Abstract
Background
With the rise of liquid biopsy in oncology, circulating miRNAs have become one of the most promising noninvasive biomarkers for early detection of hepatocellular carcinoma (HCC). However, a reliable HCC-related circulating miRNA panel and corresponding diagnostic model remain to be explored.
Methods
Five large public datasets related to intact miRNA profiles in the serum or tumors of HCC patients were included and divided into training cohorts (GSE113740 and TCGA-LIHC) and validation cohorts (GSE112264, GSE113486 and GSE106817). Compared with non-cancer controls and high-risk patients, key miRNAs dysregulated in both the serum and tumors of HCC patients were identified by differential expression analysis and overlapping analysis. The corresponding diagnostic model was constructed by LASSO logistic regression and evaluated by receiver operating characteristic curves and a nomogram with calibration plot.
Results
A distinctive panel of HCC-related circulating miRNAs, including three upregulated miRNAs (miR-184, miR-532-5p, miR-221-3p) and three downregulated miRNAs (miR-5589-5p, let-7b-3p, miR-26b-3p), were rigorously screened out, all of which displayed significant discriminability between HCC patients and controls (all P < 0.05). In addition, a reliable six-circulating miRNA-based diagnostic score was constructed and displayed robust diagnostic ability for HCC (particularly for early-stage HCC) (AUC = 0.9535, P < 0.05) compared with that of the serum α-fetoprotein test. Importantly, its efficacy was sufficiently validated in three independent datasets (AUC = 0.9780/0.9961/0.9681, all P < 0.05). Furthermore, a visual nomogram based on the diagnostic score was correspondingly established to strengthen its clinical applicability.
Conclusion
The six-circulating miRNA-based diagnostic score may be a reliable noninvasive biomarker for early-stage HCC screening and dynamic monitoring of postoperative recurrence.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Despite considerable advances in the clinical management of hepatocellular carcinoma (HCC) over the past decades, HCC remains the fourth leading cause of cancer mortality worldwide, which is mainly attributed to the unsatisfactory early detection rate and limited treatments for patients with advanced HCC [1, 2]. From a clinical perspective, enhancing early screening for HCC, particularly among high-risk patients with chronic hepatitis or cirrhosis, is vital for increasing the overall prognosis of HCC [2]. Meanwhile, timely and effective postoperative surveillance for recurrent tumors is also of great significance for patients with HCC [3].
Currently, the combined utilization of ultrasonography and serum α-fetoprotein (AFP) is a widely available, affordable and noninvasive strategy for HCC surveillance [2]. Nevertheless, their sensitivity and specificity for early-stage HCC screening are insufficient [4]. In light of this, advanced and diagnostic imaging techniques with higher accuracy and reliability, such as contrast-enhanced computed tomography (CT) and magnetic resonance imaging (MRI), have been proposed as necessary supplements for HCC surveillance among high-risk patients; however, the cost, availability and risks of radiation and contrast agents should not be ignored [2]. In addition, the inherent risk of tissue biopsy also limits its application for early-stage HCC screening [2]. Thus, the optimal and reliable means for early detection of HCC is still evolving, and benefits should be fully weighed against health costs, potential physical harms and overdiagnosis.
MicroRNAs (miRNAs), a class of small noncoding RNAs, are implicated in various biological processes mainly by posttranscriptionally regulating gene expression [5]. Correlations between miRNA dysregulation and cancer initiation or progression have been widely identified [5]. Compared to tissue biopsy, the advantages of liquid biopsy have been highlighted, such as abundant sources (particularly blood), noninvasive techniques, and dynamic monitoring abilities by frequent sampling [3]. Interestingly, alterations of tumor-specific miRNA profiles in serum reflecting cancer formation at the early phase (including HCC) have been widely reported in recent years [6,7,8,9]. As circulating miRNAs in body fluids are stable, they have become one of the most promising and fast-growing noninvasive biomarkers in the field of liquid biopsy for cancer screening [6, 7, 10, 11]. However, it is undeniable that previous studies related to HCC-related circulating miRNAs are generally limited by a small sample size, a lack of adequate external validation or a defective study design [12,13,14,15]. Thus, more reliable circulating miRNA signatures associated with HCC tumorigenesis remain to be explored, and excellent circulating miRNA-based diagnostic models are highly anticipated.
In this study, based on multiple large-scale datasets from public databases, we aimed to screen out more reliable HCC-related miRNAs with dysregulated expression in both serum and tumors by a rigorous study design. Moreover, a novel and powerful circulating miRNA-based diagnostic score for early-stage HCC screening was further established and fully validated.
Materials and Methods
Data Preparation
By thoroughly searching the Gene Expression Omnibus (GEO) database, four datasets (GSE113740, GSE112264, GSE113486 and GSE106817) related to the circulating miRNA profiles of patients with HCC were obtained. From the GSE113740 datasets, 1453 serum samples were included in this study, including 345 from HCC patients, 139 from patients with chronic liver disease (CLD) (liver cirrhosis (LC), n = 93; chronic hepatitis (CH), n = 46) and 969 from healthy individuals (non-cancer control, NC). The GSE113486 dataset provided 972 serum samples, including 40 from HCC patients, 832 from patients with other tumors (OT) and 100 from NC. The GSE112264 dataset was consisted of 1350 serum samples, including 50 from HCC patients, 1259 from OT patients and 41 from NC. Serum samples from 81 HCC patients and 2759 NC from the GSE106817 dataset were also enrolled in this study. According to the statements in the original articles regarding the four datasets, three internal control miRNAs (including miR-149-3p, miR-2861, and miR-4463) were utilized to normalize the raw miRNA signal [15,16,17,18]. Quantile normalization of the datasets was further performed in the present study. In addition, the normalized miRNA-seq data and relatively intact clinical information of 250 HCC tissues and 49 non-cancer controls were downloaded from The Cancer Genome Atlas (TCGA) database using the UCSC-Xena platform (https://xenabrowser.net/). All patients with HCC enrolled in this study were pathologically diagnosed.
Screening of HCC-Related Circulating miRNAs
To screen HCC-related circulating miRNAs, the differentially expressed miRNAs in serum samples from HCC patients compared with those of CLD patients and NC were first screened out. Then, the differentially expressed miRNAs in HCC tissues compared to NC were obtained. After that, overlapping analyses between the two datasets were performed to identify the potential miRNAs dysregulated in both the serum and tumors of HCC patients. Differential expression analysis was performed using the R package “limma”, and “fold change (FC) > 2 with adjusted P-value < 0.05” was used as the threshold.
Circulating miRNAs Selection and Model Construction
To construct a circulating miRNA-based diagnostic score using the GSE113740 dataset, LASSO-penalized logistic regression analysis was first conducted to select the optimal circulating miRNA set by using the R package “glmnet” based on tenfold cross-validation and lambda.min. The Z-score was computed for each selected miRNA as its relative expression level during model construction. Then, the selected circulating miRNAs were enrolled into a multivariate logistic regression model to calculate their coefficients (β values). The circulating miRNA-based diagnostic score was constructed using the following formula: diagnostic score = (β1 * Z-score of miRNA1) + (β2* Z-score of miRNA2) + … + (βn* Z-score of miRNAn).
Validation and Visualization of the Model
The receiver operating characteristic (ROC) curve was used to evaluate the diagnostic value of the circulating miRNA-based model for HCC. Meanwhile, for visualization of the diagnostic model, a convenient nomogram was further built using the R package “rms” and comprehensively assessed by calibration plot and concordance index (C-index). In addition, three independent datasets (GSE112264, GSE113486 and GSE106817) were utilized to externally validate the discriminability of the model for HCC.
Statistical Analysis
Continuous variables between different groups were compared by the Mann–Whitney test or one-way analysis of variance. Logistic regression model analysis was used to determine the factors associated with HCC. The optimal cutoff values of the miRNAs levels for survival analysis were calculated by X-tile software (v3.6.1), and their prognostic values in terms of overall survival and recurrence-free survival were analyzed by Kaplan–Meier analysis and log-rank test. GraphPad Prism 8.0.2, SPSS 22.0 and R 3.4.2 were applied for statistical analysis and graphing. A P-value < 0.05 was deemed statistically significant.
Results
Screening of Key HCC-Related miRNAs
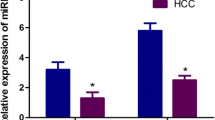
The overall design of this study was summarized in a flowchart (Fig. 1) Based on the stringent screening criteria mentioned above, 155 circulating miRNAs (including 53 upregulated miRNAs and 102 downregulated miRNAs) that could distinguish HCC patients from both CLD patients and NC were found. Then, 136 dysregulated miRNAs (including 33 upregulated miRNAs and 103 downregulated miRNAs) in HCC tissues compared with NC were screened out. Next, by performing overlapping analysis, six key miRNAs (miR-184, miR-221-3p, miR-532-5p, let-7b-3p, miR-26b-3p, miR-5589-5p) dysregulated in both serum and tumors of HCC patients were ultimately obtained (Fig. 2). The clinical implications of the dysregulated miRNAs in HCC tissues were preliminarily explored and are shown in Supplementary Table 1 and Supplementary Fig. 1.
Screening of HCC-related circulating miRNAs. (a) Identification of key miRNAs dysregulated in serum and tissues of HCC patients by differential expression analysis and overlapping analyses. (b-c) Expression levels of key miRNAs in serum (GSE113740) and liver tissues (TCGA) of different populations. CH chronic hepatitis, CLD chronic liver diseases, Down downregulation, HCC hepatocellular carcinoma, NC non-cancer control (healthy individual), NT non-cancer tissue, LC liver cirrhosis, Up upregulation
Construction of Circulating miRNA-Based Diagnostic Score
By performing LASSO-penalized logistic regression analysis using the GSE113740 dataset, it was suggested that the optimal modeling approach was combined utilization of the six circulating miRNAs (Fig. 3). The correlations between the six circulating miRNAs levels and HCC occurrence were identified by univariate logistic regression analysis (all P < 0.05). Following calculation of their coefficients using multivariate logistic regression analysis, the circulating miRNA-based diagnostic score was developed. Diagnostic score = (0.483* Z-score of miR-184) + (0.338* Z-score of miR-532-5p) + (4.437* Z-score of miR-221-3p) + (− 0.276* Z-score of miR-5589-5p) + (− 0.161* Z-score of let-7b-3p) + (− 0.421* Z-score of miR-26b-3p) (Table 1).
Internal Validation and Visualization of the Model
As shown in Fig. 4A, the diagnostic scores of HCC patients at different clinical stages (BCLC staging system, I-IV) were significantly higher than those of the non-HCC individuals (including CLD patients and NC). ROC analyses indicated the excellent discriminability of the model for HCC (HCC vs non-HCC: AUC = 0.9535, HCC vs NC: AUC = 0.9559, HCC vs CH: AUC = 0.8963, HCC vs LC: AUC = 0.9569, all P < 0.0001), which was higher than that of each of the six circulating miRNAs (Fig. 4B-C, Supplementary Fig. 2). In addition, we further evaluated the diagnostic value of the model for HCC at different clinical stages. Surprisingly, the model displayed high discriminability for both early-stage HCC and advanced HCC (HCC-stage I vs Non-HCC, AUC = 0.9490, HCC-stage II vs Non-HCC, AUC = 0.9508, HCC-stage III vs Non-HCC, AUC = 0.9547, HCC-stage IV vs Non-HCC, AUC = 0.9315, all P < 0.0001) (Fig. 4D). In contrast, the diagnostic value of serum AFP level for HCC, particularly for early-stage HCC, was relatively undesirable (HCC vs Non-HCC: AUC = 0.6840, P < 0.0001; HCC-stage I vs Non-HCC, AUC = 0.5702, P = 0.0583; HCC-stage II vs Non-HCC, AUC = 0.6810, P < 0.0001, HCC-stage III vs Non-HCC, AUC = 0.7968, P < 0.0001; HCC-stage IV vs Non-HCC, AUC = 0.8895, P < 0.0001) (Fig. 5).
Internal validation of the diagnostic ability of six-circulating miRNA-based diagnostic scores for HCC in the GSE113740 dataset. (a) Comparison of the diagnostic score in different populations. (b-c) ROC analyses evaluating the accuracy of the model for distinguishing HCC patients from non-HCC patients or specific populations (including healthy individuals (NC) and patients with chronic hepatitis (CH) or liver cirrhosis (LC)). (d) ROC analyses evaluating the diagnostic value of the model for HCC at different clinical stages (BCLC staging system)
Evaluating the diagnostic value of serum AFP levels for HCC in the GSE113740 dataset. (a) Comparison of serum AFP levels in different populations. (a) ROC analysis evaluating the accuracy of serum AFP for distinguishing HCC patients from non-HCC patients. (c) ROC analyses evaluating the diagnostic value of serum AFP for HCC at different clinical stages (BCLC staging system). AFP α-fetoprotein, CH chronic hepatitis, HCC hepatocellular carcinoma, LC liver cirrhosis
To enhance the clinical applicability of the model, a visual nomogram was developed to expediently evaluate the probability of HCC based on each diagnostic score (Fig. 6A). The C-index of the nomogram was equivalent to the AUC mentioned above (0.9535). The calibration plot showed high consistency between the nomogram-predicted probability of HCC and the actual probability of HCC, indicating that the nomogram was an excellent model for HCC diagnosis (Fig. 6B).
External Validation of the Model
To deeply assess the diagnostic power of the model, three independent datasets were utilized for external validation. As shown in Fig. 7, consistent with the results described above, ROC analyses suggested that the diagnostic score could not only effectively distinguish HCC patients from non-cancer control, but also displayed good separability between HCC patients and OT patients (GSE113486: HCC vs NC, AUC = 0.9780; HCC vs OT, AUC = 0.8602; GSE112264: HCC vs NC, AUC = 0.9961; HCC vs OT, AUC = 0.8324; GSE106817: HCC vs NC, AUC = 0.9681, all P < 0.0001).
External validation of the six-circulating miRNA-based diagnostic score. The diagnostic scores of different populations and ROC analyses evaluating the accuracy of the model for distinguishing HCC patients from non-cancer patients (NC) or patients with other tumors (OT) using three independent cohorts, including GSE113486 (a), GSE112264 (b) and GSE106817 (c)
Discussion
Circulating miRNAs in serum can remain fairly stable either by exosomal encapsulation or binding to proteins, thereby escaping RNase-mediated degradation and tolerating harsh conditions (such as variation of pH or temperature) [19]. Tumor-specific circulating miRNAs are expected to be one of the most promising and competitive noninvasive biomarkers in clinical practice, ranging in utility from early cancer screening to postoperative recurrence monitoring [10]. To date, many HCC-related single circulating miRNAs or circulating miRNA panels have been found, such as miR-122, miR-21, miR-92-3p, miR-3126-5p, miR-107, miR-320b and miR-6724-5p [12, 15, 20, 21]. However, highly reliable circulating individual miRNA or miRNA panels for early-stage HCC screening remain to be identified for the following reasons. First, the sample size in most of the previous studies was relatively small; thus, the relevant results are not sufficiently reliable [14, 22, 23]. Second, the overall study design used to explore HCC-related circulating miRNAs was relatively biased and unreasonable in some studies. For example, some studies just aimed to determine the consistent dysregulation of certain miRNAs in the serum of HCC patients according to the reported miRNAs dysregulated in HCC tissues or in the serum of patients with other tumors in previous studies [24,25,26,27]. Some studies merely focused on circulating miRNA profiles without taking miRNA profiles in HCC tissues into consideration, and there might be inadequate identification of circulating miRNAs dysregulation truly originating from pathological changes in tumors [15, 28,29,30]. Furthermore, the generally limited accuracy of single circulating miRNAs hinted at the necessity for the construction of diagnostic model based on multiple circulating miRNAs to enhance screening ability [24, 28]. Finally, previous studies were usually conducted based on single-center cohorts, so the existing circulating miRNAs and diagnostic models generally lacked powerful external validation using sufficient independent cohorts [12, 15, 29].
Given the considerations mentioned above, we included data from five large-scale reliable public datasets related to intact miRNA profiles in the serum or tumor tissues of HCC patients. Based on rigorous and comprehensive screening strategies, six key miRNAs (miR-184, miR-5589-5p, miR-532-5p, let-7b-3p, miR-221-3p, miR-26b-3p) dysregulated in both serum and tumor tissues of HCC patients were systematically identified, all of which displayed significant discriminability between HCC patients and controls (including healthy individuals and high-risk patients). Moreover, to enhance the accuracy of the above circulating miRNAs for HCC screening, a novel six-circulating miRNA-based diagnostic score was developed by appropriate statistical methods. With reference to the diagnostic efficacy of the AFP test, the robust diagnostic ability of our model for HCC (particularly for early-stage HCC) was highlighted. In addition, a visual nomogram based on the diagnostic score was correspondingly established to strengthen its clinical applicability. Most importantly, sufficient external validation was strictly carried out by utilizing three independent datasets with large sample sizes. The consistent results strongly indicated that the six-circulating miRNA-based diagnostic score might be an ideal and reliable model for noninvasive HCC screening.
In line with our findings, dysregulation of the six miRNAs in HCC tissues and their underlying biological roles or clinicopathologic implications have been revealed more or less in previous studies (Supplementary Fig. 3) [31,32,33,34,35,36]. To some extent, the close involvement of the above miRNAs with HCC initiation and progression strongly indicated that the corresponding dysregulation of circulating miRNA profiles could timely and rationally mirror the pathophysiological changes of HCC. Among the six circulating miRNAs, serum miR-221-3p exhibited the highest diagnostic accuracy for HCC. In light of the consistent findings widely demonstrated in previous studies, upregulation of circulating miR-221-3p may be an excellent and reliable indicator for HCC [37, 38]. To date, dysregulation of the other five miRNAs in the serum of HCC patients has rarely been reported. Besides, with reference to the circulating miRNAs levels in HCC, the consistent or opposite miRNA levels (including miR-221-3p, miR-532-5p, miR-26-3p, let-7b and miR-184) in the serum of patients with other cancers revealed in existing studies are worth mentioning [39,40,41,42,43,44,45,46]. This highlights that our distinctive circulating miRNA panel may encompass cancer hallmarks and heterogeneity, thereby demonstrating its excellent power to distinguish between HCC patients and non-cancer populations or OT patients. Of note, changes in certain circulating miRNA levels (such as miR-221 and miR-532) following radical cancer therapy have been observed, which indicates that our model may also be feasible for postoperative monitoring [39, 47,48,49,50]. More importantly, the mechanisms underlying the alterations of the circulating miRNAs expression in HCC in terms of their specific sources (HCC cells or non-HCC cells in tumor microenvironment) and biological roles (cell-to-cell communication) should be further elucidated in the future [10, 51].
Apart from the excellent results achieved in this study, there are still several limitations. First, detailed clinicopathological characteristics (such as tumor size, numbers and histological grade) and survival data of HCC patients were not available in the four circulating miRNA profile datasets. Thus, specific clinical implications or prognostic values of the six circulating miRNAs could not be fully analyzed. In addition, although the accuracy of the diagnostic score for HCC has been effectively validated in this study, its ability to detect early-stage HCC remains to be further evaluated compared with more powerful references (such as CT, MRI and other serologic biomarkers (including protein induced by vitamin k absence or antagonist-II, PIVKA-II)) by large prospective studies to obtain further evidence of its efficacy. Finally, the potential benefits of combining circulating miRNA-based diagnostic model with other surveillance tests (such as ultrasonography) for early-stage HCC screening need to be further studies.
In conclusion, we identified a distinctive panel of HCC-related circulating miRNAs based on reliable large-scale datasets and rigorous study design. In addition, a reliable six-circulating miRNA-based diagnostic score and its corresponding visual nomogram for HCC detection were developed and fully validated. With the development of noninvasive liquid biopsy in oncology, this model may serve as a powerful option in clinical practice for early screening of HCC and dynamic monitoring on postoperative recurrence, thereby guaranteeing the optimal timing of radical interventions.
References
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424.
Yang JD, Heimbach JK. New advances in the diagnosis and management of hepatocellular carcinoma. BMJ. 2020;371:m3544.
Ye Q, Ling S, Zheng S, Xu X. Liquid biopsy in hepatocellular carcinoma: circulating tumor cells and circulating tumor DNA. Mol Cancer. 2019;18:114.
Tzartzeva K, Obi J, Rich NE et al. Surveillance imaging and alpha fetoprotein for early detection of hepatocellular carcinoma in patients with cirrhosis: a meta-analysis. Gastroenterology. 2018;154:1706–1718.
Goodall GJ, Wickramasinghe VO. RNA in cancer. Nat Rev Cancer. 2021;21:22–36.
Otandault A, Anker P, Al Amir DZ et al. Recent advances in circulating nucleic acids in oncology. Ann Oncol. 2019;30:374–384.
Peng C, Ye Y, Wang Z et al. Circulating microRNAs for the diagnosis of hepatocellular carcinoma. Dig Liver Dis. 2019;51:621–631.
Zhao XF, Li N, Lin DD, Sun LB. Circulating MicroRNA-122 for the diagnosis of hepatocellular carcinoma: a meta-analysis. Biomed Res Int. 2020;2020:5353695.
Sorop A, Iacob R, Iacob S et al. Plasma small extracellular vesicles derived miR-21-5p and miR-92a-3p as potential biomarkers for hepatocellular carcinoma screening. Front Genet. 2020;11:712.
Anfossi S, Babayan A, Pantel K, Calin GA. Clinical utility of circulating non-coding RNAs-an update. Nat Rev Clin Oncol. 2018;15:541–563.
Heitzer E, Haque IS, Roberts CES, Speicher MR. Current and future perspectives of liquid biopsies in genomics-driven oncology. Nat Rev Genet. 2019;20:71–88.
Zhang Y, Li T, Qiu Y et al. Serum microRNA panel for early diagnosis of the onset of hepatocellular carcinoma. Medicine (Baltimore). 2017;96:e5642.
Weis A, Marquart L, Calvopina DA, Genz B, Ramm GA, Skoien R. Serum MicroRNAs as biomarkers in hepatitis C: preliminary evidence of a MicroRNA panel for the diagnosis of hepatocellular carcinoma. Int J Mol Sci. 2019;20:864.
Moshiri F, Salvi A, Gramantieri L et al. Circulating miR-106b-3p, miR-101-3p and miR-1246 as diagnostic biomarkers of hepatocellular carcinoma. Oncotarget. 2018;9:15350–15364.
Yamamoto Y, Kondo S, Matsuzaki J et al. Highly sensitive circulating microRNA panel for accurate detection of hepatocellular carcinoma in patients with liver disease. Hepatol Commun. 2020;4:284–297.
Urabe F, Matsuzaki J, Yamamoto Y et al. Large-scale circulating microRNA profiling for the liquid biopsy of prostate cancer. Clin Cancer Res. 2019;25:3016–3025.
Usuba W, Urabe F, Yamamoto Y et al. Circulating miRNA panels for specific and early detection in bladder cancer. Cancer Sci. 2019;110:408–419.
Yokoi A, Matsuzaki J, Yamamoto Y et al. Integrated extracellular microRNA profiling for ovarian cancer screening. Nat Commun. 2018;9:4319.
Shigeyasu K, Toden S, Zumwalt TJ, Okugawa Y, Goel A. Emerging role of microRNAs as liquid biopsy biomarkers in gastrointestinal cancers. Clin Cancer Res. 2017;23:2391–2399.
Jin Y, Wong YS, Goh BKP et al. Circulating microRNAs as potential diagnostic and prognostic biomarkers in hepatocellular carcinoma. Sci Rep. 2019;9:10464.
Fouda MS, Omran MM, Tarek G, Hady AAWA. Development of a novel panel based on micro-RNAs (21, 29a, 200 and 335) and alpha-fetoprotein as diagnostic biomarkers for hepatocellular carcinoma associated with hepatitis C infection. Arab J Gastroenterol. 2020;22:28–33.
Elhendawy M, Abdul-Baki EA, Abd-Elsalam S et al. MicroRNA signature in hepatocellular carcinoma patients: identification of potential markers. Mol Biol Rep. 2020;47:4945–4953.
Pascut D, Cavalletto L, Pratama MY et al. Serum miRNA are promising biomarkers for the detection of early hepatocellular carcinoma after treatment with direct-acting antivirals. Cancers (Basel). 2019;11:1773.
Sun Q, Li J, Jin B, Wang T, Gu J. Evaluation of miR-331-3p and miR-23b-3p as serum biomarkers for hepatitis c virus-related hepatocellular carcinoma at early stage. Clin Res Hepatol Gastroenterol. 2020;44:21–28.
Mohamed AA, Omar AAA, El-Awady RR et al. MiR-155 and MiR-665 role as potential non-invasive biomarkers for hepatocellular carcinoma in egyptian patients with chronic hepatitis C virus infection. J Transl Int Med. 2020;8:32–40.
Cho HJ, Kim SS, Nam JS et al. Low levels of circulating microRNA-26a/29a as poor prognostic markers in patients with hepatocellular carcinoma who underwent curative treatment. Clin Res Hepatol Gastroenterol. 2017;41:181–189.
Bhattacharya S, Steele R, Shrivastava S, Chakraborty S, Di Bisceglie AM, Ray RB. Serum miR-30e and miR-223 as novel noninvasive biomarkers for hepatocellular carcinoma. Am J Pathol. 2016;186:242–247.
Xue X, Zhao Y, Wang X, Qin L, Hu R. Development and validation of serum exosomal microRNAs as diagnostic and prognostic biomarkers for hepatocellular carcinoma. J Cell Biochem. 2019;120:135–142.
An Y, Gao S, Zhao WC et al. Novel serum microRNAs panel on the diagnostic and prognostic implications of hepatocellular carcinoma. World J Gastroenterol. 2018;24:2596–2604.
Zhu HT, Liu RB, Liang YY et al. Serum microRNA profiles as diagnostic biomarkers for HBV-positive hepatocellular carcinoma. Liver Int. 2017;37:888–896.
Gao B, Gao K, Li L, Huang Z, Lin L. miR-184 functions as an oncogenic regulator in hepatocellular carcinoma (HCC). Biomed Pharmacother. 2014;68:143–148.
Chen Z, Xiang B, Qi L, Zhu S, Li L. miR-221-3p promotes hepatocellular carcinogenesis by downregulating O6-methylguanine-DNA methyltransferase. Cancer Biol Ther. 2020;21:915–926.
Jiang ZT, Han Y, Liu XY, Lv LY, Pan JH, Liu CD. Tripterine restrains the aggressiveness of hepatocellular carcinoma cell via regulating miRNA-532-5p/CXCL2 axis. Onco Targets Ther. 2020;13:2973–2985.
He Z, Deng W, Jiang B et al. Hsa-let-7b inhibits cell proliferation by targeting PLK1 in HCC. Gene. 2018;673:46–55.
Hu X, Wang R, Ren Z et al. MiR-26b suppresses hepatocellular carcinoma development by negatively regulating ZNRD1 and Wnt/β-catenin signaling. Cancer Med. 2019;8:7359–7371.
Yu B, Liang H, Ye Q, Wang Y. Upregulation of ERCC6L is associated with tumor progression and unfavorable prognosis in hepatocellular carcinoma. J Gastrointest Oncol. 2020;11:1009–1023.
Sohn W, Kim J, Kang SH et al. Serum exosomal microRNAs as novel biomarkers for hepatocellular carcinoma. Exp Mol Med. 2015;47:e184.
Li J, Wang Y, Yu W, Chen J, Luo J. Expression of serum miR-221 in human hepatocellular carcinoma and its prognostic significance. Biochem Biophys Res Commun. 2011;406:70–73.
Rosignolo F, Sponziello M, Giacomelli L et al. Identification of thyroid-associated serum microRNA profiles and their potential use in thyroid cancer follow-up. J Endocr Soc. 2017;1:3–13.
Khare D, Goldschmidt N, Bardugo A, Gur-Wahnon D, Ben-Dov IZ, Avni B. Plasma microRNA profiling: exploring better biomarkers for lymphoma surveillance. PLoS One. 2017;12:e0187722.
Qiu X, Zhang J, Shi W et al. Circulating microRNA-26a in plasma and its potential diagnostic value in gastric cancer. PLoS One. 2016;11:e0151345.
Fu L, Li Z, Zhu J et al. Serum expression levels of microRNA-382-3p, -598-3p, -1246 and -184 in breast cancer patients. Oncol Lett. 2016;12:269–274.
Ding H, Wen W, Ding Q, Zhao X. Diagnostic valuation of serum miR-184 and miR-191 in patients with non-small-cell lung cancer. Cancer Control. 2020;27:1073274820964783.
Zou X, Zhu D, Zhang H et al. MicroRNA expression profiling analysis in serum for nasopharyngeal carcinoma diagnosis. Gene. 2020;727:144243.
Zou X, Xia T, Li M et al. MicroRNA profiling in serum: potential signatures for breast cancer diagnosis. Cancer Biomark. 2020;30:41–53.
Zou X, Wei J, Huang Z et al. Identification of a six-miRNA panel in serum benefiting pancreatic cancer diagnosis. Cancer Med. 2019;8:2810–2822.
Zedan AH, Osther PJS, Assenholt J, Madsen JS, Hansen TF. Circulating miR-141 and miR-375 are associated with treatment outcome in metastatic castration resistant prostate cancer. Sci Rep. 2020;10:227.
Swellam M, Bakr NM, El Magdoub HM, Hamza MS, El Arab ELR. Emerging role of miRNAs as liquid biopsy markers for prediction of glioblastoma multiforme prognosis. J Mol Neurosci. 2020;71:836–844.
Zhang Y, Xu D, Pan J et al. Dynamic monitoring of circulating microRNAs as a predictive biomarker for the diagnosis and recurrence of papillary thyroid carcinoma. Oncol Lett. 2017;13:4252–4266.
Benson EA, Skaar TC, Liu Y, Nephew KP, Matei D. Carboplatin with decitabine therapy, in recurrent platinum resistant ovarian cancer, alters circulating miRNAs concentrations: a pilot study. PLoS One. 2015;10:e0141279.
Wang WT, Chen YQ. Circulating miRNAs in cancer: from detection to therapy. J Hematol Oncol. 2014;7:86.
Funding
This work was supported by the Health Commission of Hubei Province Scientific Research Project (Grant Number: WJ2021Z005).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Yu, B., Zhou, S., Liang, H. et al. Development and Validation of a Novel Circulating miRNA-Based Diagnostic Score for Early Detection of Hepatocellular Carcinoma. Dig Dis Sci 67, 2283–2292 (2022). https://doi.org/10.1007/s10620-021-07031-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10620-021-07031-0