Abstract
Background
Proteasome subunit alpha type 7 (PSMA7) shows a carcinogenic effect on various human malignancies, but its role and regulatory mechanism in gastric carcinoma (GC) remain unclear.
Aims
This study aimed to explore the role and mechanism of PSMA7 in GC.
Methods
In this study, PSMA7 expressions in GC cells and tissues were detected, and relationships between PSMA7 and clinicopathological features were explored. Then, PSMA7 levels in human GC cells were intervened, and changes in cell biological behavior were observed in vitro and vivo. Key proteins and downstream factors of MAPK signaling pathway were detected after PSMA7 intervention.
Results
PSMA7 was upregulated in GC tissues and cell lines. PSMA7 overexpression was significantly associated with poor pTNM, cTNM stage, and high HP infection. PSMA7 can promote proliferation, invasion, and metastasis of GC cells in vitro and vivo. Furthermore, PSMA7 expression affected the phosphorylation level of JNK, P38, ERK and the expressions of their downstream factors Ap-1, c-myc, P53.
Conclusion
PSMA7 can promote GC proliferation, invasion, and metastasis through MAPK signaling pathway in GC cells.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Gastric cancer (GC) is the fifth most common cancer and causes the second-highest cancer-related mortality globally [1]. Since patients with early GC show no specific symptoms, the majority of GC patients have entered an advanced stage at first diagnosis [2]. Amounts of GC patients succumb to the disease due to tumor metastasis, while there are few effective targeted biomarkers for clinical treatment [3, 4]. Therefore, it is necessary to explore the cellular mechanisms of GC to provide novel targets for future treatment strategies [5, 6].
Proteasome subunit alpha type 7 (PSMA7) is a subunit of the core complex of the 20S proteasome, affecting the key composition formation of the 26S proteasome in the ubiquitin–proteasome pathway (UPP) [7]. PSMA7 gene is located in chromosome 20q13.33 that is an amplified region of tumor genes closely related to many tumors [8]. The connection between PSMA7 expression and the clinicopathological features of cancer patients suggests that PSMA7 has the potential to become a molecular target for a number of cancer treatments [9,10,11]. Our previous study has confirmed that PSMA7 may be a promising prognostic biomarker for GC patients [12].
This study aimed to confirm the influence of PSMA7 on GC biological behaviors and the molecular mechanism involved. PSMA7 expression in GC tissues and cells was detected, and the association between PSMA7 and several clinical features was confirmed. The effect of PSMA7 on cell proliferation, apoptosis, migration, and invasion was explored in vitro and vivo. We also detected the variation of MAPK pathway after PSMA7 dysregulation. Our study provides a new insight into PSMA7 in GC biological behaviors.
Methods
Clinical Specimens
Sixty pairs of fresh GC tissue samples and paracancer tissue samples were acquired from GC patients at the Department of Gastrointestinal Surgery in Xinghua People’s Hospital between December 2016 and November 2017. Patients were aged from 47 to 82 years (average age 59.2 years) including 49 males and 11 females. The study was authorized by the Institute Research Ethics Committee of Xinghua People’s Hospital (NO. XSYY2016042).
Cell Culture
The human GC cell lines MGC803, MKN28, SGC7901, MKN45, and BGC823, and the gastric epithelial cell line GES-1 were obtained from Key Laboratory of Antibody Technique of Ministry of Health, Nanjing Medical University. Cells were cultured in DMEM (Gibco, USA) with 10% fetal bovine serum (Invitrogen, USA) and 1% penicillin/streptomycin (Gibco, USA) with 5% CO2 at 37 °C.
Western Blot
Total protein was extracted from tissues or cells using RIPA lysis buffer (Beyotime, China) according to the protocol. Protein concentration was detected using BCA Protein Assay Kit (Tiangen, China). Proteins (30 μg/lane) were separated by 10% SDS-PAGE and then transferred to PVDF membranes at 90 V for 50 min. The membranes were immersed in 5% defatted milk for 120 min and were then incubated with antibodies at 4 °C overnight against the following: PSMA7(1:1000; #LS-C114781, USA), JNK (1:1,000; #9252, CST, USA), phospho-JNK (1:1500; #9255, CST, USA), p38 (1:1000; #9212, CST, USA), p-p38(1:1,000; #9211, CST, USA), Erk (1:1,000; #SC-514302, Santa Cruz, USA), p-Erk (1:1,000; #SC-81492; Santa Cruz, Santa Cruz, USA), p53 (1:500; #SAB4503015, Sigma-Aldrich, Germany), Ap-1(1:750; #ABP50669, Abbkine, USA), c-myc (1:500; #ab32072, Abcam, UK), and GAPDH (1:1,000; #5174, CST, USA). Following washing, horseradish peroxidase-conjugated secondary IgG antibody was incubated with the membrane for 120 min at ordinary temperatures.
Immunohistochemistry (IHC)
IHC staining was carried out according to the Envision technique, and detailed procedures were shown in the previous research [12].
Reverse Transcription-Quantitative Polymerase Chain Reaction (RT-qPCR)
Total RNA was extracted from human tissue samples, GC cells, and normal gastric mucosa epithelial cell using TRIzol reagent (Bio-Rad, USA). Reverse transcription was conducted using a Transcriptor First Strand cDNA Synthesis kit (Applied Biosystems, USA). The relative mRNA quantification was conducted by SYBR premix Taq (Takara, Japan) on the 7,500 real-time PCR system (Applied Biosystems, USA). RT-qPCR results were analyzed using the 2−∆∆CT method, and β-actin was utilized as the internal reference [13]. The following primers were used: PSMA7: forward, 5′-AGTGCGGAAGATCTGTGCTT-3′, reverse, 5′-TCCGTACGCGTTGTTGTAAT-3′, GAPDH: forward, 5′-GGAGCGAGATCCCTCCAAAAT-3′, reverse, 5′-GGCTGTTGTCATACTTCTCATGG-3'.
Establishment of Short Hairpin RNA (shRNA) and Stable PSMA7-Overexpression Cell Lines
BGC823 cells and MGC803 cells (1.0 × 105) were seeded into 24-well plates and incubated for 24 h. Cells were then transfected with PSMA7 shRNAs (termed shPSMA7-1, shPSMA7-2, shPSMA7-3), negative controls shRNA (shNC), PSMA7-expressing lentivirus and their negative control (NC) (GenePharma, China) using Lipofectamine 2000 (Invitrogen, USA). shRNA sequences: shPSMA7-1, 5′-CCCATTCTTCGCTCTGTGTTT-3′; shPSMA7-2, 5′-CTCCTCCATTTCCGTCTACTA-3′; shPSMA7-3, 5′-CCTCTTCCAAGTGGAGTA-3′; shNC, 5′-TTCTCCGAACGTGTCACGT-3′. DMEM with 10% FBS was replaced after 4–6 h. Puromycin (100 μg/ml; Sigma-Aldrich, Germany) was mixed in the cell culture medium 48 h later.
CCK8 Assay
Cells (5 × 103/well) were seeded into 96-well plates for 24 h. Subsequently, 10 µl CCK8 (BioTool, China) solution and 90 µl DMEM were added to each well, and the mixtures were incubated at 37 °C for 2 h. The optical density at 450 nm was then measured at 24, 48, 72, and 96 h.
Colony Formation Assay
Cells (5 × 102/well) were planted into 6-well plates. After 14 days, when a clone visible to the naked eye appeared, the plates were washed with PBS and soaked in crystal violet (Sigma-Aldrich, Germany) for 30 min. The number of colonies was determined using microscope (Leica Microsystems GmbH, Wetzlar, Germany) at × 200 magnification.
Wound-Healing Assay
Cells (4 × 105/well) were planted in 6-well plates for 24 h. A sterile 200-μl pipette tip was used to make a uniform scratch at the center of each well. Non-adherent cells of each well were washed using PBS. The wound widths were measured at 24, 48, and 72 h.
Migration and Invasion Assays
The transwell chamber was either uncoated (migration assay) or coated (invasion assay) with Matrigel. The cells (105/well) were suspended in the upper chamber with 300 μl serum-free DMEM medium. The lower chamber was filled with 700 µl DMEM containing 10% FBS. Following a 24- or 48-h incubation period, the cells on the upper surface of the filter were removed using a cotton swab. The chamber was fixed with 70% methanol for 30 min and stained with 0.1% crystal violet for 20 min. Penetrating cells were counted by an IX70 microscope in five randomized fields, and the mean cell counts were calculated.
Mice Model
A GC xenograft model was established in 4-week-old BALB/c nude mice (HFK Bioscience Co., Ltd., Beijing, China). Briefly, a single-cell suspension (5 × 106 cells in 150 µl PBS) was inoculated into the armpit of nude mouse to construct a xenograft model with six mice per group. Vital signs, including tumor growth rate and size, were observed and recorded. Tumor size was calculated using the following formula: 0.5 × length × width2. After 4 weeks, the mice were killed using cervical dislocation, and the tumor tissues were collected, weighed, fixed, and embedded; in addition, an single-cell suspension was injected into the abdominal cavity to form an animal model of cavity metastasis of GC. The general condition of nude mouse was observed every 3 days. After 4 weeks, the mice were killed to observe the metastasis of organs in the abdominal cavity. At last, an single-cell suspension was injected into the subcapsular spleen to make an animal model of liver metastasis of GC. After 4 weeks, the mice were killed to observe the number of hepatic metastasis. The tumors were also subjected to the analysis of PSMA7 expression and immunohistochemical staining analysis of Ki-67. This research was approved by the Animal Research Ethics Committees of Nanjing Medical University.
Statistical Analysis
Statistical analysis was conducted using GraphPad Prism 6.0 software (GraphPad, Inc., La Jolla, CA, USA). Quantitative data are presented as the mean ± standard deviation. Differences between groups were assessed by one-way analysis of variance (ANOVA) followed by Student–Newman–Keuls (SNK) comparisons. Categorical data were evaluated by Chi-square test. P < 0.05 was considered as statistically significant difference.
Results
PSMA7 Expression in GC Tissues and Cells
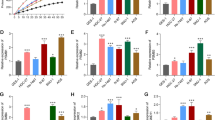
PSMA7 level in GC tissues was measured by RT-qPCR and western blot analysis. It was demonstrated that PSMA7 expression was significantly higher in GC tissues compared with paired paracancer tissues (Fig. 1a–d). IHC analysis was showed that PSMA7 was highly expressed in the cytoplasm of GC (Fig. 1e, f). In GC cells, the PSMA7 expression level was measured in GC and GES-1 cells by RT-qPCR and western blot analysis. The results reveled that PSMA7 expression in GC cells was significantly higher than that in gastric epithelial cell GES-1 (Fig. 1g–i). Among all GC cells evaluated, the highest expression of PSMA7 was observed in BGC823 cells, and MGC803 cells exhibited the lowest expression of PSMA7. Therefore, BGC823 and MGC803 cells were selected for PSMA7-knockdown and PSMA7-overexpression experiments, respectively.
PSMA7 expression in GC tissues and cell lines. a, b PSMA7 mRNA expression was detected in GC samples and paired paracancer tissues by RT-qPCR. c, d PSMA7 protein expression was detected in GC samples and paired paracancer tissues by western blot. e, f High PSMA7 protein expression was detected in the cytoplasm of GC tissues (magnification × 200). g PSMA7 mRNA expression was detected in both GC cells and the normal gastric epithelial cell line (GES-1) by RT-qPCR. h, i PSMA7 protein expression was detected in both GC cells and the normal gastric epithelial cell line (GES-1) by western blot. *P < 0.05, **P < 0.01
Correlations Between PSMA7 Expression and Clinicopathological Factors
This study further explored the relationships between PSMA7 expression and clinicopathological factors. Results showed that high PSMA7 overexpression was connected to poor pTNM (P = 0.037) and cTNM (P = 0.022) stage. PSMA7 expression was significantly higher in stage III and IV GC tissues compared with stage 0, I, and II GC tissues (P < 0.05). Furthermore, PSMA7 overexpression was associated with high HP infection (71.11%, P = 0.030). However, there is no association observed between PSMA7 level and the other clinicopathological features (Table 1).
Knockdown of PSMA7 in BGC823 and Overexpression of PSMA7 in MGC803
To further investigate the function of PSMA7 in GC, we downregulated PSMA7 in BGC823 and overexpressed PSMA7 in MGC803. Efficacy of the PSMA7 shRNA and overexpression transfections were confirmed by fluorescence microscopy (Fig. 2a, b), RT-qPCR (Fig. 2c, d), and western blot (Fig. 2e–h). During evaluation of knockdown efficiency, shPSMA7-3 showed the highest efficiency and was selected for further experiments.
The interferential and overexpressed efficacy of PSMA7 in GC cells. a, b The interferential and overexpressed efficacy of PSMA7 was detected by fluorescence microscopy (magnification × 100). c, d The interferential and overexpressed efficacy of PSMA7 was detected by RT-qPCR. e–h The interferential and overexpressed efficacy of PSMA7 was detected by western blot. *P < 0.05; **P < 0.01
PSMA7 Downregulation Inhibits GC Cell Proliferation, Migration, and Invasion
CCK8 and colony formation assays demonstrated that shPSMA7-3 significantly inhibited the proliferation of BGC823 cells (Fig. 3a–c). shPSMA7-3 increased the number of apoptotic BGC823 cells, as detected by flow cytometry (Fig. 3d, e). Wound-healing and transwell migration assays showed a significant inhibition on the migration ability of BGC823 cells in shPSMA7-3 group (Fig. 3f–i). Compared with the control group, the number of invasive cells in the shPSMA7-3 group was significantly reduced (Fig. 3h, i). These results suggested shPSMA7-3 can inhibit proliferation, migration, and invasion in BGC823 cells.
shPSMA7-3 inhibited the proliferation, migration, and invasion of BGC823 cells. a CCK8 assay and b, c plate clone assay showed the effect of shPSMA7-3 on cell proliferation. d, e Flow cytometry confirmed the effect of shPSMA7-3 on cell apoptosis. f, g Wound healing assay (magnification × 100) and h, i transwell assay (magnification × 200) showed the effect of shPSMA7-3 on migration and invasion. *P < 0.05; **P < 0.01; ***P < 0.001
PSMA7 Overexpression Promotes GC Cell Proliferation, Migration, and Invasion
CCK8 and colony formation assays confirmed that the overexpression of PSMA7 could promote the proliferation of MGC803 cells (Fig. 4a–c). Flow cytometry revealed that overexpression of PSMA7 inhibited the apoptosis of MGC803 cells (Fig. 4d, e). Wound-healing assay and transwell assay also demonstrated significant increases in the migration ability of MGC803 with PSMA7 upregulation (Fig. 4f–i). The number of invasive cells in the PSMA7-overexpression group was significantly more than the control group (Fig. 4h, i). As aforementioned, the current data confirmed that PSMA7 overexpression can promote proliferation, migration, and invasion in MGC803 cells.
PSMA7 overexpression promoted the proliferation, migration, and invasion of MGC803 cells. a CCK8 assay and plate clone assay, b, c showed the effect of PSMA7 overexpression on cell proliferation. d, e Flow cytometry confirmed the effect of PSMA7 overexpression on cell apoptosis. f, g Wound healing assay (magnification × 100) and h, i transwell assay (magnification × 200) showed the effect of PSMA7 overexpression on migration and invasion. *P < 0.05; **P < 0.01; ***P < 0.001
PSMA7 shRNA Inhibits Xenograft Growth and PSMA7 Overexpression Promotes Xenograft Growth
Nude mice were used to perform xenograft tumor assays with BGC823 and MGC803 cells stably transfected with shPSMA7-3, PSMA7, and their negative control lentiviruses shNC and NC. Dissected tumor samples and tumor growth curves revealed that the mean tumor size was smaller in the shPSMA7-3 transfected group than that in the shNC group (Fig. 5a–e). Furthermore, IHC staining of Ki-67 demonstrated that the tumor proliferation of the shPSMA7-3-transfected group was significantly inhibited compared with the controls (Fig. 5f). By contrast, the overexpression of PSMA7 promoted the tumor growth in vivo (Fig. 5g–k). The number of Ki-67-positive cells in the PSMA7-overexpression group was more than that in the NC group (Fig. 5l). These results demonstrated that PSMA7 could promote GC growth in vivo.
PSMA expression regulated proliferation of GC cells in vivo. a–d shPSMA7-3 inhibited xenograft tumor growth of BGC823 cells in vivo (n = 6). e The tumor weight of shPSMA7-3 group was less than shNC group. f Representative photographs of shPSMA7-3 and Ki-67 IHC staining in the indicated tumors (magnification × 400). g–j PSMA7 overexpression promoted xenograft tumor growth of MGC803 cells in vivo (n = 6). k The tumor weight of PSMA7 group was more than NC group. (L) Representative photographs of PSMA7 and Ki-67 IHC staining in the indicated tumors (magnification × 400). *P < 0.05, **P < 0.01
PSMA7 Overexpression Promotes Abdominal and Hepatic Metastasis Formation
In order to further verify the function of PSMA7 on tumor metastasis, NC group and PSMA7 group cells were injected into the abdominal cavity and the spleen subcapsular membrane, respectively. In the PSMA7-overexpression group, the nodules could be touched and observed in the abdomen at 2-3 w, with uneven surface and hard texture (Fig. 6a). PSMA7-overexpression group showed more tumors in the abdominal cavity, with lymph node enlargement and a small amount of ascites (Fig. 6a, b). In the hepatic metastasis model, the number of hepatic metastatic tumors in the PSMA7 group was significantly higher than that in the NC group (fig. 6c). These results confirmed the promotion role of PSMA7 in GC metastasis.
PSMA7 overexpression promoted abdominal and hepatic metastasis formation in vivo. a In the PSMA7-overexpression group, the abdominal wall of the nude mice was obviously distended and thinner, and gray tumors were seen through the wall. b The volume of intraperitoneal tumors was larger and the number of tumors was more in the PSMA7-overexpression group. c PSMA7 overexpression promoted the formation of hepatic metastatic tumors. *P < 0.05, **P < 0.01
PSMA7 Promotes the Activation of the MAPK Signaling Pathway
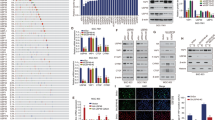
Protein–protein interaction (PPI) network showed that PSMA7 may indirectly affect the MAPK signaling pathway through NOD1, which is a substrate of PSMA7 (Fig. 7a). NOD1 was also found to have a close connection to ERK and JNK in GC (Fig. 7b). Western blotting demonstrated that PSMA7 knockdown significantly decreased the phosphorylation of JNK, Erk1/2, and p38, while PSMA7 overexpression promoted the phosphorylation of JNK, Erk1/2, and p38 (Fig. 7c, d). Furthermore, PSMA7 shPSMA7-3 significantly decreased the expression levels of AP-1, c-myc, and p53 that are important downstream molecules of the MAPK pathway, and the overexpression of PSMA7 significantly increased the protein expression levels of AP-1, c-myc, and p53 (Fig. 7e, f).
PSMA7 expression was associated with MAPK pathway. a PPI network around PSMA7. b PSMA7 showed a significant relationship with ERK and JNK. c, d shPSMA7-3 significantly decreased the phosphorylation of JNK, Erk1/2, and p38. PSMA7 overexpression significantly increased the phosphorylation of JNK, Erk1/2, and p38. e shPSMA7-3 expression also decreased AP-1, c-myc, and p53 protein expression. f PSMA7 overexpression significantly increased AP-1, c-myc, and p53 protein expression. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as internal control. *P < 0.05, **P < 0.01
Discussion
As a potential prognostic marker, PSMA7 has been confirmed to be highly expressed in various tumors, including colorectal cancer [14], cervical cancer [7], and lung cancer [15]. In this study, we confirmed that PSMA7 expression was increased in GC tissues compared to paired paracancer tissues in fresh stomach tissue. The clinicopathological correlation analysis showed that PSMA7 expression was associated with pTNM, cTNM stage, and HP infection. Therefore, PSMA7 is closely connected with GC progression.
GC is highly malignant and possesses considerable metastatic potential [16]. Malignant biological behaviors, including proliferation, migration, and invasion, are the predominant causes of tumor metastasis [17]. In human myeloid leukemia cells, researchers have found that down-regulation of PSMA7 expression can inhibit the proliferation of K562 cells, but does not affect apoptosis [18]. In human colon cancer cells, it was also found that interference with PSMA7 did not affect RKO cell apoptosis [19]. However, interference with PSMA7 in lung adenocarcinoma has little effect on the cell proliferation, but affects cell apoptosis [20]. Therefore, PSMA7 has different biological functions in different tumor cells. Through cell proliferation assay and flow cytometry, we confirmed that PSMA7 could promote cell proliferation and inhibit cell apoptosis in GC. Cell invasion and metastasis are the specific reflection of malignant degree and the key factor of tumor recurrence and metastasis [21]. Our results showed that the cell proliferation, migration, and invasion were inhibited after interfering with PSMA7 and were promoted after the overexpression of PSMA7, which is consistent with the study in colon cancer [19].
In order to further verify the effect of PSMA7 in vivo, subcutaneous tumor formation, intraperitoneal implantation, and liver metastatic model in nude mice were conducted, respectively. Subcutaneous tumorigenesis showed that the growth rate of gastric tumors was significantly slowed down after interfering with PSMA7. The growth rate of PSMA7-overexpression group was significantly accelerated comparing with the control. Additionally, immunohistochemistry demonstrated a positive correlation between Ki67 expression and PSMA7 expression. Intraperitoneal implantation metastasis is a common cause of postoperative recurrence and death of gastric cancer. Once intraperitoneal metastasis occurs, the invasion of tumor cells is increased, and the five-year survival rate is less than 20% [22]. The size and number of intraperitoneal tumors in the PSMA7-overexpression group were significantly higher than those in the control group. Liver metastasis rate of advanced GC is as high as 44%, and liver is the most common site of hematogenous metastasis [22]. In the hepatic metastasis model, we found the number of liver metastasis increased significantly after overexpression of PSMA7, thus confirming that the overexpression of PSMA7 can promote the GC metastasis in vivo.
MAPK is an important regulator of malignant biological behaviors in human cancers, including cell proliferation, migration, and invasion [23,24,25]. Based on available studies, PPI network was constructed and demonstrated that PSMA7 seems to indirectly interact with ERK and JNK, which are commonly studied members of the MAPK signaling pathway [26, 27]. NOD1 is a newly identified substrate of PSMA7, which is also involved in proteasome-dependent degradation [28]. NOD1 binds to ligands and triggers the secretion of pro-inflammatory cytokines by activating the MAPK pathway in colorectal cancer [29]. Pearson correlation analysis further confirmed that NOD1 is associated with ERK and JNK in GC. Therefore, PSMA7 probably regulates GC cell biological functions through MAPK pathway. In this study, MAPK-associated proteins were evaluated by western blotting, confirming that upregulation of PSMA7 could promote phosphorylation level of p38/Erk/JNK. The downstream targets Ap-1, c-myc, and p53, which regulate the proliferation, apoptosis, and invasion [30,31,32], were then investigated. PSMA7 overexpression was also confirmed to upregulate the expression of these genes. These results suggested that PSMA7 promotes the proliferation and metastasis of GC cells through MAPK signaling pathway.
Conclusion
In summary, our study indicates that PSMA7 serves as a key role in the malignant biological behaviors of gastric cancer cells, including proliferation, invasion, and metastasis. Furthermore, PSMA7 was identified to regulate these biological behaviors of GC cells via the MAPK signaling pathway. Therefore, PSMA7 may be a potential target for new therapeutic methods for GC patients.
References
Choi RS, Lai WYX, Lee LTC, Wong WLC, Pei XM, Tsang HF, Leung JJ, Cho WCS, Chu MKM, Wong EYL. Current and future molecular diagnostics of gastric cancer. Expert Revi Mol Diagn 2019;19:863–874
Venerito M, Vasapolli R, Rokkas T, Malfertheiner P. Gastric cancer: epidemiology, prevention, and therapy. Helicobacter 2018;23:e12518
Cao W, Wei W, Zhan Z, Xie D, Xie Y, Xiao Q. Regulation of drug resistance and metastasis of gastric cancer cells via the microRNA647-ANK2 axis. Int J Mol Med 2018;41:1958–1966
Bando E, Makuuchi R, Irino T, Tanizawa Y, Kawamura T, Terashima M. Validation of the prognostic impact of the new tumor-node-metastasis clinical staging in patients with gastric cancer. Gastric Cancer 2018;22:123–129
Zhang RR, Zhou WB, Pan ZG, Pathology DO. Study on the mechanism of RNA interference Rac1 to inhibit the proliferation and promote apoptosis of human gastric cancer cell line. J Clin Exp Med 2017;16:1279–1282
Cheng Z, Zhang D, Gong B, Wang P, Liu F. CD163 as a novel target gene of STAT3 is a potential therapeutic target for gastric cancer. Oncotarget 2017;8:87244–87262
Ren CC, Yang L, Liu L, Chen YN, Cheng GM, Zhang XA, Liu H. Effects of shRNA-mediated silencing of PSMA7 on cell proliferation and vascular endothelial growth factor expression via the ubiquitin-proteasome pathway in cervical cancer. J Cell Physiol 2018;234:5851–5862
Hu XT, Chen W, Wang D, Shi QL, Zhang FB, Liao YQ, Jin M, He C. The proteasome subunit PSMA7 located on the 20q13 amplicon is overexpressed and associated with liver metastasis in colorectal cancer. Oncol Rep. 2008;19:441–446
Tan JY, Huang X, Luo YL. PSMA7 inhibits the tumorigenicity of A549 human lung adenocarcinoma cells. Mol Cell Bioch. 2012;366:131–137
Honma K, Takemasa I, Matoba R, Yamamoto Y, Takeshita F, Mori M, Monden M, Matsubara K, Ochiya T. Screening of potential molecular targets for colorectal cancer therapy. Int J Gen Med. 2009;2:243–257
Krüger M, Beger C, Welch PJ, Barber JR, Manns MP, Wongstaal F. Involvement of proteasome α-subunit PSMA7 in hepatitis C virus internal ribosome entry site-mediated translation. J Hepatol. 2001;34:8357–8364
Xia S, Tang Q, Wang X, Zhang L, Jia L, Wu D, Xu P, Zhang X, Tang G, Yang T, Feng Z, Lu L. Overexpression of PSMA7 predicts poor prognosis in patients with gastric cancer. Oncol Let. 2019;18:5341–5349
Zheng Z, Dong XJ. Clinical value of serum trophoblast cell surface protein 2 (TROP2) antibody in non-small cell lung cancer patients. Bio Bioch Ind Exp Re Sus Che. 2016;21:739–742
Hu XT, Chen W, Wang D, Shi QL, Zhang FB, Liao YQ, Jin M, He C. High expression of proteasome subunit PSMA7 in colorectal cancer is significantly correlated with liver metastasis. Chin J Oncol 2008;30:515
Cai CJ, Ye SJ, Zhu W, Sun LY, Wan HS, Feng ZH, Ma L, Li L. Study on invasion and metastasis-associated genes of lung cancer related with NM23-H1 gene. J Sichuan Uni Med Sci Edi 2010;41:941
Carcas LP. Gastric cancer review. J Carcinogen 2014;13:14
Sheng W, Chen Y, Tu C et al. ANGPTL2 expression in gastric cancer tissues and cells and its biological behavior. World J Gastroenterol 2016;22:10364–10370
Qin T, Fan CQ, Zhu N, Shen Y, Chen MH. Knockdown of proteasome subunit α7 with small interfering RNA inhibits cell proliferation of K562 cell line. Acta Aca Med Sin 2013;35:601–606
Hu X, Chen W, Zhang F et al. Depletion of the proteasome subunit PSMA7 inhibits colorectal cancer cell tumorigenicity and migration. Oncol Rep 2009;22:1247–1252
Tan J, Huang X, Luo Y et al. PSMA7 inhibits the tumorigenicity of A549 human lung adenocarcinoma cells. Mol Cell Biochem 2012;366:131–137
Zhu T, Hu X, Wei P, Shan G. Molecular background of the regional lymph node metastasis of gastric cancer. Oncol Lett 2018;15:3409–3414
Saito H, Kihara K, Kuroda H, Matsunaga T, Tatebe S, Ikeguchi M. Surgical outcomes for gastric cancer patients with intraperitoneal free cancer cell, but no macroscopic peritoneal metastasis. J Sur Oncol 2011;104:534–537
Evans MK, Brown MC, Geradts J, Bao X, Robinson TJ, Jolly MK, Vermeulen PB, Palmer GM, Gromeier M, Levine H. XIAP regulation by MNK links MAPK and NFκB signaling to determine an aggressive breast cancer phenotype. Cancer Res 2018;78:1726–1738
Jiang K, Lu Q, Li Q, Ji Y, Chen W, Xue X. Astragaloside IV inhibits breast cancer cell invasion by suppressing Vav3 mediated Rac1/MAPK signaling. Int Immunopharmacol 2017;42:195–202
Sun Q, Liang Y, Zhang T, Wang K, Yang X. ER-α36 mediates estrogen-stimulated MAPK/ERK activation and regulates migration, invasion, proliferation in cervical cancer cells. Biochem Biol Res Commun 2017;487:625–632
Lanna A, Gomes DCO, Muller-Durovic B, McDonnell T, Escors D, Gilroy DW, Lee JH, Karin M, Akbar AN. A sestrin-dependent Erk-Jnk-p38 MAPK activation complex inhibits immunity during aging. Nat Immunol 2017;18:354–363
Cicenas J, Zalyte E, Rimkus A, Dapkus D, Noreika R, Urbonavicius S. JNK, p38, ERK, and SGK1 inhibitors in cancer. Cancers 2017;10:1
Yang L, Tang Z, Zhang H, Kou W, Lu Z, Li X, Li Q, Miao Z. PSMA7 directly interacts with NOD1 and regulates its function. Cell Physiol Biochem. 2013;31:952–959
Wang Z, Liu M, Nie X, Zhang Y, Chen Y, Zhu L, Chen X, Chen L, Chen H, Zhang J. NOD1 and NOD2 control the invasiveness of trophoblast cells via the MAPK/p38 signaling pathway in human first-trimester pregnancy. Placenta 2015;36:652–660
Sankpal NV, Mayfield J, Willman M, Fleming TP, Gillanders WE. Activator protein 1 (AP-1) contributes to EpCAM-dependent breast cancer invasion. Breast Cancer Res 2011;13:1–13
He X, Tan X, Wang X, Jin H, Liu L, Ma L, Yu H, Fan Z. C-Myc-activated long noncoding RNA CCAT1 promotes colon cancer cell proliferation and invasion. Tumor Biol 2014;35:12181–12188
Ramadoss S, Guo G, Wang CY. Lysine demethylase KDM3A regulates breast cancer cell invasion and apoptosis by targeting histone and the non-histone protein p53. Oncogene 2017;36:47–59
Acknowledgments
We thank all patients enrolled in the study.
Funding
This project was supported by the National Natural Science Foundation of China (Grant numbers 81201596, 81773100) and Special Fund of Clinical Medicine in Jiangsu Province (BL2013038).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors declare no potential conflicts of interest.
Ethics approval
The study was ratified by the Institute Research Ethics Committee of Xinghua People’s Hospital. Written informed consents were obtained from all study participants. Animal studies were conducted as per the principles and procedures approved by the Ethics Committee of Nanjing Medical University.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Xia, S., Ji, L., Tang, L. et al. Proteasome Subunit Alpha Type 7 Promotes Proliferation and Metastasis of Gastric Cancer Through MAPK Signaling Pathway. Dig Dis Sci 67, 880–891 (2022). https://doi.org/10.1007/s10620-021-06903-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10620-021-06903-9