Abstract
Background
Long non-coding RNAs (lncRNAs) have been increasingly uncovered to participate in multiple human cancers, including pancreatic cancer (PC). However, the underlying mechanisms of most of the lncRNAs have not been fully understood yet.
Aims
In this study, we probed the role and latent mechanism of LINC01420 in PC.
Methods
Several online tools were applied. Gene expression was evaluated by qRT-PCR or Western blot. Both in vitro and in vivo assays were conducted to probe LINC01420 function in PC. ChIP, RIP, and luciferase reporter assays were performed to determine relationships between genes.
Results
The bioinformatics analyses revealed LINC01420 was highly expressed in PC tissues. Besides, LINC01420 was pronouncedly upregulated in PC cell lines and its depletion controlled PC cell proliferation and EMT in vitro and hindered tumor growth in vivo. Importantly, KRAS was proved to mediate LINC01420-facilitated PC cell proliferation. Further, we explained that KRAS transcription was regulated by MYC, while LINC01420 enhanced the binding of MYC to KRAS promoter in the nucleus of PC cells. Intriguingly, LINC01420 boosted MYC expression in the cytoplasm of PC cells by sponging miR-494-3p.
Conclusion
This study illustrated that LINC01420 accelerates PC progression through releasing miR-494-3p-silenced MYC in cytoplasm and upregulating MYC-activated KRAS in nucleus, unveiling LINC01420 as a latent therapeutic strategy for PC patients.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Pancreatic cancer (PC) is one of the deadliest human malignancies all over the world [1]. In the past decades, a good deal of molecules have been uncovered to be implicated in the pathogenesis of PC, including growth factors, oncogenes, anti-tumor genes, and so on [2, 3]; however, few of them come into clinical application. Hence, due to the limited progress in the diagnostics and therapeutics of PC, most patients with PC have already had distant metastasis or a high risk of complications when diagnosed [4]. Meanwhile, 5-year survival time of PC patients postoperation is still disappointing [5]. Hence, more effective regulators involving in PC tumorigenesis and development are helpful to develop useful diagnostic and therapeutic targets for PC patients.
Long non-coding RNAs (lncRNAs) are a class of RNA transcripts with longer than 200 nucleotides in length, and the importance of lncRNAs in the carcinogenesis and progression of various human cancers have already been revealed by accumulating evidences over decades [6]. Lately, the varied function of lncRNAs has been uncovered based on their different localizations [7,8,9]. For example, the cytoplasmic lncRNA CASC9 facilitates hepatocellular carcinoma survival through an AKT-dependent way [10]. LncRNA CCAT1 in the cytoplasm promotes esophageal squamous cell carcinoma (ESCC) cell growth and migration by miR-7/HOXB13 signaling [11]. The cytoplasmic lncRNA LINK-A facilitates triple-negative breast cancer by targeting HIF1α signaling [12]. Also, lncRNA AC026904.1 acts as an enhancer RNA in the nucleus, while UCA1 functions as a competitive endogenous RNA (ceRNA) in the cytoplasm in the development of breast cancer [13]. Moreover, lncRNA CASC9 contributes to ESCC metastasis through targeting LAMC2 via interacting with the CREB-binding protein in the nucleus [14]. LINC01420, also called NBDY (negative regulator of P-body association), is a newly identified oncogene in nasopharyngeal carcinoma [15]; nevertheless, its expression pattern and role in PC remain elusive.
In the current study, we aimed to investigate the function and the potential mechanism of LINC01420 in PC progression.
Materials and Methods
Cell Culture
The human normal pancreatic ductal epithelial cell line (HPDE6-C7) and five PC cell lines (PANC-1, SW1990, HPAC, CFAPC-1, and BxPC-3) as well as HEK-293T cells were all provided by the Institute of Biochemistry and Cell Biology of the Chinese Academy of Sciences (Shanghai, China). Cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; HyClone, USA) with 10% fetal bovine serum (FBS; Invitrogen Life Technologies, Carlsbad, CA, USA), 100 U/ml penicillin, and 100 μg/ml streptomycin in a humid incubator containing 5% CO2 at 37 °C.
Cell Transfection
Two short hairpin RNAs (shRNAs) specially targeting LINC01420 (shLINC01420#1 and shLINC01420#2) and its scramble control shRNA (shCtrl) were obtained from GenePharma (Shanghai, China). In addition, miR-494-3p mimics and NC mimics were also generated by GenePharma (Shanghai, China). The pcDNA3.1 vectors (Invitrogen) containing the amplified cDNA of KRAS, MYC or LINC01420 were, respectively, used to overexpress KRAS, MYC and LINC01420, with the empty vector used as negative control. The above plasmids were transfected appropriately into PC cells or HEK-293T cells by using Lipofectamine 2000 (Invitrogen) following the manufacturer’s instruction.
RNA Extraction and Quantitative Real-Time PCR (qRT-PCR)
Total RNA was extracted from cells by using an RNeasy Mini kit (Qiagen, #74104) following the manufacturer’s protocols. Then cDNAs were obtained from isolated RNAs with a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, #4368813). Subsequently, the real-time PCR was carried out on a QuantStudio 6 Flex Real-Time PCR System (Applied Biosystems) by the use of the Power SYBR Green PCR Master Mix (Applied Biosystems, #4367659). Gene expression relative to GAPDH was assessed with 2−ΔΔCt methods. All qRT-PCR analyses were performed in triplicate and repeated at least three times.
Cell Proliferation Assays
To evaluate cell viability, the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazolium bromide (MTT) assay was conducted in PANC-1 and BXPC-3 cells. In short, cells maintained in complete medium were placed in a 96-well plate and cultured for 24, 48, 72, and 96 h. Thereafter, 20 μl of 0.5 mg/ml MTT was supplemented into each well, and then cells underwent 4 h of incubation with MTT at 37 °C, followed by subsequent incubation with 100 μl of dimethyl sulfoxide (DMSO) for 1 h. Eventually, the absorbance of each well was evaluated at 490 nm by the use of a microplate reader. To assess the cell proliferative ability, EdU assays were performed using the Cell Light EdU DNA imaging kit (Invitrogen, Carlsbad, CA, USA) based on the manufacturer’s guide. All experiments were conducted in triplicate for at least three repeats.
Western Blot
Total protein was isolated with 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred onto polyvinylidene difluoride (PVDF) membranes (Roche). Afterward, the membranes were blocked and incubated with primary antibodies overnight at 4 °C, followed by further incubation with secondary antibodies (Santa Cruz Biotech, CA, USA) for 2 h at 37 °C. At length, the blots were visualized with the bands detected using Bioimaging Systems (UVP, Upland, CA, USA). The primary antibodies included anti-E-cadherin, anti-N-cadherin, anti-Vimentin (Abcam, MA, USA), anti-KRAS, anti-AKT, anti-p-AKT, anti-ERK, anti-p-ERK (Cell Signaling Technology, Inc., Danvers, MA, USA), and anti-GAPDH (Abcam, MA, USA). GAPDH served as a loading control.
RNA Immunoprecipitation (RIP) Assay
The Magna RNA-binding protein immunoprecipitation kit (Millipore, Billerica, MA, USA) was employed in RIP assays in the light of the protocols of manufacturer. Antibodies targeting MYC and normal IgG (negative control) were applied in such experiments. The immune-precipitated RNAs were purified using proteinase K and then examined by qRT-PCR.
Luciferase Reporter Assay
HEK-293T cells seeded in a 24-well plate with a density of 1 × 105 cells/well were co-transfected with pGL3-KRAS promoter and MYC or co-treated with pGL3-MYC promoter and shCtrl or shLINC01420#1 using Lipofectamine 2000 (Invitrogen). With respect to the identification of relationship between LINC01420 and miR-494-3p, the sequences of LINC01420-WT/Mut were subcloned into the pmirGLO dual-luciferase vector (Promega, Madison, WI, USA), followed by the co-transfection of these recombinant plasmids with miR-494-3p mimics or miR-NC into HEK-293T cells by the use of Lipofectamine 2000. Similarly, the pmirGLO dual-luciferase vector (Promega, Madison, WI, USA) subcloned with the sequences of MYC-WT/Mut was co-transfected with miR-NC, miR-494-3p mimics, or 494-3p mimics + LINC01420 into HEK-293T cells. Two days later, the transfected cells were collected followed by the measurement of luciferase activities with the Dual-Luciferase Reporter Assay System (Promega, Wisconsin, WI, USA). All transfection experiments were carried out in triplicate for three times.
Subcellular Fractionation Assay
The cytoplasmic and nuclear fractionation of PANC-1 and BXPC-3 cells was processed by the use of a PARISTM kit (ThermoFisher, #AM1921) in line with the manufacture’s recommendations. Then, the isolated RNA from each fraction was determined using qRT-PCR.
In Vivo Experiments
Six BALB/c nude mice at 5 weeks old obtained from SLAC Laboratory Animal (Shanghai, China) were applied in this study. Then, shCtrl- or shLINC01420#1-transfected BXPC-3 cells were subcutaneously injected into the left flank of these mice (two groups, 3 mice for each group). The volumes of tumors in the above two groups were measured every 4 days according to following formula: V = 0.5 × D × d2, where V means volume, D is longitudinal diameter, and d represents latitudinal diameter. After 4 weeks, mice were killed and tumors were weighted. Thereafter, these tumors were embedded with paraffin and finally used for immunohistochemical (IHC) staining.
Immunohistochemical (IHC) Staining
The IHC assay was conducted following previous protocol [16]. The primary antibody against Ki67 (Cat. #9027, 1:1600 dilution, Cell Signaling Technology) and horseradish peroxidase (HRP)-conjugated secondary antibody were used here. The pictures were obtained using a Leica DMIL LED microscope.
Statistical Analysis
The differences were determined by the Student’s t test (between two groups) or one-way ANOVA (among at least three groups) through analyzing all data by using SPSS 15.0 software (SPSS, Inc., Chicago, IL, USA). All the experiments were repeated for at least 3 times. All results were presented as mean ± standard deviation (SD), and P < 0.05 was thought as statistically significant.
Results
LINC01420 Is Remarkably Upregulated in PC
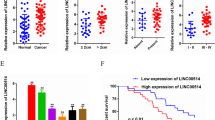
To explore the potential role of LINC01420 in PC, several online tools were firstly applied to evaluate the expression pattern of LINC01420 in PC. According to the bioinformatics analyses, we found that LINC01420 expression in 8555 normal pancreatic samples was generally expressed at low level based on the data from UCSC (http://genome.ucsc.edu/cgi-bin/hgc?hgsid=715371743_8yfcS3no0SKdr8zxqH8nfbfZ5EcB&c=chrX&l=56755691&r=56844813&o=56755691&t=56844813&g=gtexGene&i=LINC01420) (Fig. 1a). Accordantly, the low expression of LINC01420 in normal pancreatic tissues from 95 human individuals was also indicated by NCBI (https://www.ncbi.nlm.nih.gov/gene/?term=LINC01420) (Fig. 1b). In contrast, TCGA database (http://gepia2.cancer-pku.cn/#analysis) suggested a relative high level of LINC01420 in PC tissues in comparison with normal pancreatic tissues (Fig. 1c). By experimental data, we proved that LINC01420 was observably upregulated in five PC cell lines (PANC-1, BXPC-3, SW1990, HPAC, and CFPAC-1) relative to the human normal pancreatic duct epithelial cell line HPDE6-C7, with PANC-1 and BXPC-3 cells exhibiting the highest LINC01420 expression (Fig. 1d). Altogether, these data strongly indicate the upregulation of LINC01420 in PC.
LINC01420 was highly expressed in PC. a LINC01420 expression profile in 53 normal tissues from 570 donors that obtained from UCSC. b The profile of LINC01420 level in 27 tissues from 95 normal individuals was obtained from NCBI. c The data from TCGA showed that LINC01420 was upregulated in PC tissues compared with normal tissues. d qRT-PCR result of LINC01420 expression in five PC cell lines and the normal HPDE6-C7 cells. *P < 0.05, **P < 0.01
Silencing LINC01420 Hampers Cancer Cell Proliferation and PC EMT
To investigate the precise role of LINC01420 in PC development, loss-of-function assays were implemented in PANC-1 and BXPC-3 cells which showed highest LINC01420 expression endogenously. Compared to the shCtrl-transfected control groups, LINC01420 was successfully and notably silenced in both PANC-1 and BXPC-3 cells under the transfection of either shLINC01420#1 or shLINC01420#2 (Fig. 2a). Meanwhile, the result of MTT assays demonstrated that LINC01420 inhibition by either shLINC01420#1 or shLINC01420#2 led to noticeably confined viability of both PANC-1 and BXPC-3 cells (Fig. 2b). Based on the results that more effective suppression induced by shLINC01420#1 was noticed in the above two PC cells, we then chose shLINC01420#1-transfected PC cells to continue our study. LINC01420 depletion resulted in apparent impairment on the proliferative capacities of these two PC cells (Fig. 2c, d). Moreover, the impact of LINC01420 on epithelial-to-mesenchymal transition (EMT), a crucial driver for cancer metastasis [17], was also evaluated here. Results indicated that inhibition of LINC01420 hindered EMT process in PC, since the level of epithelial marker E-cadherin was increased while those of mesenchymal markers N-cadherin and Vimentin were decreased after LINC01420 silencing (Fig. 2e). Collectively, our results show that LINC01420 serves as a contributor in PC development via proliferation EMT promotion.
Knockdown of LINC01420 suppressed PC cell proliferation. a qRT-PCR result of LINC01420 level in PANC-1 and BXPC-3 cells upon the transfection of shCtrl, shLINC01420#1 or shLINC01420#2. b The viability of the above PC cells was assessed by MTT assays. c, d EdU assays were conducted to determine the proliferation capacity of PANC-1 and BXPC-3 cells with or without LINC01420 inhibition. e Western blot analysis of EMT-related proteins in PC cells with or without LINC01420 silence. *P < 0.05, **P < 0.01
KRAS Is Involved in LINC01420-Promoted PC Cell Proliferation and EMT
Next, we further investigated underlying mechanism underlying LINC01420-affected PC development. In this situation, KRAS, a proto-oncogene highly related to PC that has been proved by MalaCards (https://www.malacards.org/card/pancreatic_cancer#related_genes), caught our attention. Besides, the TCGA database (http://gepia2.cancer-pku.cn/#index) suggested that KRAS was also distinctly highly expressed in PC tissues compared with the normal pancreatic tissues (Fig. 3a). Meanwhile, the data from TCGA also revealed a high prognostic value of KRAS for PC patients (Fig. 3b). Interestingly, TCGA further suggested that KRAS expression was strongly correlated with LINC01420 level in PC tissues (Fig. 3c). Therefore, we intensely suspected that LINC01420 had a regulatory effect on KRAS and it might function in PC through a KRAS-mediated pathway.
KRAS was involved in LINC01420-affected PC cell proliferation. a–c The expression of KRAS in PC tissues and normal tissues (a), the association of KRAS with the prognosis of PC patients (b), and the correlation of KRAS expression and LINC01420 level in PC tissues (c) were all provided by TCGA database. d Relative expression of KRAS in LINC01420-silenced PANC-1 and BXPC-3 cells was tested using qRT-PCR. e qRT-PCR result of KRAS level in PANC-1 cells upon different transfections. f The protein levels of KRAS and its downstream proteins were estimated by conducting Western blot. g, h Cell proliferation in the indicated PANC-1 cells was determined through performing MTT and EdU assays. i The level of E-cadherin, N-cadherin, and Vimentin in indicated PC cells was assessed by western blot. *P < 0.05, **P < 0.01
To prove the above speculation, we then probed whether LINC01420 could modulate KRAS expression in PC cells. As a result, KRAS expression was strikingly decreased in LINC01420-silenced PANC-1 and BXPC-3 cells (Fig. 3d). However, such inhibitory effect was then revived in response to forced KRAS expression (Fig. 3e). Additionally, the protein level of KRAS and the phosphorylation levels of its downstream proteins were all restrained in face of LINC01420 depletion but recovered in the context of KRAS overexpression (Fig. 3f), verifying that LINC01420 influenced KRAS signaling in PC through directly regulating KRAS expression. More importantly, both the confined viability and proliferative ability in PANC-1 cells upon LINC01420 suppression were potentiated under KRAS upregulation (Fig. 3g, h). Meanwhile, LINC01420 inhibition-suppressed EMT was apparently reversed in the context of KRAS upregulation (Fig. 3i). In sum, we revealed that LINC01420 promotes PC progression through targeting KRAS pathway.
LINC01420 Contributes to KRAS Expression Through Interacting with MYC
Further, we explored the detailed mechanism by which LINC01420 affected KRAS expression. Firstly, the online TRRUST version 2 (https://www.grnpedia.org/trrust/result_tonly.php?gene=KRAS&species=human&confirm=0) and UCSC implied that MYC was the transcription factor (TF) of KRAS, which has been recognized previously [18]. Besides, a recent report explained that the interaction of lncRNAs with MYC intensifies the occupancy of MYC on its target promoters [19]. Also, the online RPISeq (http://pridb.gdcb.iastate.edu/RPISeq/) predicted a high potential for LINC01420 to interact with MYC protein. Hence, we deduced that LINC01420 might regulate KRAS through a similar manner.
In order to validate our inference above, our priority was to confirm the transcriptional regulation of MYC on KRAS in PC cells. As expected, a notable binding of MYC to KRAS promoter was revealed in all the four PC cells (Fig. 4a and Fig. S1A). Importantly, it was proved that the luciferase activity of KRAS promoter was considerably enhanced in response to MYC upregulation (Fig. 4b). Next, over half of LINC01420 was shown to locate in the nucleus of the two PC cells, which further intensified the potential of LINC01420-MYC interaction in nucleus (Fig. 4c). Unsurprisingly, we observed the apparent presence of LINC01420 in the immunoprecipitates of anti-MYC rather than that of anti-IgG in these four PC cells (Fig. 4d and Fig. S1B). Intriguingly, the amount of KRAS promoter precipitated by MYC antibody was sharply erased in face of LINC01420 knockdown in four kinds of PC cells (Fig. 4e, f and Fig. S1C-D). All in all, we draw a conclusion that LINC01420 upregulates KRAS in PC through facilitating the binding of MYC to KRAS promoter via interacting with MYC protein.
LINC01420 facilitated MYC occupation on KRAS promoter through interaction with MYC. a, b RIP and luciferase reporter assays were carried out to confirm the transcriptional regulation of MYC on KRAS. c The levels of LINC01420 in the cytoplasm and the nucleus of both PANC-1 and BXPC-3 cells were analyzed by qRT-PCR.d The interaction of LINC01420 and MYC in two PC cells was confirmed using RIP assay. e, f The impact of LINC01420 on MYC bound to KRAS promoter was assessed by using RIP assays in both PANC-1 and BXPC-3 cells. *P < 0.05, **P < 0.01, ***P < 0.001
LINC01420 Boosts MYC Expression in PC by Sponging miR-494-3p
In depth, we wondered whether LINC01420 could impact on MYC expression in PC cells in the meantime. Interestingly, it was unmasked that the expression of MYC was markedly declined in LINC01420-silenced PC cells (Fig. 5a and Fig. S1E). However, we revealed LINC01420 had no evident effect on MYC transcription in PC cells since the luciferase activity of MYC promoter was almost unaffected upon LINC01420 inhibition (Fig. 5b). Given that nearly 40% of LINC01420 was distributed in cytoplasm of PC cells (Fig. 4c), we then guessed that LINC01420 might regulate MYC expression via a recently emerged competing endogenous RNAs (ceRNAs) mechanism [7]. Fortunately, miR-494-3p was predicted through bioinformatics analysis to bind to both LINC01420 and MYC mRNA (Fig. 5c). Moreover, the competitive relationship between LINC01420 and MYC mRNA in the interaction with miR-494-3p was further verified by luciferase reporter assays, as the luciferase activity of both LINC01420-WT and MYC-WT was reduced under miR-494-3p upregulation, while miR-494-3p mimics-decreased luciferase activity of MYC-WT was partially revived in the context of LINC01420 overexpression (Fig. 5d, e). Furthermore, we also uncovered that the level of MYC mRNA that was hindered by miR-494-3p mimics was recovered in all the four PC cells in face of ectopic expression of LINC01420 (Fig. 5f and Fig. S1F). Based on these findings, we conclude that LINC01420 enhances MYC expression in PC by miR-494-3p sequestration.
LINC01420 elevated MYC expression in PC by competitively interacting with miR-494-3p. a, b The effects of LINC01420 inhibition on MYC expression and transcription were, respectively, estimated by qRT-PCR (a) and luciferase reporter assay (b). c Bioinformatics analysis suggested miR-494-3p as the shared miRNA to interact with both LINC01420 and MYC mRNA. d, e Luciferase reporter assay was performed to confirm the ceRNA mechanism among LINC01420, miR-494-3p, and MYC. f The expression of MYC in PC cells under different conditions was detected via qRT-PCR. *P < 0.05, **P < 0.01
LINC01420 Facilitates PC Tumor Growth In Vivo
Last but not least, we carried out in vivo experiments to further certify the in vitro observations above. As seen from Fig. 6a, tumors originated from LINC01420-depleted BXPC-3 cells were smaller in size than those from shCtrl-transfected control cells. Also, LINC01420 knockdown led to lightened tumor weight in comparison with control group (Fig. 6b). Then, we delineated that the expression mRNA levels of MYC and KRAS were both lessened in tumors with silenced LINC01420 expression (Fig. 6c). More importantly, the staining of Ki67 was remarkably abrogated in tumors with LINC01420 suppression (Fig. 6d), directly proving the impairment of LINC01420 on cell proliferation in PC tumors. Hence, we testified that LINC01420 aggravates PC tumor growth in vivo.
Knockdown of LINC01420 suppressed tumor growth in vivo. a Representative images and the growth curve of in vivo xenografts obtained from two groups with or without LINC01420 silence. b Mean weight of tumors in indicated two groups. c qRT-PCR result of the expression levels of LINC01420, MYC, and KRAS in tumors with or without LINC01420 depletion. d IHC assay was carried out to measure Ki67 staining in tumors from these two groups. **P < 0.01
Discussion
Recently, mounting researches have indicated the participation of lncRNAs in the initiation and progression of a myriad of human cancers including PC [20, 21]. As an example, lncRNA GLS-AS controls GLS-mediated metabolism and inhibits PC progression [22]. LncRNA-BX111 aggravates PC metastasis and progression via regulating ZEB1 transcription [23]. LncRNA HOTTIP mediates PC stem cell properties through regulating HOXA9 [24]. In the present study, we elucidated that LINC01420, a recently identified oncogene in nasopharyngeal carcinoma [15], was also highly expressed in PC and functioned as a facilitator for both the in vitro PC cell proliferation and EMT process and in vivo tumor growth. The precise role of LINC01420 was, for the first time, disclosed by our paper.
KRAS proto-oncogene (KRAS) is a member of the small GTPase superfamily that is implicated in various malignancies, including lung adenocarcinoma, mucinous adenoma, colorectal carcinoma, and PC [25,26,27,28]. Shockingly, KRAS mutants are proved as a key driver of PC and its mutation occurs in more than 95% PC patients [29]. Moreover, Mueller et al. [30] revealed that the dosage of KRAS could affect PC phenotypes, and Kamerkar et al. [31] suggested that KRAS could be an effective therapeutic target for PC. Besides, the association of lncRNA, KRAS, and PC has also been uncovered before [32]. For instance, lncRNA-NUTF2P3-001 promotes PC tumorigenesis by targeting the miR-3923/KRAS axis [33]. LncRNA MALAT1 serves as a ceRNA to regulate KRAS expression by absorbing miR-217 in PC [34]. Thus, we sought to probe the possible correlation between LINC01420 and KRAS in PC. Currently, we elaborated that LINC01420-facilitated PC cell proliferation was mediated by KRAS-dependent pathway. Even more strikingly, our results explained that nuclear LINC01420 regulated KRAS expression in PC through interacting with MYC, a TF of KRAS validated previously [18]. LINC01420 promoted the binding of MYC to KRAS promoter, therefore stimulating KRAS expression at transcriptional level. Such interaction between LINC01420 and MYC was similar with a latest report [19].
Recently, lncRNAs in the cytoplasm have been increasingly suggested as a ceRNA to regulate gene expression at post-transcriptional level by competitively binding with miRNAs [35]. Here, we proved that MYC could be regulated by LINC01420 through post-transcriptional manner rather than transcriptional manner. Experimental data unveiled that the cytoplasmic LINC01420 positively modulated MYC expression in PC cells through acting as a sponge of miR-494-3p, a miRNA that exerts different function in diverse carcinomas [36,37,38]. Also, the negative regulation of miR-494 in PC has previously been reported by Li et al. [39].
Conclusively, this study disclosed that LINC01420 sponges miR-494-3p to elevate MYC expression in the cytoplasm and interacts with c-MYC protein to trigger KRAS expression in the nucleus, thus exerting a tumor promotion effect on PC tumorigenesis. Our study displayed the high potential of LINC01420 as an effective therapeutic molecular target for PC treatment. Yet, whether LINC01420 has the clinical application value needs to be further confirmed in future studies. Recently, tumor microenvironment has become a hotpot in cancer research. Further studies will also focus on searching feasible axes underlying LINC01420 in tumor microenvironment, e.g., stromal components.
References
Murr MM, Sarr MG, Oishi AJ, van Heerden JA. Pancreatic cancer. CA Cancer J Clin. 1994;44:304–318.
Li D, Xie K, Wolff R, Abbruzzese JL. Pancreatic cancer. Lancet. 2004;363:1049–1057.
Wolfgang CL, Herman JM, Laheru DA, et al. Recent progress in pancreatic cancer. CA Cancer J Clin. 2013;63:318–348.
Yachida S, Jones S, Bozic I, et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467:1114–1117.
Jooste V, Dejardin O, Bouvier V, et al. Pancreatic cancer: wait times from presentation to treatment and survival in a population-based study. Int J Cancer. 2016;139:1073–1080.
Cheetham SW, Gruhl F, Mattick JS, Dinger ME. Long noncoding RNAs and the genetics of cancer. Br J Cancer. 2013;108:2419–2425.
Noh JH, Kim KM, McClusky WG, Abdelmohsen K, Gorospe M. Cytoplasmic functions of long noncoding RNAs. Wiley Interdiscip Rev RNA IF4.838. 2018;9:e1471.
Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152:1298–1307.
Sun M, Kraus WL. From discovery to function: the expanding roles of long noncoding RNAs in physiology and disease. Endocr Rev. 2015;36:25–64.
Noh JH, Gorospe M. AKTions by cytoplasmic lncRNA CASC9 promote hepatocellular carcinoma survival. Hepatology (Baltimore, Md.). 2018;68:1675–1677.
Zhang E, Han L, Yin D, et al. H3K27 acetylation activated-long non-coding RNA CCAT1 affects cell proliferation and migration by regulating SPRY4 and HOXB13 expression in esophageal squamous cell carcinoma. Nucleic Acids Res. 2017;45:3086–3101.
Lin A, Li C, Xing Z, et al. The LINK-A lncRNA activates normoxic HIF1α signalling in triple-negative breast cancer. Nat Cell Biol. 2016;18:213–224.
Li GY, Wang W, Sun JY, et al. Long non-coding RNAs AC026904.1 and UCA1: a “one-two punch” for TGF-β-induced SNAI2 activation and epithelial-mesenchymal transition in breast cancer. Theranostics. 2018;8:2846–2861.
Liang Y, Chen X, Wu Y, et al. LncRNA CASC9 promotes esophageal squamous cell carcinoma metastasis through upregulating LAMC2 expression by interacting with the CREB-binding protein. Cell Death Differ. 2018;25:1980–1995.
Yang L, Tang Y, He Y, et al. High expression of LINC01420 indicates an unfavorable prognosis and modulates cell migration and invasion in nasopharyngeal carcinoma. J Cancer. 2017;8:97–103.
Mallakin A, Taneja P, Matise LA, Willingham MC, Inoue K. Expression of Dmp1 in specific differentiated, nonproliferating cells and its regulation by E2Fs. Oncogene. 2006;25:7703–7713.
Lo HC, Zhang XHF. EMT in metastasis: finding the right balance. Dev Cell. 2018;45:663–665.
Mongroo PS, Noubissi FK, Cuatrecasas M, et al. IMP-1 displays cross-talk with K-Ras and modulates colon cancer cell survival through the novel proapoptotic protein CYFIP2. Cancer Res. 2011;71:2172–2182.
Wang Z, Yang B, Zhang M, et al. lncRNA epigenetic landscape analysis identifies EPIC1 as an oncogenic lncRNA that interacts with MYC and promotes cell-cycle progression in cancer. Cancer Cell. 2018;33:706–720.
Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1:391–407.
Knoll M, Lodish HF, Sun L. Long non-coding RNAs as regulators of the endocrine system. Nat Rev Endocrinol. 2015;11:151–160.
Deng S-J, Chen H-Y, Zeng Z, et al. Nutrient stress-dysregulated antisense lncRNA GLS-AS impairs GLS-mediated metabolism and represses pancreatic cancer progression. Cancer Res. 2019;79:1398–1412.
Deng S-J, Chen H-Y, Ye Z, et al. Hypoxia-induced LncRNA-BX111 promotes metastasis and progression of pancreatic cancer through regulating ZEB1 transcription. Oncogene. 2018;37:5811–5828.
Fu Z, Chen C, Zhou Q, et al. LncRNA HOTTIP modulates cancer stem cell properties in human pancreatic cancer by regulating HOXA9. Cancer Lett. 2017;410:68–81.
Slebos RJ, Kibbelaar RE, Dalesio O, et al. K-ras oncogene activation as a prognostic marker in adenocarcinoma of the lung. N Engl J Med. 1990;323:561–565.
Ichikawa Y, Nishida M, Suzuki H, et al. Mutation of K-ras protooncogene is associated with histological subtypes in human mucinous ovarian tumors. Cancer Res. 1994;54:33–35.
Abubaker J, Bavi P, Al-Haqawi W, et al. Prognostic significance of alterations in KRAS isoforms KRAS-4A/4B and KRAS mutations in colorectal carcinoma. J Pathol. 2009;219:435–445.
Ying H, Kimmelman AC, Lyssiotis CA, et al. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell. 2012;149:656–670.
Iacobuzio-Donahue CA, Herman JM. Autophagy, p53, and pancreatic cancer. N Engl J Med. 2014;370:1352–1353.
Mueller S, Engleitner T, Maresch R, et al. Evolutionary routes and KRAS dosage define pancreatic cancer phenotypes. Nature. 2018;554:62–68.
Kamerkar S, LeBleu VS, Sugimoto H, et al. Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature. 2017;546:498–503.
Zhang L, Yu S, Wang C, Jia C, Lu Z, Chen J. Establishment of a non-coding RNAomics screening platform for the regulation of KRAS in pancreatic cancer by RNA sequencing. Int J Oncol. 2018;53:2659–2670.
Li X, Deng S-J, Zhu S, et al. Hypoxia-induced lncRNA-NUTF2P3-001 contributes to tumorigenesis of pancreatic cancer by derepressing the miR-3923/KRAS pathway. Oncotarget. 2016;7:6000–6014.
Liu P, Yang H, Zhang J, et al. The lncRNA MALAT1 acts as a competing endogenous RNA to regulate KRAS expression by sponging miR-217 in pancreatic ductal adenocarcinoma. Sci Rep. 2017;7:5186.
Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–352.
Kim WK, Park M, Kim Y-K, et al. MicroRNA-494 downregulates KIT and inhibits gastrointestinal stromal tumor cell proliferation. Clin Cancer Res. 2011;17:7584–7594.
Zhan M-N, Yu X-T, Tang J, et al. MicroRNA-494 inhibits breast cancer progression by directly targeting PAK1. Cell Death Dis. 2017;8:e2529–e2529.
Lim L, Balakrishnan A, Huskey N, et al. MicroRNA-494 within an oncogenic microRNA megacluster regulates G1/S transition in liver tumorigenesis through suppression of mutated in colorectal cancer. Hepatology (Baltimore, Md.). 2014;59:202–215.
Li L, Li Z, Kong X, et al. Down-regulation of MicroRNA-494 via loss of SMAD4 increases FOXM1 and β;-catenin signaling in pancreatic ductal adenocarcinoma cells. Gastroenterology. 2014;147:485–497.
Acknowledgment
We appreciate all the contributors during our study.
Funding
None.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants performed by any of the authors. All applicable international, national, and/or institutional guidelines for the care and use of animals were followed.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Fig. 1 Mechanistic investigation in SW1990 and HPAC cells.
(A) The binding of MYC protein to KRAS promoter in SW1990 and HPAC cells was validated by ChIP assay. (B) The interaction between LINC01420 and MYC protein in these two PC cells was proved by RIP assay. (C-D) ChIP assay uncovered the inhibition of LINC01420 silence on MYC occupation in KRAS promoter, (E-F) qRT-PCR result of MYC expression in SW1990 and HPAC cells under diverse transfections. * P < 0.05, ** P < 0.01. (TIFF 8015 kb)
Rights and permissions
About this article
Cite this article
Zhai, H., Zhang, X., Sun, X. et al. Long Non-coding RNA LINC01420 Contributes to Pancreatic Cancer Progression Through Targeting KRAS Proto-oncogene. Dig Dis Sci 65, 1042–1052 (2020). https://doi.org/10.1007/s10620-019-05829-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10620-019-05829-7