7 cells after transfection with control, oe-DDX17, oe-DDX17 + kd-PTEN, and oe-DDX17 + 740Y-P. B, C Percentage of apoptotic cells was detected by flow cytometry in control or sorafenib-treated HCCLM3 and Huh7 cells after transfection with control, oe-DDX17, oe-DDX17 + kd-PTEN, and oe-DDX17 + 740Y-P. D, E The expression of Bcl-2 and Bax was detected by RT-qPCR assay in HCCLM3 and Huh7 cells. F–H The expression of E-cadherin, N-cadherin and Vimentin was detected by RT-qPCR analysis in HCCLM3 and Huh7 cell. Data are expressed as the mean ± SD. Statistical analyses used one-way ANOVA analysis (A–H). *P < 0.05; **P < 0.01; ***P < 0.001
Discussion
HCC is one of the leading causes of cancer-related death in human worldwide [32]. However, current surgical treatments and combination therapies have limited effectiveness. Therefore, it is crucial to delve into the molecular mechanisms underlying HCC pathogenesis and identify potential target genes for therapeutic purposes. In recent years, there are numerous studies focusing on the role of circRNAs in cancers. Notably, circDDX17 has been reported to be a tumor suppressor in colorectal cancer [12], and circDDX17 reduces cell proliferation, invasion and EMT process in prostate cancer cells [13]. However, there are no studies about circDDX17 on HCC initiation and progression. In this report, we revealed that cricDDX17 was downregulated in HCC, and overexpression of circDDX17 induced apoptosis and inhibited EMT and sorafenib resistance of HCC. Taken together, we highlighted that the tumor suppressor function of circDDX17 in HCC progression.
The number of studies has confirmed that miRNAs can be involved in regulating cancer progression as diagnostic biomarkers and therapeutic targets [33]. In our study, we focused on miR-21-5p, which has been proved to be a potential binding miRNA for circDDX17 by dual-luciferase reporter assay and RIP assay. Furthermore, miR-21-5p has been identified as an oncogene in multiple human cancers. For instance, inhibition of miR-21-5p blocks tumorigenesis of lung cancer cells [18]. And colorectal cancer-secreted exosomes miR-21-5p induces angiogenesis and vascular permeability [34]. Additionally, HCC cells-derived exosomes miR-21 promotes cell proliferation, migration and reduces cell apoptosis [35]. Besides, circRNAs function as sponges for miRNAs, inhibiting their activity [36]. For example, circ_0008259 competitively binds to miR-21-5p, resulting in the upregulation of PDCD4 expression in osteosarcoma cells [37]. Similarly, circ_SAR1A impedes cell proliferation, migration, invasion and EMT and enhances cell apoptosis of lung cancer cells via sponging miR-21-5p. Here, we confirmed that circDDX17 sponged miR-21-5p to inhibit miR-21-5p expression, and upregulation of miR-21-5p ameliorated the malignant phenotype of HCC induced by overexpression of circDDX17. Based on the above studies, we demonstrated the effect of circDDX17 and miR-21-5p on HCC.
PTEN is commonly recognized as a tumor suppressor in various types of cancer [38]. Inhibition of PTEN exacerbates tumorigenesis by activating the PI3K/AKT signaling pathway [39]. For example, OTUD1 stabilizes PTEN to increase tyrosine kinase inhibitors sensitive in renal caner [40]. In addition, PTEN can be targeted by multiple different miRNAs. For instance, miR-425-5p inhibits PTEN and activates the PI3K/AKT pathway to augment lung cancer development [41]. In our data, we found that miR-21-5p repressed PTEN expression and activated the PI3K/AKT pathway, and inhibition of PTEN and activation of PI3K/AKT by 740Y-P can abolished the tumor-suppressing effects of circDDX17 overexpression. Our results shed light on the regulatory role of circDDX17 on miR-21-5p and PTEN/PI3K/AKT.
In conclusion, we found that circDDX17 expression in HCC was remarkably decreased and its upregulation hampered HCC tumorigenesis. Mechanically, we demonstrated the regulatory role of circDDX17 in HCC apoptosis, sorafenib resistance and EMT by modulation of the miR-21-5p/PTEN/PI3K/AKT axis. Collectively, targeting circDDX17/miR-21-5p/PTEN/PI3K/AKT may be a potential therapeutic intervention for HCC patients, which may serve as potential biomarkers and targets for HCC prognosis and therapy. Nevertheless, further investigation is required to determine the efficacy of targeting the circDDX17/miR-21-5p/PTEN/PI3K/AKT axis as a strategy for managing HCC and chemotherapy resistance in clinical settings.
Data availability
All data in our study are including in this article.
References
Wang W, Wei C. Advances in the early diagnosis of hepatocellular carcinoma. Genes Dis 2020; 7: 308–319. https://doi.org/10.1016/j.gendis.2020.01.014.
Nia A, Dhanasekaran R. Genomic landscape of HCC. Curr Hepatol Rep 2020; 19: 448–461. https://doi.org/10.1007/s11901-020-00553-7.
Kim DW, Talati C, Kim R. Hepatocellular carcinoma (HCC): beyond sorafenib-chemotherapy. J Gastrointest Oncol 2017; 8: 256–265. https://doi.org/10.21037/jgo.2016.09.07.
Wang T, Xu L, Jia R et al. miR-218 suppresses the metastasis and EMT of HCC cells via targeting SERBP1. Acta Biochim Biophys Sin (Shanghai) 2017; 49: 383–391. https://doi.org/10.1093/abbs/gmx017.
Llovet JM, Castet F, Heikenwalder M et al. Immunotherapies for hepatocellular carcinoma. Nat Rev Clin Oncol 2022; 19: 151–172. https://doi.org/10.1038/s41571-021-00573-2.
Li X, He J, Ren X et al. Circ_0003998 enhances doxorubicin resistance in hepatocellular carcinoma by regulating miR-218-5p/EIF5A2 pathway. Diagn Pathol 2020; 15: 141. https://doi.org/10.1186/s13000-020-01056-1.
Li P, Song R, Yin F et al. circMRPS35 promotes malignant progression and cisplatin resistance in hepatocellular carcinoma. Mol Ther 2022; 30: 431–447. https://doi.org/10.1016/j.ymthe.2021.08.027.
Chen L, Wang C, Sun H et al. The bioinformatics toolbox for circRNA discovery and analysis. Brief Bioinform 2021; 22: 1706–1728. https://doi.org/10.1093/bib/bbaa001.
Meng H, Niu R, Huang C et al. Circular RNA as a novel biomarker and therapeutic target for HCC. Cells. 2022. https://doi.org/10.3390/cells11121948.
Chen H, Liu S, Li M et al. Circ_0003418 inhibits tumorigenesis and cisplatin chemoresistance through Wnt/β-catenin pathway in hepatocellular carcinoma. Onco Targets Ther 2019; 12: 9539–9549. https://doi.org/10.2147/ott.S229507.
Liu T, Li Y, Chen S et al. CircDDX17 enhances coxsackievirus B3 replication through regulating miR-1248/NOTCH receptor 2 axis. Front Microbiol 2022; 13: 1012124. https://doi.org/10.3389/fmicb.2022.1012124.
Li XN, Wang ZJ, Ye CX et al. RNA sequencing reveals the expression profiles of circRNA and indicates that circDDX17 acts as a tumor suppressor in colorectal cancer. J Exp Clin Cancer Res 2018; 37: 325. https://doi.org/10.1186/s13046-018-1006-x.
Lin Q, Cai J, Wang QQ. The significance of circular RNA DDX17 in prostate cancer. Biomed Res Int 2020;2020:1878431. https://doi.org/10.1155/2020/1878431.
Peng HH, Wen YG. CircDDX17 acts as a competing endogenous RNA for miR-605 in breast cancer progression. Eur Rev Med Pharmacol Sci 2020; 24: 6794–6801. https://doi.org/10.26355/eurrev_202006_21668.
Xu P, Li X, Liang Y et al. PmiRtarbase: a positive miRNA-target regulations database. Comput Biol Chem 2022; 98: 107690. https://doi.org/10.1016/j.compbiolchem.2022.107690.
Xu WP, Liu JP, Feng JF et al. miR-541 potentiates the response of human hepatocellular carcinoma to sorafenib treatment by inhibiting autophagy. Gut 2020; 69: 1309–1321. https://doi.org/10.1136/gutjnl-2019-318830.
Qin J, Luo M, Qian H et al. Upregulated miR-182 increases drug resistance in cisplatin-treated HCC cell by regulating TP53INP1. Gene 2014; 538: 342–347. https://doi.org/10.1016/j.gene.2013.12.043.
Tang J, Li X, Cheng T et al. miR-21-5p/SMAD7 axis promotes the progress of lung cancer. Thorac Cancer 2021; 12: 2307–2313. https://doi.org/10.1111/1759-7714.14060.
Koleśnik M, Malm M, Drop B et al. miRNA-21-5p as a biomarker in EBV-associated oropharyngeal cancer. Ann Agric Environ Med 2023; 30: 77–82. https://doi.org/10.26444/aaem/156852.
Asadirad A, Khodadadi A, Talaiezadeh A et al. Evaluation of miRNA-21-5p and miRNA-10b-5p levels in serum-derived exosomes of breast cancer patients in different grades. Mol Cell Probes 2022; 64: 101831. https://doi.org/10.1016/j.mcp.2022.101831.
Huang G, Liang M, Liu H et al. CircRNA hsa_circRNA_104348 promotes hepatocellular carcinoma progression through modulating miR-187-3p/RTKN2 axis and activating Wnt/β-catenin pathway. Cell Death Dis 2020; 11: 1065. https://doi.org/10.1038/s41419-020-03276-1.
Carnero A, Blanco-Aparicio C, Renner O et al. The PTEN/PI3K/AKT signalling pathway in cancer, therapeutic implications. Curr Cancer Drug Targets 2008; 8: 187–198. https://doi.org/10.2174/156800908784293659.
Braglia L, Zavatti M, Vinceti M et al. Deregulated PTEN/PI3K/AKT/mTOR signaling in prostate cancer: still a potential druggable target? Biochim Biophys Acta Mol Cell Res 2020; 1867: 118731. https://doi.org/10.1016/j.bbamcr.2020.118731.
Pérez-Ramírez C, Cañadas-Garre M, Molina M et al. PTEN and PI3K/AKT in non-small-cell lung cancer. Pharmacogenomics 2015; 16: 1843–1862. https://doi.org/10.2217/pgs.15.122.
Zhou Y, Mu L, Liu XL et al. Tetrandrine inhibits proliferation of colon cancer cells by BMP9/PTEN/PI3K/AKT signaling. Genes Dis 2021; 8: 373–383. https://doi.org/10.1016/j.gendis.2019.10.017.
Liu J, Nie C. KDM5B regulates the PTEN/PI3K/Akt pathway to increase sorafenib-resistance in hepatocellular carcinoma. Anticancer Drugs 2022; 33: 840–849. https://doi.org/10.1097/cad.0000000000001329.
Wang T, Chen Z, Chen H et al. Brusatol inhibits the growth of renal cell carcinoma by regulating the PTEN/PI3K/AKT pathway. J Ethnopharmacol 2022; 288: 115020. https://doi.org/10.1016/j.jep.2022.115020.
Chang M, Wu M, Li H. Curcumin combined with glycyrrhetinic acid inhibits the development of hepatocellular carcinoma cells by down-regulating the PTEN/PI3K/AKT signalling pathway. Am J Transl Res 2017; 9: 5567–5575.
Xing X, Guo S, Zhang G et al. miR-26a-5p protects against myocardial ischemia/reperfusion injury by regulating the PTEN/PI3K/AKT signaling pathway. Braz J Med Biol Res 2020; 53: e9106. https://doi.org/10.1590/1414-431x20199106.
Ni J, Chen Y, Fei B et al. microRNA-301a promotes cell proliferation and resistance to apoptosis through PTEN/PI3K/Akt signaling pathway in human ovarian cancer. Gynecol Obstet Investig 2021; 86: 108–116. https://doi.org/10.1159/000513070.
Panda AC. Circular RNAs act as miRNA sponges. Adv Exp Med Biol 2018; 1087: 67–79. https://doi.org/10.1007/978-981-13-1426-1_6.
Akinyemiju T, Abera S, Ahmed M et al. The burden of primary liver cancer and underlying etiologies from 1990 to 2015 at the global, regional, and national level: results from the global burden of disease study 2015. JAMA Oncol 2017; 3: 1683–1691. https://doi.org/10.1001/jamaoncol.2017.3055.
Yang W, Xiao W, Cai Z et al. miR-1269b drives cisplatin resistance of human non-small cell lung cancer via modulating the PTEN/PI3K/AKT signaling pathway. Onco Targets Ther 2020; 13: 109–118. https://doi.org/10.2147/ott.S225010.
He Q, Ye A, Ye W et al. Cancer-secreted exosomal miR-21-5p induces angiogenesis and vascular permeability by targeting KRIT1. Cell Death Dis 2021; 12: 576. https://doi.org/10.1038/s41419-021-03803-8.
Cao LQ, Yang XW, Chen YB et al. Exosomal miR-21 regulates the TETs/PTENp1/PTEN pathway to promote hepatocellular carcinoma growth. Mol Cancer 2019; 18: 148. https://doi.org/10.1186/s12943-019-1075-2.
Piwecka M, Glažar P, Hernandez-Miranda LR et al. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science. 2017. https://doi.org/10.1126/science.aam8526.
Guan K, Liu S, Duan K et al. Hsa_circ_0008259 modulates miR-21-5p and PDCD4 expression to restrain osteosarcoma progression. Aging (Albany NY) 2021; 13: 25484–25495. https://doi.org/10.18632/aging.203769.
Ortega-Molina A, Serrano M. PTEN in cancer, metabolism, and aging. Trends Endocrinol Metab 2013; 24: 184–189. https://doi.org/10.1016/j.tem.2012.11.002.
Papa A, Pandolfi PP. The axis in cancer. Biomolecules. 2019. https://doi.org/10.3390/biom9040153.
Liu W, Yan B, Yu H et al. OTUD1 stabilizes PTEN to inhibit the PI3K/AKT and TNF-alpha/NF-kappaB signaling pathways and sensitize ccRCC to TKIs. Int J Biol Sci 2022; 18: 1401–1414. https://doi.org/10.7150/ijbs.68980.
Zhou JS, Yang ZS, Cheng SY et al. miRNA-425-5p enhances lung cancer growth via the PTEN/PI3K/AKT signaling axis. BMC Pulm Med 2020; 20: 223. https://doi.org/10.1186/s12890-020-01261-0.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
Xiaochuan Zhang conceived and wrote this paper; Wenyu Wang conducted the sample collection, investigation and data collation; Shanshan Mo and Xiaochuan Zhang conducted the experiment; Xueying Sun supervised and revised the article. All authors have read and agreed to the final version of the article.
Corresponding author
Ethics declarations
Conflict of interest
We declared that all authors have no conflicts of interests.
Ethical approval
This study was approved by the Ethics Committee of The First Affiliated Hospital of Harbin Medical University, and was conducted in accordance with the Declaration of Helsinki. All patients signed a written informed consent form. All animal experiments were performed in accordance with ARRIVE guidelines, and were approved by the Ethics Committee of the First Affiliated Hospital of Harbin Medical University.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, X., Wang, W., Mo, S. et al. DEAD-Box Helicase 17 circRNA (circDDX17) Reduces Sorafenib Resistance and Tumorigenesis in Hepatocellular Carcinoma. Dig Dis Sci 69, 2096–2108 (2024). https://doi.org/10.1007/s10620-024-08401-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10620-024-08401-0