Abstract
Background and Aim
Biliary atresia (BA) is a pediatric liver disease with unknown underlying etiology. MicroRNAs (miRNAs) represent a family of small noncoding RNAs. Among them, miR-19b has been suggested to function in the diseased liver. We therefore decided to investigate its potential role in BA.
Methods
We used infant-derived specimens to analyze miR-19b expression in a tissue- and cell-specific fashion, predicted interaction with genes, and finally performed a functional study in vitro.
Results
Patients with BA showed significantly lower miR-19b level in liver compared with controls, and pediatric end-stage liver disease (PELD) score was inversely correlated with miR-19b level. In vitro, miR-19b was significantly downregulated in activated hepatic stellate cells (HSCs) and exerted inhibitory effects on HSC activation, as confirmed by decreased alpha-smooth muscle actin (α-SMA) and type I collagen expression. Moreover, one mRNA target gene (TGFβR2) was identified. Computational prediction of miR-19b binding to the 3′-untranslated region (UTR) of TGFβR2 was validated by luciferase reporter assay. Furthermore, miR-19b mimic negatively regulated transforming growth factor-beta (TGF-β) signaling components, as demonstrated by decreased drosophila mothers against decapentaplegic homolog 3 (SMAD3) expression and blocking of TGF-β-induced expression of a1(I) and a2(I) procollagen miRNAs.
Conclusions
Our data indicate that miR-19b may be involved in BA-related fibrosis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Biliary atresia (BA), one of the commonest causes of neonatal cholestasis, results from fibroinflammatory obstruction of both intra- and extrahepatic bile ducts, having unknown etiology. Loss of the large bile ducts that drain the liver results in severe, life-threatening cholestasis, and patients present with hyperbilirubinemia and acholic stools, typically by four weeks of age. The only intervention currently available to restore bile flow from the native liver is Kasai portoenterostomy, which is successful in less than 80% of cases. Of patients with successful short-term outcome after Kasai procedure, 50% will ultimately require liver transplant for chronic complications [1]. Unfortunately, even after surgical intervention, biliary cirrhosis progresses in the vast majority of infants.
MicroRNAs (miRNAs, miRs) are small noncoding RNAs that negatively regulate target gene expression through base pairing with the 3′-untranslated region (UTR), inducing messenger RNA (mRNA) cleavage or translational repression. With multiple and diverse targets, miRNAs exert control over key cellular developmental processes, including differentiation and proliferation. The specific contribution of select miRNAs in hepatic disease development and progression has been described [2], and miRNAs are dysregulated in numerous pathologies, including experimental BA [3]. Among them, miRNA-19b (miR-19b) is recognized as a novel regulator of profibrotic TGF-β signaling [4]. Although the field continues to advance, no studies have determined whether miR-19b is involved in BA pathogenesis, which is characterized by inflammatory and fibrotic obliteration of extrahepatic bile ducts.
In this work, we analyzed miR-19b expression in human BA tissue. Functional analysis indicated that miR-19b is required for activated hepatic stellate cells (HSCs), suggesting a similar critical role in mammalian fibrogenesis. This is supported by our identification of several miR-19b target genes that are known or suspected regulators of profibrotic signaling. The results reveal the roles of miR-19b in BA pathogenesis, providing a potential therapeutic approach for treating BA.
Methods
Human Tissues
Liver tissues were obtained from the liver surgery archives at Renji Hospital (Shanghai, China) and Shanghai Children’s Medical Center, with approval from the Institutional Review Board. Biliary atresia samples (n = 109) were either from diagnostic biopsies or liver explant tissue. Among patients, 43 underwent Kasai portoenterostomy and 66 liver transplantation. For controls, we examined eighty-one histologically normal liver samples (resected tissue adjacent to the mass) from children with hepatoblastoma or metabolic disease. Detailed information on these patients is listed in Tables 1 and 2. Tissues were fixed in 10% neutral buffered formalin, dehydrated through serial alcohol washes using an automated processor, and cleared with xylene before embedding in paraffin. Histologic staging was applied to determine fibrosis severity. Histologic staging from F0 to F4 was performed by two pathologists using the METAVIR scoring system, with stage F0 indicating no fibrosis and stage F4 representing cirrhosis (no fibrosis F0; early fibrosis F1–F2; late fibrosis F3–F4). Fibrosis severity was also assessed by hematoxylin and eosin (HE) staining of peritumoral sections and of BA samples. Parental consent was obtained for patients under 18 years of age, and informed consent for miRNA expression analysis was obtained from all patients prior to surgery; the study has been approved by the National Institutional Animal Care and Use Committee.
Primary Cell Isolation, Culture
Male Sprague–Dawley (SD) rats (>500 g; SLRC Laboratory Animal Company, Shanghai) were used for these studies. All experiments were reviewed and approved by the National Medical Center Institutional Animal Care and Use Committee. Primary rat HSCs were isolated by pronase/collagenase perfusion digestion followed by density gradient centrifugation as reported [5]. Cell purity (>95%) and viability were confirmed by autofluorescence and trypan blue staining. HSCs were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 U/mL penicillin, and 100 Lg/mL streptomycin. Culture medium was replaced every 48 h unless otherwise described, and cells were incubated at 37 °C with 5% CO2. Primary hepatocytes, Kupffer and liver sinusoidal endothelial cells (LSEC) were isolated by density gradient centrifugation as described [6, 7]. Moreover, normal rat cholangiocytes (NRCs) were isolated and grown in DMEM culture medium as previously described [8, 9].
Transient Transfection
Activated HSCs (day 6) were transfected with mature miR-19b mimic and negative control probes (scramble, SCR) (Dharmacon, Lafayette, CO) using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. Untransfected HSCs were used as normal control. Briefly, cells were plated at density of 1–4 × 105 cells/mL in standard culture medium following isolation. Cells were washed three times with Opti-MEM I Reduced-Serum Medium prior to addition of transfection complexes. Lipofectamine–mimic complexes (miR-19b, UGUGCAAAUCCAUGCAAAACUGA; SCR, UUGUACUACACAAAAGUACUG) were incubated for 20 min and added to HSCs in Opti-MEM at final concentration of 25, 50, and 75 nM. After 6 h, transfection medium was aspirated and replaced with standard culture medium supplemented with 5% FBS. After 48 h, transfection cells were harvested and prepared for further analysis.
miRNA Target Prediction
Putative targets of the miRNA were identified using Target Scan and miRanda. Genes listed by both prediction algorithms were adopted as putative target genes for individual miRNAs.
Identification of Direct Targets of miR-19b
DNA from the 3′-UTR of TGFβR2 was cloned into pSiCheck2 (Promega, Madison, USA). For miR-19b overexpression, 3 × 104 293T cells were seeded into 24-well plates with 900 ng expression plasmid and 100 ng dual luciferase reporter plasmid per well, using 3 μL FugeneHD (Roche, Basel, Switzerland). For antisense oligonucleotide (ASO) assays, 2 μL Lipofectamine 2000 (Invitrogen, Carlsbad, CA) was used to cotransfect 200 ng reporter plasmid with antisense oligonucleotide (Regulus Therapeutics, as described above) at 20 nM in 24-well format using the same seeding density. After 16 h, cells were rinsed once with 1× phosphate-buffered saline (PBS), and medium was replaced with fresh medium. After 24 h of additional outgrowth, the cells were rinsed once with 1× PBS and lysed in 150 μL 1× Passi lysis buffer (Promega, Madison, USA), and firefly and Renilla luciferase activities were measured from a 10-μL aliquot on a GloMax Multi luminometer using Stop and Glo reagents (Promega, Madison, USA), according to the manufacturer’s instructions. Relative light units were calculated as the ratio of Renilla to firefly luciferase activity, and the reporters were normalized using control expression plasmids or ASOs to correct for nonspecific effects of the differences between the experimental UTRs and the empty pSiCheck2. Values from empty pSiCheck2 samples for a given control treatment were used to correct for nonspecific effects of the treatment on the normalizer.
Quantitative Real-Time Polymerase Chain Reaction and Immunoblotting
For miR analysis, total RNA was isolated from tissues using the miRNeasy Mini Kit (Qiagen, Valencia, CA) with modifications. Tissues were homogenized on ice using an Omni TH homogenizer (Omni International, Kennesaw, GA) with soft tissue probes. Following chloroform extraction, two 350- and 500-μL washes were performed using RWT and RPE buffer, respectively, prior to elution in 50 μL RNase-free water. First-strand complementary DNA (cDNA) synthesis was performed using MicroRNA Reverse Transcription Kit primed with miR-specific primer (Applied Biosystems, Foster City, CA). Real-time quantitative reverse-transcription polymerase chain reaction (qRT-PCR) was performed using MicroRNA assays (Applied Biosystems), following the manufacturer’s recommendations, with an ABI Prism 7500 sequence detection system using Universal Master Mix (Applied Biosystems). For mRNA analysis, total RNA was isolated using Trizol reagent, and cDNA synthesis performed as previously described. Expression was measured using the CFX96 real-time PCR detection system with 50 ng cDNA, gene-specific oligonucleotide primers, and IQ SYBR Green Supermix (BIO RAD, Hercules, CA). The \(2^{{ -\Delta C_{\text{t}} }}\) method was used to calculate relative mRNA expression levels compared with β-actin or U6. Proteins were isolated, subjected to sodium dodecyl sulfate (SDS) polyacrylamide gel electrophoresis (PAGE), and transferred to nitrocellulose membranes as previously described [10]. Bradford assays were used to measure protein concentration, and Ponceau S staining to verify equal protein loading. After blocking, membranes were incubated with primary antibodies (β-actin, p-SMAD3, TGFβR2, SMURF-1, TIMP-2, and TGIF-1; Santa Cruz Biotechnology, Inc., Santa Cruz, CA; type I collagen, Meridian Life Sciences, Saco, ME) overnight at 4 °C followed by incubation with horseradish peroxidase (HRP)-conjugated secondary antibodies. Chemiluminescence was used to visualize immunoreactivity as previously described [10].
Statistical Analysis
Data are presented as mean ± standard error (SE) as determined from at least three independent experiments unless otherwise stated. Statistical significance between experimental groups was assessed using unpaired two-sample Student’s t test, and P values <0.05 were considered significant. Results of clinical data and miRNA tissue concentrations from patients are expressed as median and range, due to skewed distributions of most variables in patients. Differences between controls and patients or between patient subgroups were analyzed by Mann–Whitney U test, and correlations between tissue levels of miR-19b and pediatric end-stage liver disease (PELD) score were determined by calculating the Spearman rank correlation coefficient. Receiver-operating characteristic (ROC) curve analysis and the derived c statistic provide global and standardized appreciation of the accuracy of a marker or a composite score for predicting an event. This statistic allows simple comparison of the accuracy of different prognostic scores within the same population. The ROC curve plots sensitivity versus 1 − specificity. A c statistic of 0.5 means that discrimination is due to chance alone, whereas a c statistic of 1 means that the score perfectly predicts the outcome (a goal never achieved in clinical practice). Therefore, the accuracy of a score increases as the c statistic increases from 0.5 to 1 [11]. All analyses were performed using the Statistical Package for the Social Sciences (SPSS) version 18 (SPSS Institute, Chicago, IL, USA).
Results
Low Tissue Levels of miR-19b Are Characteristic for BA Patients
Recently, plasma levels of miRNAs have emerged as potential biomarkers for various pathological conditions such as cystic fibrosis liver disease and cancer [12, 13]. We therefore hypothesized that dysregulation of members of the miR-17-92 family in fibrotic livers might be associated with significant change in miR-19b tissue levels. To test this hypothesis, we isolated miRNAs from tissues of 109 patients with BA at different stages and compared miR-19b levels with histologically normal liver samples. miR-19b tissue levels were significantly downregulated in BA patients compared with controls (Fig. 1a). Next, we examined miR-19b expression at different time points. There was decreased liver expression of miR-19b at the time of both Kasai portoenterostomy and transplantation compared with controls (Fig. 1b). This was in accordance with the analysis by the METAVIR scoring system, which indicated that tissues from patients at both early stage (with no or minimal fibrosis, F0, F1–F2) and late stage (with severe fibrosis, F3–F4) demonstrated lower miR-19b levels than controls (Fig. 1c). In addition, patients with advanced liver cirrhosis (Child stages B and C) displayed significantly lower miR-19b levels than patients with early cirrhosis (Child A) (Fig. 1d). Furthermore, pediatric end-stage liver disease (PELD) score was inversely correlated with miR-19b tissue level (Fig. 1e). In comparison with normal liver, low miR-19b levels might correlate with presence of BA, as shown by a c-statistic of 0.803 in receiver-operating characteristic curve analysis (Fig. 1f). Finally, representative morphology of excised masses from normal peritumoral section and a BA sample is shown by HE staining in Fig. S1.
Low tissue miR-19b levels are characteristic of advanced liver fibrosis in BA patients. a Box-whisker plot indicating significant downregulation of miR-19b level in tissue samples of BA patients compared with controls. Tissue levels of miR-19b were determined by qPCR. The bold line indicates the median per group, the box contains 50% of the values, horizontal lines show the minimum and maximum values of the calculated nonoutliers, and plus signs indicate outliers. b Tissue levels of miR-19b determined by qPCR in samples of patients at different time points (at Kasai portoenterostomy and transplantation) and c at different fibrosis stage. d Results also revealed stage-dependent downregulation of miR-19b in liver tissue of BA patients. e Correlation analysis of tissue levels of miR-19b and PELD score revealing correlation between these two parameters with correlation coefficient R = −0.413 (Spearman rank correlation test, P < 0.001). f Receiver-operating characteristic curve analysis displaying the diagnostic power of miR-19b level in predicting BA (area under curve, AUC 0.803). Data presented as mean ± SE. *P < 0.05; **P < 0.01
miR-19b Is Expressed in HSCs and Downregulated During Their Activation
Since the principal cells responsible for promoting accumulation of extracellular matrix proteins during hepatic fibrogenesis are activated hepatic stellate cells (HSCs), we performed qPCR analysis on RNA extracts to evaluate miR-19b expression in primary HSCs from livers of SD rat. To characterize miR-19 expression in different hepatic cell compartments, hepatocytes, endothelial cells, and Kupffer cells from rat livers were also isolated. Since cholangiocytes are widely accepted as one of the most important pacemakers of fibrosis in several chronic liver diseases [14, 15], NRCs were also isolated to evaluate miR-19b expression level. As depicted in Fig. 2a, miR-19b showed high expression in HSCs compared with other cell types, suggesting a specific function in these cells. qRT-PCR verified a significant decrease of miR-19b in activated compared with quiescent HSCs (Fig. 2b). Moreover, miR-19b expression was recorded over 14 days in culture, with a significant decrease ultimately observed (Fig. 2c).
miR-19b is expressed in HSCs and downregulated during their activation. a Hepatocytes, HSCs, Kupffer cells, liver sinusoidal endothelial cells, and cholangiocytes were isolated from livers or bile ducts of rat, and relative miR-19b expression compared with U6 was determined by qPCR. b Rat HSCs were harvested and miR-19b expression determined in quiescent (n = 7) and activated (day 14, n = 6) HSCs as assessed by qRT-PCR. c qRT-PCR analysis of miR-19b expression levels over days in culture as compared with U6. Data presented as mean ± SE. **P < 0.01
HSC Transdifferentiation Is Inhibited by miR-19b
Next, we tested whether miR-19b can modulate activation of HSCs and extracellular matrix (ECM) protein production. Activated HSCs were transfected with miR-19b (19b) or miRNA mimic negative control (SCR), and following 48 h of transfection, RNA and protein were analyzed. Forced expression of miR-19b dampened mRNA level of both α-SMA (a marker of activated HSCs) (Fig. 3a) and type I collagen (fibrillar collagen). The results were confirmed by Western blot, which indicated drastically decreased expression of these two proteins (Fig. 3b, c). Furthermore, miR-19b restored glial fibrillary acidic protein (GFAP), a marker of quiescent HSCs (Fig. 3d).
HSC transdifferentiation is blunted by miR-19b. Activated HSCs (day 6) were transiently transfected with miR-19b mimic (25–75 nM) or miR mimic negative control (SCR). Untransfected HSCs were used as normal control, and a α-SMA gene expression was measured by qRT-PCR at 48 h (n = 4). Meanwhile, type I collagen was also measured. Representative immunoblot of α-SMA (b) and type I collagen (c) protein expression 48 h posttransfection (miR-19b, 75 nM) (n = 3). β-Actin was used as invariant control. d Day 6 HSCs were transiently transfected with miR-19b mimic or SCR (75 nM), and GFAP expression was assessed by qRT-PCR at 48 h (n = 4). Expression compared with β-actin. Data presented as mean ± SE. *P < 0.05
TGFβR2 Is Direct Target of miR-19b
Based on previous literature and in silico analyses (TargetScan and miRanda prediction databases), we identified four fibrosis-related mRNAs, viz. TGIF1, SMURF1, TGFβR2, and TIMP2, as possible targets for miR-19b. Activated HSCs were transfected with miR-19b (19b) or miRNA mimic negative control (SCR), and following 48 h of transfection, RNA and protein were analyzed. As shown in Fig. 4a, b, overexpression of miR-19b resulted in significant decrease in expression of TGFβR2, whereas expression of other fibrosis-related genes (TGIF1, SMURF1, and TIMP2) was not affected by transfection of miR-19b (data not shown). To test whether TGFβR2 is a direct target of miR-19b regulation, we cloned the 3′-UTRs of the gene into a reporter plasmid (pSiCheck2) and performed dual luciferase assays in HSC cells, while overexpressing miR-19b (Fig. 4c). When miR-19b was overexpressed, the reporters containing the TGFβR2 3′-UTRs were significantly downregulated relative to empty reporter. These data strongly suggest direct regulation of both genes by miR-19b. Conversely, when reporters were cotransfected with antisense oligonucleotide (ASO) directed against miR-19b, the normalized Renilla luciferase activity was increased relative to control vector (Fig. 4d). Taken together, these data strongly suggest direct regulation of TGFβR2 by miR-19b.
Dual luciferase reporter assays: TGFβR2 is direct target of miR-19b. Day 6 HSCs were transiently transfected with miR-19b mimic (25–75 nM) or miR mimic negative control (SCR). Untransfected HSCs were used as normal control, and a TGFβR2 gene expression was measured by qRT-PCR after 48 h of transfection. b Representative immunoblot of TGFβR2 protein expression 48 h posttransfection (miR-19b, 75 nM). β-Actin was used as invariant control. c Luciferase reporter assays to detect repression of TGFβR2 3′-UTR sequences containing candidate miR-19b target sites by exogenous miR-19b. Results expressed relative to control reporter without additional UTR sequences. d Luciferase reporter assays to detect de-repression of TGFβR2 3′-UTR sequences containing candidate miR-19b target sites by locked nucleic acid antisense oligonucleotide-mediated miR-19b inhibition. Results expressed relative to control oligonucleotide. Data presented as mean ± SE. *P < 0.05; **P < 0.01
miR-19b Negatively Regulates Profibrotic TGF-β Signaling and Decreases Expression of TGF-β Target Genes
Fibrotic TGF-β signaling propagates through the SMAD family of transcriptional activators, which like TGFβR2, SMAD2, and SMAD3, are upregulated following fibrotic liver injury [16]. Downregulation of TGF-β signaling can impact expression of downstream SMAD3 and SMAD7 [17, 18]. Although SMAD2/3 3′-UTRs do not harbor putative miR-19b binding sites (as predicted by TargetScan and miRanda), mRNA expression of SMAD3 was significantly downregulated after 24 and 48 h of miR-19b transfection (Fig. 5a). More importantly, to determine whether downstream TGF-β signaling was impacted by disrupting TGFβR2, phosphorylation of SMAD3 was assessed. Compared with SCR and untransfected controls, cells transfected with miR-19b showed marked decrease in p-SMAD3 (Fig. 5b). Similarly, effects of increasing miR-19b on downstream TGF-β signaling target procollagen mRNA and protein were also measured. Overexpression of miR-19b resulted in decreased mRNA expression of both procollagen Col α1(I) (Fig. 5c) and Col α2(I) (Fig. 5d), with more significant effects observed on transcription of Col α2(I).
miR-19b exerts inhibitory effects on TGF-β signaling and TGF-β target genes. Day 6 HSCs were transiently transfected with miR-19b mimic (25–75 nM) or miR mimic negative control (SCR). Untransfected HSCs were used as normal control, and a SMAD gene expression was determined by qRT-PCR at 24 and 48 h (n = 3). b Representative immunoblot of phosphorylated SMAD3 (p-SMAD3). β-Actin was used as invariant control. Col a1(I) (c) and Col a2(I) (d) gene expression was assessed by qRT-PCR at 24 h (n = 4). Data presented as mean ± SE. *P < 0.05; **P < 0.01
Discussion
Although miR-19b has been postulated to be important in experimental BA, its role in human BA has not been shown. This study first analyzed miR-19b expression in human BA liver in comparison with control livers. We identified, for the first time using clinical specimens from infants, miR-19b as a candidate miRNA with potential roles in liver fibrosis resulting from BA. Our results indicate that miR-19b exerts inhibitory effects on HSC activation through suppression of fibrotic TGF-β signaling and that TGFβR2, with roles in fibrosis, is a direct target of miR-19b.
miRNAs are increasingly recognized as regulators of liver physiology and diseases [19]. By regulating the levels of specific mRNAs and their posttranscriptional regulation, miRNAs have been linked to pathogenesis of viral hepatitis, nonalcoholic fatty liver disease and steatohepatitis, fibrogenesis, and hepatocellular carcinoma. For fibrogenesis, several studies in settings of chronic liver disease with underlying fibrosis have shown strong correlations between specific miR expression patterns and responses to drug treatments as well as disease progression/prognostic outcome [2]. Plasma levels of miR-122 are elevated in both hepatitis B virus (HBV) and hepatitis C virus (HCV) patients as well as in models of alcohol- and drug-induced liver damage, reinforcing a role for miRs as biomarkers [20]. A recent report identified miR-19b as a novel inhibitor of HSC-mediated fibrogenesis [4]. BA is characterized by inflammatory and fibrotic obliteration of extrahepatic bile ducts leading to severe cholestasis and biliary cirrhosis. Therefore, miR-19b also might play a crucial role in biliary cirrhosis. In line with this hypothesis, we found significantly decreased levels of miR-19b in tissue samples from BA infants. The molecular process that leads to lower tissue levels of miR-19b in patients with BA is not clear. It was previously demonstrated that miRNAs are packed into exosomes, which can be exchanged between cells without loss of function of the included miRNAs [21]. This raises the question of whether miRNAs may play a role as extracellular messengers mediating intercellular communication. Despite the currently unknown mechanism of miRNA regulation in tissue, the striking regulation of miR-19b in the tissue of BA patients might have implications for clinical aspects of BA. BA at different stages is associated with decreased expression of miR-19b. Even early-stage BA without fibrosis is also detected with this microRNA. Thus, other congenital cholestatic diseases could also be studied to reinforce the specificity of our findings, although the rarity of many of these diseases may limit such work currently. Therefore, larger patient cohorts as diseased controls will have to be analyzed to further test the potential of miR-19b levels in tissue as biomarkers for detection or monitoring of BA.
miR profiling in human and murine liver fibrosis, and additional published in vitro manipulation studies, have highlighted a role for miRs in fibrosis. Although fibrosis underlies most chronic liver diseases, including HBV, HCV, alcoholic liver disease (ALD), and BA, Roderburg et al. [22] found miR expression in human samples to be highly variable among patients with viral versus alcohol-induced fibrosis, indicating that the role of miRs in fibrosis may be stimulus specific. Therefore, future studies are needed to carefully analyze miR-19b expression in diseases with other etiologies, e.g., HBV, HCV, and ALD.
TGF-β is recognized as the most potent fibrogenic cytokine regulating HSC collagen production by way of autocrine and paracrine signaling [23], and its receptor, TGFβR2, is believed to promote TGF-β signaling. Inhibition of TGF-β receptors (soluble TGFβR2 and knockout models) and/or signaling components decreases HSC activation and dramatically blunts chronic hepatic wound healing in experimental animal models [24, 25]. In the current study, regulation of TGFβR2 expression by sites within its 3′-UTR has been described. Forced expression of mature miR-19b in activated HSCs significantly reduced expression of TGFβR2 through direct binding to the 3′-UTR, which is vital for efficient activation of downstream profibrotic gene expression. Expression of procollagen mRNAs and secreted type I collagen were also markedly reduced by miR-19b. One limitation of our approach to detecting miR-19b targets is the use of in vitro study only, because there may be target genes expressed specifically in animals. Furthermore, confirmation of a functional role for miR-19b in BA will require successful inhibition of the miRNA in the context of the mouse model. Thus, in vivo experiments are still required to verify the function of mi-19b.
Overall, our study indicates that miR-19b is a novel regulator of fibrotic TGF-β signaling and reveals that it might has potential roles in clinical aspects of BA. What’s important, it hasn’t been proven that miR-19b causes biliary atresia Instead, it just associates with fibrosis severity. However, there are several limitations to our study. Despite analysis of the association between miR-19b and fibrosis in BA, we did not analyze the biliary component of this disease. Although we have investigated miR-19b expression in BA livers and histologically normal livers adjacent to resected tumors, experiments have yet to be performed in controls with diseases such as progressive family intrahepatic cholestasis (PFIC) or alpha-1 antitrypsin deficiency. Thus, further studies are needed to evaluate the diagnostic utility of this miRNA in larger populations. Future work will include development and testing of diagnostic and prognostic logistical regression models using serum miR-19b levels in larger populations and in combination with other biochemical parameters in an effort to facilitate early detection and improved patient outcome.
References
Sokol RJ, Shepherd RW, Superina R, Bezerra JA, Robuck P, Hoofnagle JH. Screening and outcomes in biliary atresia: summary of a National Institutes of Health workshop. Hepatology. 2007;46:566–581.
Lakner AM, Bonkovsky HL, Schrum LW. microRNAs: fad or future of liver disease. World J Gastroenterol. 2011;28:2536–2542.
Hand NJ, Horner AM, Master ZR, et al. MicroRNA profiling identifies miR-29 as a regulator of disease-associated pathways in experimental biliary atresia. J Pediatr Gastroenterol Nutr. 2012;54:186–192.
Lakner AM, Steuerwald NM, Walling TL, et al. Inhibitory effects of microRNA 19b in hepatic stellate cell-mediated fibrogenesis. Hepatology. 2012;56:300–310.
Lakner AM, Walling TL, McKillop IH, Schrum LW. Altered aquaporin expression and role in apoptosis during hepatic stellate cell activation. Liver Int. 2011;31:42–51.
Schrum LW, Black D, Iimuro Y, Rippe RA, Brenner DA, Behrns KE. c-Jun does not mediate hepatocyte apoptosis following NFkappaB inhibition and partial hepatectomy. J Surg Res. 2000;88:142–149.
Knolle PA, Uhrig A, Hegenbarth S, et al. IL-10 down-regulates T cell activation by antigen-presenting liver sinusoidal endothelial cells through decreased antigen uptake via the mannose receptor and lowered surface expression of accessory molecules. Clin Exp Immunol. 1998;114:427–433.
Huang BQ, Masyuk TV, Muff MA, Tietz PS, Masyuk AI, Larusso NF. Isolation and characterization of cholangiocyte primary cilia. Am J Physiol Gastrointest Liver Physiol. 2006;291:G500–G509.
Vroman B, LaRusso NF. Development and characterization of polarized primary cultures of rat intrahepatic bile duct epithelial cells. Lab Investig. 1996;74:303–313.
Zhao D, Long XD, Lu TF, et al. Metformin decreases IL-22 secretion to suppress tumor growth in an orthotopic mouse model of hepatocellular carcinoma. Int J Cancer. 2015;1:2556–2565.
Tacke F, Fiedler K, Trautwein C. A simple clinical score predicts high risk for upper gastrointestinal hemorrhages from varices in patients with chronic liver disease. Scand J Gastroenterol. 2007;42:374–382.
Cook NL, Pereira TN, Lewindon PJ, Shepherd RW, Ramm GA. Circulating microRNAs as noninvasive diagnostic biomarkers of liver disease in children with cystic fibrosis. J Pediatr Gastroenterol Nutr. 2015;60:247–254.
Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;29:10513–10518.
Clouston AD, Powell EE, Walsh MJ, Richardson MM, Demetris AJ, Jonsson JR. Fibrosis correlates with a ductular reaction in hepatitis C: roles of impaired replication, progenitor cells and steatosis. Hepatology. 2005;41:809–818.
Fabris L, Cadamuro M, Guido M, et al. Analysis of liver repair mechanisms in Alagille syndrome and biliary atresia reveals a role for notch signaling. Am J Pathol. 2007;171:641–653.
Kitamura Y, Ninomiya H. Smad expression of hepatic stellate cells in liver cirrhosis in vivo and hepatic stellate cell line in vitro. Pathol Int. 2003;53:18–26.
Bauge C, Cauvard O, Leclercq S, Galera P, Boumediene K. Modulation of transforming growth factor beta signalling pathway genes by transforming growth factor beta in human osteoarthritic chondrocytes: involvement of Sp1 in both early and late response cells to transforming growth factor beta. Arthritis Res Ther. 2011;13:R23.
Yingling JM, Blanchard KL, Sawyer JS. Development of TGF-beta signalling inhibitors for cancer therapy. Nat Rev Drug Discov. 2004;3:1011–1022.
Wang XW, Heegaard NH, Orum H. MicroRNAs in liver disease. Gastroenterology. 2012;142:1431–1443.
Zhang Y, Jia Y, Zheng R, et al. Plasma microRNA-122 as a biomarker for viral-, alcohol-, and chemical-related hepatic diseases. Clin Chem. 2010;56:1830–1838.
Gibbings DJ, Ciaudo C, Erhardt M, Voinnet O. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat Cell Biol. 2009;11:1143–1149.
Roderburg C, Urban GW, Bettermann K, et al. Micro-RNA profiling reveals a role for miR-29 in human and murine liver fibrosis. Hepatology. 2011;53:209–218.
Dooley S, Delvoux B, Streckert M, et al. Transforming growth factor beta signal transduction in hepatic stellate cells via Smad2/3 phosphorylation, a pathway that is abrogated during in vitro progression to myofibroblasts. TGFbeta signal transduction during transdifferentiation of hepatic stellate cells. FEBS Lett. 2001;27:4–10.
Dooley S, Delvoux B, Lahme B, Mangasser-Stephan K, Gressner AM. Modulation of transforming growth factor beta response and signaling during transdifferentiation of rat hepatic stellate cells to myofibroblasts. Hepatology. 2000;31:1094–1106.
George J, Roulot D, Koteliansky VE, Bissell DM. In vivo inhibition of rat stellate cell activation by soluble transforming growth factor beta type II receptor: a potential new therapy for hepatic fibrosis. Proc Natl Acad Sci USA. 1999;26:12719–12724.
Acknowledgments
We thank Dr. Xia for collection of liver specimens. We also thank the infants and their families and the nursing and medical records staffs of Department of Liver Surgery, Renji Hospital and Shanghai Children’s Medical Center for supporting this collaborative study.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Dong Zhao and Yi Luo contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
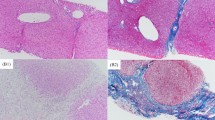
Fig. S1
Representative photomicrographs (200×) of HE-stained masses from a normal peritumoral section and a BA sample. (TIFF 5373 kb)
Rights and permissions
About this article
Cite this article
Zhao, D., Luo, Y., Xia, Y. et al. MicroRNA-19b Expression in Human Biliary Atresia Specimens and Its Role in BA-Related Fibrosis. Dig Dis Sci 62, 689–698 (2017). https://doi.org/10.1007/s10620-016-4411-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10620-016-4411-z