Abstract
Cave-obligate aquatic organisms are difficult to monitor for conservation due to cryptic diversity, unknown subterranean hydrological connectivity, and accessibility to habitats. Conservation management practices have benefitted from evolutionary data; however, the evolutionary and biogeographic histories of most cave-obligate organisms are unknown. The modes and patterns leading to most cave-obligate organism distributions are also uncertain. The Southern Cavefish (Typhlichthys subterraneus, Amblyopsidae) is the largest ranging cavefish in the world but represents a species complex of which the distribution and relatedness within remains unclear. To explore modes of cave-adaptive evolution, we performed population genomic analyses on a dataset of single nucleotide polymorphisms harvested from ultraconserved elements. We found five to eight strongly delineated genetic clusters. Little to no genetic exchange occurred between clusters, indicating high genetic distinctiveness and low connectivity, a concerning result for the fitness and conservation of these fishes. Genetic clusters did not correspond to caves nor to other geographic boundaries examined. Unfortunately, one of the geographic units most easily communicated for conservation– caves– do not match the biological units of interest. Our results support multiple independent colonization events from a widespread surface ancestor with a small degree of cave connectivity among, but not between, clusters. We suggest whole cave system conservation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Caves and associated karst groundwater aquifers provide a unique ecosystem that produces some of the most bizarre, yet fascinating, organisms known to science. Cave-obligate organisms have adapted to life in complete darkness with loss of pigment and eyes and enhanced non-visual sensory perception (Trontelj et al. 2012; Soares and Niemiller 2013, 2020). Additionally, cave-adapted organisms are indicator species that show signs of toxic environmental conditions before the effects can reach humans (Danielopol 1981; Malard et al. 1996; Doran et al. 1999; Culver et al. 2000). Even with this significance, the conservation of cave life has trailed surface counterparts, in part due to difficulties in monitoring (Gibert et al. 2009). Caves and karst groundwater aquifers support unique assemblages of species; yet, these ecosystems and their inhabitants have long been overlooked in conservation science, lagging in research and protection compared to surface ecosystems (Niemiller et al. 2018, 2019; Mammola et al. 2019; Gladstone et al. 2022).
In addition to monitoring challenges, cryptic diversity, uncertain species distributions, and unknown anthropogenic impacts make assessing conservation needs for cave organisms difficult (Foster and Chilton 2003; Culver et al. 2009; Gibert et al. 2009; Trontelj et al. 2009; Niemiller et al. 2018, 2019; Mammola et al. 2019). Molecular investigations of cave-obligate taxa have uncovered genetically distinct lineages that lack morphological characters to distinguish them (Niemiller et al. 2012; Devitt et al. 2019). This unseen cryptic diversity complicates our ability to assess the distributions of lineages and, in turn, limits assessment of extinction risk. However, it is most likely that these animals have high endemism, and individual genetic lineages are at great conservation risk (Trontelj et al. 2009; Niemiller et al. 2012, 2013b; Devitt et al. 2019). Accurate estimation of population and range sizes are key components to conservation assessments (e.g., IUCN Red List and NatureServe).
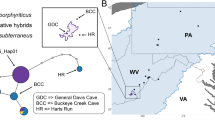
The Southern Cavefish (Typhlichthys subterraneus; Fig. 1), native to cave systems in southeastern North America, is an excellent study organism to address questions related to the evolutionary and biogeographic history of cave-obligate organisms. The Southern Cavefish has the largest range of any cavefish (Proudlove 2010; Niemiller et al. 2012); however, this species is comprised of several distinct genetic lineages (i.e., it is a species complex), including an Ozark group currently recognized as the Salem Plateau Cavefish (T. eigenmanni, included in this study) (Swofford 1982; Niemiller and Poulson 2010; Niemiller et al. 2012). The Southern Cavefish species complex is distributed across at least five principal karst aquifers and six surface HUC subregions, and localities in new karst regions are still being discovered (Fig. 2) (Niemiller and Poulson 2010; Niemiller et al. 2012, 2016). Multiple Southern Cavefish lineages have been assessed as Vulnerable, Critically Imperiled, or Imperiled based IUCN Red List assessments (Niemiller et al. 2013b).
A) Typhlichthys fishes are relatively small (~ 30–100 mm) blind, depigmented, cave-obligate fishes that live in streams, pools, and aquifers in North America. Photo by P. Chakrabarty. Map of sampling localities colored by genetic clusters designated by B) STRUCTURE with C) DAPC with principal aquifers in the background. Localities with individuals grouping in different genetic clusters are displayed as pie charts, representing the percentage of individuals assigned to each genetic cluster
Genetic clusters designated by A) STRUCTURE and B) DAPC analysis. Individuals grouped into different clusters by DAPC than those in STRUCTURE are highlighted by different colors. Ts = Typhlichthys subterraneus, Te = T. eigenmanni. Cave codes can be found in Table S1. C) Adapted multispecies coalescent phylogenetic reconstruction from Hart et al. (2020) with clades colored by STRUCTURE genetic clustering (bottom-up color blocks) and DAPC genetic clustering (top-down color blocks)
A looming question in speleobiology has been the feasibility of subterranean dispersal by cave-obligate organisms (Culver et al. 2009; Gibert et al. 2009). Most cave-obligate species appear to have small, restricted ranges (Culver 1970, 1976; Culver et al. 2009); however, there are a small percentage of cave-obligate organisms (like the Southern Cavefish) that have broader distributions including spiders (e.g., Phanetta subterranea), pseudoscorpions (e.g., Hesperochernes mirabilis), flies (e.g., Spelobia tenebrarum), springtails (e.g., Pseudosinella hisuta), copepods (e.g., Caecidotea bicrenata), and planarians (e.g., Sphalloplana percoeca Niemiller et al. 2013c; Slay et al. 2016). Can subterranean dispersal best explain the seemingly broad distributions of some cave-obligate species, such as the Southern Cavefish? A “population” can be classically defined as a group of organisms occupying the same time and place so that interbreeding is possible (Tarsi and Tuff 2012); the term “place” is an integral part to this definition, and not always straightforward as it relates to cave organisms. Traditionally, a “cave” may be thought of as an isolated location or even an island (Culver 1970, 1976), leading to the assumption that a cave itself is a population; however, connectivity between caves can be unknown and create uncertainty. With aquatic cave-obligate organisms, we have a unique way to address the question of cave connectivity with aquifers. All karst caves are or were related to a water source, such as an aquifer (Gilli 2011). Cave vertebrates are known to inhabit aquifers (e.g., the Edward’s Aquifer) (Longley 1981; Krejca and Reddell 2019). Aquifers may connect multiple caves through water-filled conduits, possibly allowing for interbreeding among aquatic cave-obligate organisms between caves, creating populations larger than a single cave. The definition of populations and population size is crucial for some conservation management assessments (e.g., NatureServe). Evolutionary relationships among the Southern Cavefish lineages have been examined using allozyme alleles, mitochondrial genes, and legacy nuclear loci, and have brought cavefishes into the conservation spotlight (Swofford 1982; Niemiller et al. 2012, 2013b; Burress et al. 2017).
Swofford’s (1982) allozyme analyses indicated differentiation in geographically close groups of the widespread Southern Cavefish though sample size prevented further investigation into genetic subdivision. Niemiller et al. (2012) examined cryptic diversity within the Southern Cavefish using de novo species delimitation on nuclear and mitochondrial markers, interrogating the differences in species delimitation with different datasets (i.e., differing the number of individuals and genes per dataset). These analyses provided multiple species designation schemes, but overall found up to 15 putative cryptic species within the Southern Cavefish, thus designating it as a species complex (Niemiller et al. 2012). In that study, a genealogical sorting index suggested mixed answers to exclusive ancestry of delimited species. Hierarchical analysis of molecular variance (AMOVA) with each cave as a population in Niemiller et al. (2012) also indicated genetic structuring among surface hydrological basins, subbasins, and ecoregions for both nuclear and mitochondrial loci examined. Due to strong phylogenetic distinctiveness, Typhlichthys eigenmanni (the Salem Plateau Cavefish) was resurrected for sites located west of the Mississippi River in Missouri and Arkansas (Niemiller et al. 2012). The conservation implications of cryptic species within the Southern Cavefish were explored by Niemiller et al. (2013b). One of the lineage designation schemes from Niemiller et al. (2012) was used to assess individual lineage conservation risks using the IUCN Red List and NatureServe criteria (i.e., ten lineages), with one lineage designated as “Critically Endangered”, and most others assessed as at risk for extinction. Morphological differences among lineages as designated by Niemiller et al. (2012) as well as among geographic areas were explored by Burress et al. (2017) to determine if geometric morphometrics could provide support for species description (i.e., 15 lineages); however, no morphological differences among lineages or geographic areas were found.
Genome-wide studies have uncovered interesting population structure not previously found in methods using legacy markers (e.g., Coghill et al. 2014; Devitt et al. 2019). Contemporary population genomic methods analyzing thousands of single-nucleotide polymorphisms (SNPs) can reveal unidentified genetic structure, allowing for better informed conservation assessments (Devitt et al. 2019). With SNPs mined from genome-wide UCE data presented in Hart et al. (2020), we re-evaluated genetic structure within the most wide-ranging cavefish species complex. Our goal was to explore population genomic structure and test geographic population genetic hypotheses (i.e., surface hydrological basin and aquifer connectivity). First, we performed genetic cluster analysis in a Bayesian framework (STRUCTURE) and with Discriminant Analysis of Principal Components (DAPC), then estimated diversity of genetic clusters using pairwise FST and genetic distances. We performed hierarchical analyses of molecular variance (AMOVA) using surface hydrological basin and subbasin (HUC 6 and HUC 8) as well as principal aquifers for our geographic levels to examine potential association between genetic structure and hydrology. Lastly, phylogenetic relationships from Hart et al. (2020) were adapted to further discuss the evolutionary history of the species complex.
Methods
Samples and data analysis
An extensive sampling effort across ten years of work went into the collection of tissues for the Southern Cavefish species complex. Sampling was performed to maximize the range of samples, minimize impact of sampling on putative populations, and to find new localities that could fill gaps in species distribution. The entirety of the Typhlichthys species range was sampled. Tissues were obtained from museum genetic repositories or from specimens captured in the field following IACUC and agency approved protocols (Table S1; IACUC Protocol 19–091). All sequence data were archived samples from Hart et al. (2020; PRJ610650), where UCE loci were generated. UCE loci were chosen due to the level of sequence conservation, which allows for comparisons across taxa at a deeper level, but that also contains variable sites for more shallow inference in adjacent flanking regions (Faircloth et al. 2012; Harvey et al. 2016; Alda et al. 2019). All UCE phylogenetic reconstructions are adapted from Hart et al. (2020). The SNP dataset presented herein were mined from the UCE dataset in Hart et al. (2020). SNPs were mined for Typhlichthys population genomic estimates using the seqcap_pop pipeline (Harvey et al. 2016)(SI 2; Zenodo #).
Genetic cluster analysis
Genetic clusters were estimated from the SNP dataset using the Bayesian statistical program STRUCTURE (Porras-Hurtado et al. 2013) and by Discriminant Analysis of Principal Components (DAPC) (Jombart et al. 2010). STRUCTURE is a model-based method in that it jointly infers parameters for each K and the cluster membership of each individual by assuming each individual from each cluster is a random draw from a parametric model (Pritchard 2000; Falush et al. 2003, 2007; Porras-Hurtado et al. 2013). For the STRUCTURE analysis, burn-in was set at 100,000 with 10,000,000 Markov Chain Monte Carlo (MCMC) replicates after burn-in. We conducted runs exploring a range of values of K from one to 17 populations based on previous investigation indicating as many as 15 genetic lineages (Niemiller et al. 2012). Ten runs were performed for each K.
We used both STRUCTURE HARVESTER and CLUMPAK to merge runs, visualize STRUCTURE data, and to determine the best fitting K using the ∆K Method (Evanno et al. 2005; Earl and vonHoldt 2011). To determine the most likely number of genetic clusters, STRUCTURE estimates the posterior probability of each K by estimating the log likelihood of the data at each MCMC step. These log likelihoods are averaged, and half of their variance is subtracted to the mean; this is known as ‘Ln P(D)’. Once the most likely K is reached Ln P(D) plateaus, and an ad hoc quantity based on the second order rate of change of the likelihood function provides a clear peak at the real K value, a quantity known as ∆K (Evanno et al. 2005).
DAPC was performed using the adegenet v 2.1.4 package in R Studio v. 1.2.1335 with R version 3.6.0 (Jombart 2008). First, the input file from STRUCTURE was imported as a genind object. Then, the dudi.pca function was performed for Principal Components Analysis. The find.clusters function was used to obtain BIC scores for the best fit number of clusters. In this, the best number of clusters was eight. Cross-validation was then performed using xvalDapc with 1000 repetitions and the lowest Mean Squared Error was produced by eight PCs and seven discriminant functions. These eight PCs and seven discriminant functions were then used in the dapc function.
Lastly, we have adapted previously constructed phylogenomic hypotheses from Hart et al. (2020) to visualize the evolutionary relationships of proposed Typhlichthys genetic clusters; these relationships have not been previously detailed.
Genetic diversity estimates and geographic genetic structuring
Analyses for genetic diversity estimates were conducted in R (v.3.9.0) using the SNP dataset with both STRUCTURE (n = 5) and DAPC (n = 8) groupings. We obtained overall (i.e., averaged across loci) statistics including the global fixation index (Nei 1973), observed heterozygosity (HO), overall gene diversity (HT), and amount of gene diversity (DST) using the basic.stats function in the package hierfstat (Goudet 2005). Genetic variance was estimated using pairwise FST according to Weir and Cockerham (1984) with the pairwise.WCfst function. We used the function boot.ppfst to produce upper and lower confidence limits with 100 bootstrap replicates for our pairwise FST.
We calculated both Nei’s standard genetic distance and Cavalli-Sforza and Edwards Chord distance using the genet.dist function specifying method as “DS” and “DCH”, respectively. D statistics, or genetic distances, are measures of genetic divergence between populations by using allele frequency data. Genetic structuring estimates based on geography were obtained using analysis of molecular variance (AMOVA). For these analyses, we used the poppr package (Kamvar et al. 2014, 2015) in R with the function poppr.amova using the ade4 AMOVA implementation. We used quasi-Euclidean distances as a correction method for non-Euclidean distances. To obtain significance values, the function randtest was used with 999 iterations.
Niemiller et al. (2012) found that genetic variation in Typhlichthys based on mitochondrial and nuclear loci was correlated with surface hydrological boundaries, including basins (HUC6) and subbasins (HUC8), with most of the variation partitioned by subbasin (HUC8). Hydrologic Unit Codes (HUCs) are a system of divided hydrologic units nested within one another by size of the geographic region, such that the higher the number, the more subunits (i.e., HUC8 is nested within HUC6). Each subunit has its own unique HUC that consists of the number of digits in the HUC level (i.e., HUC6 basins have six-digit codes) (Seaber et al. 1987).
We additionally wanted to examine if genetic variation was correlated with sub-surface hydrological boundaries, as aquifers may provide a means for interconnection among caves. Thus, we examined genetic variation with both the STRUCTURE and DAPC datasets using hierarchical AMOVAs grouping by HUC(6) basin, HUC(8) subbasin, and principal karst aquifer (Seaber et al. 1987; Miller 2000).
Results
Our two genetic cluster analyses found K = 5 and K = 8 to be the best fit (STRUCTURE and DAPC, respectively). The genetic clusters designated by STRUCTURE are denoted with a “P” and DAPC with a “G” for the rest of the paper. We found instances where individuals from the same cave grouped with different genetic clusters in both analyses. We estimated low genetic diversity and high genetic structure with both STRUCTURE and DAPC groups. We recovered no relationship between the geographic levels tested (aquifers, HUC6s, and HUC8s) again for groups from both STRUCTURE and DAPC. Clades in the phylogenies and groups in the splits network corresponded to STRUCTURE groups and most DAPC groups, though one DAPC group was non-monophyletic as another group was nested within it.
Genetic cluster analyses
Using Evanno’s ∆K, we found the most likely number of genetic clusters from the SNP dataset using STRUCTURE was five (Tables S3 and S4; ∆K = 30.94). We had clearly delineated genetic clusters with little to no admixture between clusters (Fig. 2A). No individual had over 16% ancestry assignment to multiple clusters (e.g., TsSHL02, 84% ancestry assigned to P5; Table S5); however, we intriguingly found five caves in which individuals were assigned to multiple genetic clusters (e.g., Hering and Bobcat caves in Madison Co., Alabama; Jacque’s Cave in Putnam Co. Tennessee; Fig. 2A, S1A; Table S5). For example, Bobcat Cave individual TsBOB05 (P1) does not group with TsBOB03 (P5) or TsBOB06 (P5) while TsBOB01(P1) shares 7.7% genetic composition with the genetic cluster of TsBOB03 and TsBOB06 (Fig. 2A&B, S1A).
We found the most support for eight genetic clusters using DAPC. Individuals were assigned to nearly the same genetic clusters between the STRUCTURE analysis and the DAPC analysis (Fig. 2B, S1B); however, three genetic clusters from STRUCTURE were split into two distinct clusters each in the DAPC (e.g., STRUCTURE group P3 = DAPC group G3 and G7; Fig. 2A&B, S1A&B, Table S3). All individuals were assigned with 100% of their ancestry to one genetic cluster in the DAPC analysis (i.e., there was no admixture among clusters; Fig. 2B, S1B). Notably, the Salem Plateau Cavefish (Typhlichthys eigenmanni) was recovered as its own distinct cluster in the DAPC analysis (G7; Fig. 2B). Additionally, individuals from one lineage defined by Niemiller et al. (2012; lineage A) consistently grouped together in both the STRUCTURE and DAPC analyses (Group P4/G4 in STRUCTURE and DAPC; Fig. 2A&B).
We then compared the alternative genetic clustering scheme for each analysis (i.e., we examined STRUCTURE with K = 8 and DAPC with K = 5, each other’s best K). Interestingly, the results were not identical between STRUCTURE K = 5 and DAPC K = 5 nor with STRUCTURE K = 8 and DAPC K = 8. The Salem Plateau Cavefish (Typhlichthys eigenmanni) was not recovered as its own genetic cluster in DAPC K = 5; rather, the Salem Plateau Cavefish grouped with DAPC group G5 (Fig. S2A-D). STRUCTURE K = 8 did recover what was DAPC group G6 in DAPC K = 8, but additional clusters (P7 and P8) did not align with DAPC clusters G7 or G8 (Fig. S2A-D).
Genetic diversity estimates and geographic genetic structuring
With our STRUCTURE (n = 5) groupings, we found low estimates for observed heterozygosity (HO = 0.0129), observed gene diversities (HS = 0.051), overall gene diversity (HT = 0.108), and gene diversity among samples (DST = 0.057). The overall genetic structure was high, at 0.531(Nei’s FST)(Nei 1973). All pairwise FST estimates were high (Tables S6 & S7). The largest estimate was between P2 and P4 (FST = 0.695). Genetic distances were low between groups (Table S8). The genetic clusters with the greatest genetic distance according to both Nei’s DS and Cavalli-Sforza and Edwards Chord distance (DCH) were P1 and P4 (DS = 0.100, DCH = 0.120; Table S8).
Using the genetic clusters as designated by DAPC (n = 8), we found similar results with observed gene diversities as slightly lower (HS = 0.039) and gene diversity among samples slightly higher (DST = 0.068). Overall genetic structure based on Nei’s FST (1973) was even closer to 1 (FST = 0.636; Tables S9 & S10). Pairwise estimates for FST were again similar between STRUCTURE and DAPC analyses, in that most were high, except for G5 vs. G8 (FST = 0.383). The greatest genetic distance was G4 and G8 with both Nei’s DS (DS = 0.138) and Cavalli-Sforza and Edwards Chord distance (DCH = 0.113; Table S11).
We found no relationship between genetic structure as delimited by STRUCTURE (i.e., five clusters) and HUC6 basin (p = 0.311, phi = 0.021; Table 1 A), HUC8 subbasin (p = 0.263, phi = 0.047; Table 1B), or aquifer (p = 0.718, phi = -0.053; Table 1C). We additionally found no relationship between DAPC (i.e., eight) genetic clusters and HUC6 basin (p = 0.304, phi = 0.019; Table 2 A), HUC8 subbasin (p = 0.034, phi = 0.338; Table 2B), or aquifer (p = 0.745, phi = -0.059; Table 2 C).
Spatial geography did not appear to be correlated with levels of divergence among related genetic clusters. The most closely related group to the Salem Plateau Cavefish (DAPC group G7) was not the closest geographically: G8 from DAPC (STRUCTURE group P5) was physically closer to the Salem Plateau Cavefish (~ 234 straight-line miles apart), yet the DAPC group G3 (STRUCTURE group P3) found in north-central Tennessee into Kentucky was most closely related phylogenetically (~ 279 miles apart; Figs. 1B, amp and C and 2 A-C, S1-7).
Phylogenomic hypotheses: Tree and Network
Genetic clusters from the STRUCTURE analysis were found to be clades in the multispecies coalescent analysis. Multispecies coalescent analysis recovered genetic clusters P1 and P2 as sister lineages, with P5 as the most closely related, followed by P4. P3 is the sister-group to the rest of the Typhlichthys genetic clusters (Fig. 2C, S6). We recovered the relationship between P1, P2, and P5 with low support (< 0.7 bootstrap support). As with the genetic cluster analysis, individuals from the same cave in some instances were recovered in different clades (e.g., Hering Cave; Fig. 2C, S6). Short internal branch lengths indicate relatively few expected numbers of substitutions between clades.
When matching the phylogeny to the DAPC clustering dataset, six out of eight genetic clusters were clades; however, group G8 was nested within group G5, recovering group G5 as non-monophyletic (Fig. 2C & S6). Other than this relationship, groups from STRUCTURE that were split by DAPC were recovered as sister to one another (e.g., group P1 in STRUCTURE became groups G1 and G6 in DAPC and were reciprocally monophyletic as were groups G3 and G7, which were P3 in STRUCTURE; Fig. 2C, S6).
Most clusters were identifiable as groups within the splits network with both STRUCTURE and DAPC grouping, except for DAPC group G8 found nested within DAPC Group G5/STRUCTURE group P5 (Fig. S7). We found the most reticulation at the base of the clusters rather than among the tips, indicating co-ancestry or reticulate events deeper in the phylogeny. More shallow reticulate events are seen in DAPC group G6 (part of DAPC G1/STRUCTURE group P1) and G7 (DAPC group G3/STRUCTURE group P3; Fig. S7). We did recover both deep and some shallow reticulation between the Salem Plateau Cavefish (DAPC group G7) and DAPC group G3/STRUCTURE group P3 (Fig. S7).
Discussion
We utilized population genomics analyses over both broad and shallow geographic scales and across complicated genetic relationships within the Southern Cavefish species complex to examine modes of cave-adaptive evolution. We found high genetic distinctiveness and low genetic exchange between groups using two genetic cluster analyses. Low heterozygosity, high genetic structuring, and short branch lengths suggest recent and incomplete divergence within the Southern Cavefish species complex- all of which are concerning in a conservation context. Our results support the Southern Cavefish species complex’s wide distribution as a consequence of a widespread surface ancestor that had existing genetic structuring and independently invaded the subterranean habitat multiple times. These results could also support some level of undiscovered hydrologic connectivity within genetic clusters that is not related to principal aquifers. We discovered that a cave is not a proxy for a population within the Southern Cavefish species complex. This work is applicable to other at-risk groundwater fauna, such as salamanders, crayfishes, amphipods, isopods, etc., many of which co-occur to varying degrees with cavefishes. Preserves for caves with the most genetic diversity should be prioritized, but groundwater recharge area protection could save whole cave systems.
Genetic diversity in the Southern Cavefish species complex
Sister relationships of Typhlichthys genetic clusters are somewhat inconclusive due to low support values and short internodes in our multispecies coalescent analysis. The low support and short internodes may be due to large amounts of non-bifurcating signal as shown in our splits network analysis; these results have not been recovered in previous examinations (Niemiller et al. 2012; Burress et al. 2017). Niemiller et al. (2012) performed species delimitations with multilocus (3, 6, and 9 gene) datasets on the Southern Cavefish species complex and found substantially more genetic structure than that found in the current study (delimiting up to 15 lineages). We found that results were not identical when the best K was applied to the opposite method (i.e., STRUCTURE K = 5 and DAPC K = 8), and new clusters may suggest additional population structure. STRUCTURE guidelines recommend that users go with the smallest number of genetic clusters that explain the data, thus why we chose to explore K = 5 and 8 and not larger K values (Pritchard 2000; Falush et al. 2003, 2007).
We found just one lineage from Niemiller et al. (2012) that aligned with our genetic cluster P4/G4. This genetic cluster is also the most genetically distinct based on our genetic diversity estimates using both grouping schemes. We additionally recovered the Salem Plateau Cavefish (Typhlichthys eigenmanni) nested within the Southern Cavefish (T. subterraneus). Our results with regards to the Salem Plateau Cavefish are consistent with previously molecular examinations into the species complex (Niemiller et al. 2012; Burress et al. 2017).
Our low genetic diversity estimates (HO, HS, HT, DST) indicate small effective population sizes, which can have significant impacts on the survival and fitness of a population (Ellstrand and Elam 1993). We found that genetic structuring did not have a relationship with either surface or subsurface geographical boundaries; this was a surprising result as a previous investigation found that genetic structuring aligned with surface hydrological boundaries within the Southern Cavefish species complex (Niemiller et al. 2012). Though UCEs and SNPs mined from them have been shown to be informative at both higher-order and population level scales (Faircloth et al. 2012; Harvey et al. 2016), there is a possibility that these loci, due to their conserved nature, may not be variable enough to detect population structure in this group. Follow-up investigations utilizing whole genome sequencing would be beneficial to further understand the nuances of these cavefish populations.
Short branch lengths on both phylogenetic reconstructions in conjunction with extremely low genetic distances yet high genetic structuring suggest incomplete lineage sorting (as opposed to introgression or hybridization among genetic groups) as an explanation for the non-bifurcating and complex relationships among Typhlichthys. Additionally, due to its linear relationship with time, our very low DS estimates support recent diversification (Tables S8 & S11). Support for recent diversification of the species complex was also found in Niemiller et al. (2013a), in which the authors estimated diversification began around one to three million years ago.
The Salem Plateau Cavefish (T. eigenmanni) was recovered as a clade and as a genetic cluster in our phylogenomic and one of our population genomic analyses (DAPC, G7; Fig. 2B, S1). Genetic distances were also relatively higher with regards to the Salem Plateau Cavefish with the rest of the Southern Cavefish genetic clusters (Table S8 & S11). Despite these analyses, the Salem Plateau Cavefish remains nested within the Southern Cavefish (Fig. S7). We recommend retaining the Salem Plateau Cavefish under a separate name while continuing to consider it a part of the Southern Cavefish species complex.
Cave is Not a Proxy for Population
The presence of individuals in the same cave belonging to different genetic clusters is a surprising finding, as previous studies of groundwater organisms in the region typically only find one lineage in a cave. A few exceptions to this include multiple species of cave crayfishes (Cambarus spp.), that are known to co-occur in a few caves in Northern Alabama (interestingly, also co-occurring with Typhlichthys in these caves) (Culver 1970; Culver et al. 2000), and among the amblyopsids themselves. One of the caves with multiple Typhlichthys lineages is Hering Cave located east of Huntsville, AL. The authors previously predicted that two morphotypes occur in Hering cave that differ in expressed pigment (patches vs. more evenly distributed color) and in head shape (wide with a blunt snout versus narrow with a more pointed snout; JWA pers. obs.). These differences may be artifacts as pigmentation is generally only expressed after exposure to light (Poulson 1963) and allometry obscures shape differences (Burress et al. 2017); however, presence of two lineages within a cave with no apparent admixture would support the separation of genetic clusters into species. An additionally interesting result is the Hering Cave individuals grouping with P2/G2 and P3/G3 (STRUCTURE and DAPC, respectively), despite most localities assigned to these genetic clusters not being spatially close to this cave. One locality, Bobcat Cave (BOB), is found in two of the three additional genetic clusters designated by DAPC, and these clusters are not sister groups (Fig. 2B&C, S1, S7). Additional research with more robust sampling is needed on caves that contain multiple genetic clusters.
The finding of multiple genetic clusters within the same cave at first seems counterintuitive; however, there may be physically boundaries and/or sexual selection preventing gene flow. There is already syntopy (i.e., species found at the same locality) among cavefishes in the Amblyopsidae, including the Alabama Cavefish (Speoplatyrhinus poulsoni) and the Southern Cavefish in Key Cave, Alabama (Kuhajda and Mayden 2001). Additionally, the Northern Cavefish (Amblyopsis spelaea) and the Southern Cavefish (T. subterraneus) are found in syntopy in Mammoth Cave, Kentucky (Helf and Olson 2017; Niemiller et al. 2021). As for physical boundaries, the Southern Cavefish lives in high-gradient streams that are shallower in vertical cave depth than the Northern Cavefish, which occurs in base-level habitats, separating them (Helf and Olson 2017). With regards to the Alabama Cavefish and the Southern Cavefish, these fishes are in the same pools (Kuhajda and Mayden 2001). The species may have once been separated similarly to the Northern Cavefish and the Southern Cavefish in Mammoth Cave, and connectivity could have been created via a conduit or cave breakdown.
Broader implications for subterranean speciation
We found five extremely well-defined genetic clusters using STRUCTURE analysis and eight with DAPC, yet connectivity within genetic clusters is still somewhat unclear. Little genetic admixture between genetic clusters was found, such that no individual had more than 16% genetic makeup from a cluster other than their primary assigned cluster in STRUCTURE. Our DAPC analysis supported no admixture between eight genetic clusters. These results suggest high genetic distinctiveness for each individual genetic cluster, with minimal to no genetic exchange occurring among clusters. Though we found little to no connectivity among genetic clusters, there may be connectivity within clusters across boundaries including aquifers. We found that multiple caves showed homogeneous genetic structuring, indicating either historical structuring or current and on-going gene flow between these caves. We additionally recovered relatively shallow reticulation events among genetic clusters including between the Salem Plateau Cavefish (DAPC group G7) and it’s most closely related Southern Cavefish genetic group (group P3/G3 in STRUCTURE and DAPC), indicating gene flow between the groups at some point.
Our result that the genetic clusters do not align with geographic and hydrological boundaries and finding shallow reticulation events suggests within-group genetic exchange across boundaries. Within-group genetic exchange across hydrological boundaries is also supported by the higher levels of non-bifurcating signal within genetic clusters in comparison to between clusters, potentially supporting a hierarchical island model (Excoffier et al. 2009). Hierarchical island model support within cave-obligate vertebrates has also been found within Texas cave salamanders (Eurycea spp.) (Devitt et al. 2019), perhaps pointing to a larger pattern among cave-obligate organisms. An additional explanation for cross-boundary genetic exchange could be subsurface stream capture. Stream capture occurs when an eroding lowland stream diverts part of the drainage water of a higher-land stream (Lauder 1997). A stream capture event within a subterranean system with streams on two levels corresponding with different aquifers may be a way for gene flow to occur. Evidence of dispersal events associated with stream capture and other cave-obligate fauna has been found along the southern United States Cumberland Plateau (part of the Interior Low Plateau karst region) and the Appalachian karst Ridge and Valley region (Fig. S8; Culver 2000; Burress et al. 2017), including amphipods (Crangonyx attennatus), the Tennessee Cave Salamander (Gyrinophilus palleucus), and isopods (Caecidotea spp.)(Niemiller et al. 2008; Niemiller et al. 2019).
The extremely low evidence of admixture suggests isolation of genetic clusters, in that physical connectivity (i.e., subterranean conduits or surface connections) between genetic clusters may be minimal to absent. However, high genetic structuring may indicate some subterranean connectivity among genetic clusters that is unrelated to primary aquifers. We did not find a connection between principal karst aquifers and genetic structure, but these principal aquifers represent only the uppermost level of groundwater containment. The three-dimensional complexity, both spatially and temporally (e.g., water level fluctuations creating and removing connections over time), of aquifers means that it is difficult to physically study the interconnectivity of aquifers. Instead of using geography to understand species genetic structure, we may be able to use population genomics to connect geography. Patterns of species distributions are often used to either affirm or predict connections between geographic areas (Lujan et al. 2011), thus, phylogenomics and population genomics of groups like Typhlichthys may be useful in determining how aquifers are connected.
The only other cavefish for which population genomic studies have been conducted is the model Mexican Blind Cave Tetra Astyanax mexicanus (Bradic et al. 2012b, 2013; Coghill et al. 2014; Fumey et al. 2018; Herman et al. 2018). Support for cave-to-cave migration has been found in the species using microsatellite loci (Bradic et al. 2012a). Genetic exchange occurred between caves in the same geographic region (i.e., caves in the El Abra or Guatemala region). Additionally, with regards to the El Abra region, migration rates decreased with increased geographic distance (Bradic et al. 2012a). Further evidence suggests introgression within one cave between two independent invasion event populations (one “old” and one “new”) (Strecker et al. 2012; Herman et al. 2018). The migration and introgression results for the Mexican Blind Cave Tetra support previous hypotheses about underground connections between caves.
One limitation we encountered was the total number of populations we could test for in STRUCTURE. If there is in fact no cave connectivity and each cave is truly a population, we would have had to test over a K of 40 in STRUCTURE, which is not computationally feasible. Instead, we designated genetic clusters as “populations” and performed downstream analyses on these. Additionally, we acknowledge that the best K according to likelihood ratio or BIC criteria may not actually be the true K.
Support for climate-relict hypothesis
A second interpretation for cross-boundary genetic signal could be a wide-spread surface ancestor invading caves multiple times, and subsequently going extinct. The curious current distribution of the Southern Cavefish species complex may become clearer when comparing it to the Mexican Blind Cave Tetra. Multiple independent colonization events have occurred in the cave tetra, and the species has both surface and cave populations that can interbreed (Bradic et al. 2013; Coghill et al. 2014). If the surface populations of the Mexican Blind Cave Tetra were to go extinct, the cave tetra’s distribution may look a bit like the Southern Cavefish species complex currently does (Fig. S4 A&B).
Additional support for multiple independent colonization events within the Southern Cavefish species complex comes from allozyme analyses (Swofford 1982) as well as from analyses of the eye gene rhodopsin (Niemiller et al. 2013a), in which some genetic lineages possess different loss of function mutations. These results are consistent with the climate-relict hypothesis as an explanation for subterranean colonization and speciation (Holsinger 1988, 2000; Ashmole 1993; Niemiller et al. 2008). The climate-relict hypothesis expounds that, as climate fluctuations occurred during the late Pliocene and Pleistocene, surface ancestors that were adapted to cool and moist temperate environments retreated to subterranean habitats; the surface populations then became extirpated due to inhospitable surface conditions, facilitating allopatric speciation of the subterranean populations (Holsinger 1988, 2000; Ashmole 1993; Niemiller et al. 2008). Additional support for this hypothesis includes the biogeography of other co-occurring cave-obligate aquatic species, including the Tennessee Cave Salamander, that have independently invaded the Ozark karst region located in the central United States and the Interior Low Plateau (Fig. S8) (Niemiller et al. 2008, 2019). Lastly, a few freshwater surface fish species distributions also mirror that of the Southern Cavefish (i.e., disjunct distributions in the Tennessee-Cumberland area and the Ozarks; Fig. S8): the Whitetail Shiner (Cyprinella galactura), the Telescope Shiner (Notropis telescopus), and the Northern Studfish (Fundulus catenatus). The distributions of these surface freshwater fishes may also be linked to climatic changes (i.e., maximum glacial advances) (Starnes and Etnier 1986; Mayden 1988).
Groundwater Fauna Conservation
Our results indicate that geographic units do not correspond to the biological units of interest (i.e., genetic clusters). Evolutionarily significant units (ESUs) are a useful way to designate a group of organisms that do not easily correspond to a species definition (i.e., no morphological or geographic differentiation for definition). These units are used to classify a group of organisms that are reproductively isolated, leading to adaptive differences such that the group may be considered a separate evolutionary component of the species (Moritz 1994; Fraser and Bernatchez 2001). Previous work has been done to designate delimited lineages of the Southern Cavefish as ESUs (Niemiller et al. 2013b). The genetic clusters found in this investigation could also be considered ESUs. Our genetic cluster designations also provide a new hypothesis from which morphological examinations can occur. With new clustering data come new opportunities for comparisons. Future work will examine if our genetic clusters and ESUs will be echoed in morphological or physiological data, potentially leading to species designations.
This project is the largest application of these techniques to an endemic North American cave-obligate species complex that extends across many hydrological boundaries both above and below ground. We found evidence within Typhlichthys for high genetic distinctiveness, no genetic exchange between clusters, and extremely low heterozygosity, all aspects concerning to the maintenance of healthy populations in conservation (Allendorf and Leary 1993). This result is unsurprising, as apparently wide-ranging cave-obligate organisms have often been found to contain cryptic diversity with high levels of genetic structuring (Danielopol et al. 2000; Trontelj et al. 2009; Niemiller et al. 2012; Devitt et al. 2019). Issues of uncertain evolutionary relationships, population structures, and species distributions are ongoing problems in cave biology and for conservation efforts across continents (Ferreira et al. 2007; Culver et al. 2009; Zakšek et al. 2009; Bichuette and Trajano 2010; Trajano et al. 2016; Sciuto et al. 2017; Devitt et al. 2019; Gladstone et al. 2022).
Groundwater is vital not only to humans and groundwater fauna, but aquifers support surface ecosystems including wetlands, rivers, and lakes (Hancock et al. 2005; Kløve et al. 2011); despite this, groundwater depletion continues at unsustainable levels, in particular due to irrigation for food supply and due to global climate change causing prolonged droughts (Gleeson et al. 2012; Wada et al. 2012; Famiglietti and Rodell 2013; Grogan et al. 2017; Dalin et al. 2019). Some attention in the United States has been generated with groundwater levels supplying California seeing the greatest depletions (Famiglietti et al. 2011; Scanlon et al. 2012), yet in-depth investigations into groundwater depletion across the nation are desperately warranted. As the southeastern United States is considered a hotspot for subterranean biodiversity (Culver et al. 2013; Zagmajster et al. 2019), the groundwater depletions as monitored by the Gravity Recovery and Climate Experiment are particularly concerning (Famiglietti and Rodell 2013). Additionally, the boundaries and characteristics for individual aquifers across the southeastern U.S. are far less detailed than those of other aquifers (Devitt et al. 2019), creating even more difficulty for groundwater management and conservation of groundwater fauna.
Protecting single caves alone is not adequate to preserve the genetic diversity, as our results have shown, but we would recommend the creation of cave preserves first for the caves with apparent multiple genetic clusters, coupled with the management of the aquifer recharge area around the cave or hypothesized cave system. As for managing the recharge area, it is possible that an entire cave system could be protected from groundwater pollution, effectively managing multiple caves at once. Unfortunately, recharge area delimitation is highly time- and effort-intensive. But future collaborations with resource managers and cave biologists will provide the community with greater information on aquifer recharge areas.
Data accessibility statement
Raw sequence data is archived in the NCBI Sequence Repository Archive (PRJ610650). Code for SNP mining will be archived on Dryad. Specimens accession numbers are available in the Supplemental Information.
References
Alda F, Tagliacollo VA, Bernt MJ, Waltz BT, Ludt WB, Faircloth BC et al (2019) Resolving deep nodes in an Ancient Radiation of Neotropical Fishes in the Presence of conflicting signals from incomplete lineage sorting. Syst Biol 68:573–593
Allendorf FW, Leary RF (1993) Heterozygosity and fitness in natural populations of animals. In: Conservation Biology: The Science of Scarcity and Diversity (ed. Soulé, ME). Sinauer Associates, Inc., pp. 57–76
Ashmole NP (1993) Colonization of the underground environment in volcanic islands. Mémoires de Biospélogie 20:1–11
Bichuette ME, Trajano E (2010) Conservation of Subterranean Fishes. In: Biology of Subterranean Fishes (eds. Trajano, E, Bichuette, ME & Kapoor, BG). CRC Press Boca Raton, FL, pp. 65–80
Bradic M, Beerli P, Garcia-de Leon FJ, Esquivel-Bobadilla S, Borowsky RL (2012a) Gene flow and population structure in the mexican blind cavefish complex (Astyanax mexicanus). BMC Evol Biol 12:9
Bradic M, Beerli P, Garcia-de Leon FJ, Esquivel-Bobadilla S, Borowsky RL (2012b) Gene flow and population structure in the mexican blind cavefish complex (Astyanax mexicanus). BMC Evol Biol 12:1–16
Bradic M, Teotonio H, Borowsky RL (2013) The population genomics of repeated evolution in the blind cavefish Astyanax mexicanus. Mol Biol Evol 30:2383–2400
Burress PBH, Burress ED, Armbruster JW (2017) Body shape variation within the Southern Cavefish, Typhlichthys subterraneus (Percopsiformes: Amblyopsidae). Zoomorphology 136:365–377
Coghill LM, Hulsey D, Chaves-Campos C, Garcia de Leon J, F.J., Johnson SG (2014) Next generation phylogeography of cave and surface Astyanax mexicanus. Mol Phylogenet Evol 79:368–374
Culver DC (1970) Analysis of simple Cave Communities I. Caves as Islands Evolution 24:463–474
Culver DC (1976) The evolution of aquatic Cave Communities. Am Nat 110:945–957
Culver DC, Master LL, Christman MC, Hobbs HHI (2000) Obligate Cave Fauna of the 48 Contiguous United States. Conserv Biol 14:386–401
Culver DC, Pipan T, Schneider K (2009) Vicariance, dispersal and scale in the aquatic subterranean fauna of karst regions. Freshw Biol 54:918–929
Dalin C, Taniguchi M, Green TR (2019) Unsustainable groundwater use for global food production and related international trade. Global Sustain 2:1–11
Danielopol D (1981) Distribution of ostracods in the groundwater of the North Western Coast of Euboea (Greece). Int J Speleol 11:91–103
Danielopol DL, Pospisil P, Rouch R (2000) Biodiversity in groundwater: a large-scale view. Trends Ecol Evol 15:223–224
Devitt TJ, Wright AM, Cannatella DC, Hillis DM (2019) Species delimitation in endangered groundwater salamanders: implications for aquifer management and biodiversity conservation. Proc Natl Acad Sci U S A 116:2624–2633
Doran NE, Kiernan K, Swain R, Richardson AMM (1999) Hickmania Troglodytes, the Tasmanian Cave Spider, and its potential role in Cave Management. J Insect Conserv 3:257–262
Earl DA, vonHoldt BM (2011) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361
Ellstrand NC, Elam DR (1993) Population genetic consequences of small population size: implications for plant conservation. Annual Review of Ecology, Evolution and Systematics, pp 217–252
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Excoffier L, Hofer T, Foll M (2009) Detecting loci under selection in a hierarchically structured population. Heredity (Edinb) 103:285–298
Faircloth BC, McCormack JE, Crawford NG, Harvey MG, Brumfield RT, Glenn TC (2012) Ultraconserved elements anchor thousands of genetic markers spanning multiple evolutionary timescales. Syst Biol 61:717–726
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:20
Falush D, Stephens M, Pritchard JK (2007) Inference of population structure using multilocus genotype data: dominant markers and null alleles. Mol Ecol Notes 7:4
Famiglietti JS, Rodell M (2013) Water in the Balance Science 340:1300–1302
Famiglietti JS, Lo M, Ho SL, Bethune J, Anderson KJ, Syed TH et al (2011) Satellites measure recent rates of groundwater depletion in California’s Central Valley. Geophys Res Lett 38:1–4
Ferreira D, Malard F, Dole-Olivier M-J, Gibert J (2007) Obligate groundwater fauna of France: diversity patterns and conservation implications. Biodivers Conserv 16:567–596
Foster SS, Chilton PJ (2003) Groundwater: the processes and global significance of aquifer degradation. Philos Trans R Soc Lond B Biol Sci 358:1957–1972
Fraser DJ, Bernatchez L (2001) Adaptive evolutionary conservation: towards a Unified Concept for defining conservation units. Mol Ecol 10:2741–2752
Fumey J, Hinaux H, Noirot C, Thermes C, Retaux S, Casane D (2018) Evidence for late pleistocene origin of Astyanax mexicanus cavefish. BMC Evol Biol 18:1–19
Gibert J, Culver DC, Dole-Olivier M-J, Malard F, Christman MC, Deharveng L (2009) Assessing and conserving groundwater biodiversity: synthesis and perspectives. Freshw Biol 54:930–941
Gilli E (2011) Karstology: Karsts, Caves and Springs - Elements of Fundamental and Applied Karstology. Taylor & Francis Group, Tampa, Florida
Gladstone NS, Niemiller ML, Hutchins B, Schwartz B, Czaja A, Slay ME et al (2022) Subterranean freshwater gastropod biodiversity and conservation in the United States and Mexico. Conservation Biology, p 36
Gleeson T, Alley WM, Allen DM, Sophocleous MA, Zhou Y, Taniguchi M et al (2012) Towards sustainable groundwater use: setting long-term goals, backcasting, and managing adaptively. Ground Water 50:19–26
Goudet J (2005) HIERFSTAT, a package for R to compute and test hierarchical F-statistics. Mol Ecol Notes, 184–186
Grogan DS, Wisser D, Prusevich A, Lammers RB, Frolking S (2017) The use and re-use of unsustainable groundwater for irrigation: a global budget. Environ Res Lett 12:1–10
Hart PB, Niemiller ML, Burress ED, Armbruster JW, Ludt WB, Chakrabarty P (2020) Cave-adapted evolution in the north american amblyopsid fishes inferred using phylogenomics and geometric morphometrics. Evolution 74:936–949
Harvey MG, Smith BT, Glenn TC, Faircloth BC, Brumfield RT (2016) Sequence capture versus Restriction Site Associated DNA sequencing for shallow systematics. Syst Biol 65:910–924
Helf KL, Olson RA (2017) In: Mammoth C (ed) Subsurface aquatic Ecology of Mammoth Cave. Springer Cham, Switzerland, pp 209–226. Hobbs, HHI, Olson, R, Winkler, E & Culver, DC)
Herman A, Brandvain Y, Weagley J, Jeffery WR, Keene AC, Kono TJY et al (2018) The role of gene flow in rapid and repeated evolution of cave-related traits in mexican tetra, Astyanax mexicanus. Mol Ecol 27:4397–4416
Holsinger JR (1988) Troglobites: the evolution of cave-dwelling organisms. Am Sci 76:85–105
Holsinger JR (2000) Ecological derivation, colonization, and speciation. In: Wilkens H, Culver (eds) Ecosystems of the World. Elsevier Oxford, pp 399–415. DC & Humphreys, WF)
Jombart T (2008) Adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24:1403–1405
Kamvar ZN, Tabima JF, Grunwald NJ (2014) Poppr: an R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ, 2, e281
Kamvar ZN, Brooks JC, Grunwald NJ (2015) Novel R tools for analysis of genome-wide population genetic data with emphasis on clonality. Front Genet 6:208
Krejca J, Reddell J (2019) Biology and Ecology of the Edwards Aquifer. In: Sharp (ed) The Edwards Aquifer: past, Present, and future of a Vital Water Resourse: Geological Society of America Memoir 215. JM, Green, RT & Schindel, GM). The Geological Society of America
Kuhajda BR, Mayden RL (2001) Status of the federally endangered Alabama cavefish, Speoplatyrhinus poulsoni (Amblyopsidae), in Key Cave and surrounding caves. Alabama Environ Biology Fishes 62:215–222
Longley G (1981) The Edwards Aquifer: Earth’s most diverse Groundwater Ecosystem? Int J Speleol 11:123–128
Lujan NK, Armbruster JW, Albert JS (2011) The Guiana Shield. In: Historical Biogeography of Neotropical Freshwater Fishes (eds. Albert, JS & Reis, RE). University of California Press, p. 224
Malard F, Mathieu J, Reygribellet J, Lafont M (1996) Biomonitoring groundwater contamination: application to a karst area in Southern France. Aquat Sci 58:158–187
Mammola S, Cardoso P, Culver DC, Deharveng L, Ferreira RL, Fišer C et al (2019) Scientists’ Warning on the Conservation of Subterranean Ecosystems, vol 69. BioScience, pp 641–650
Mayden RL (1988) Vicariance biogeography, parsimony, and evolution in north american freshwater fishes. Syst Zool 37:329–355
Miller JA (2000) Ground Water Atlas of the United States. (ed. Interior, USDot) Reston, Virginia
Moritz C (1994) Defining ‘Evolutionarily significant units’ for Conservation. TREE 9:373–375
Nei M (1973) Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci U S A 70:3321–3323
Niemiller ML, Poulson TL (2010) Subterranean fishes of North America: Amblyopsidae. Biology of Subterranean Fishes. Science Publishers Enfield, New Hampshire, p 111. Trajano, E, Bichuette, ME & Kapoor, BG)
Niemiller ML, Fitzpatrick BM, MIller BT (2008) Recent divergence with gene flow in Tennessee cave salamanders (Plethodontidae: Gyrinophilus) inferred from gene geneaologies. Mol Ecol 17:2258–2275
Niemiller ML, Near TJ, Fitzpatrick BM (2012) Delimiting species using multilocus data: diagnosing cryptic diversity in the southern cavefish, Typhlichthys subterraneus (Teleostei: Amblyopsidae). Evolution 66:846–866
Niemiller ML, Fitzpatrick BM, Shah P, Schmitz L, Near TJ (2013a) Evidence for repeated loss of selective constraint in rhodopsin of amblyopsid cavefishes (Teleostei: Amblyopsidae). Evolution 67:732–748
Niemiller ML, Graening GO, Fenolio DB, Godwin JC, Cooley JR, Pearson WD et al (2013b) Doomed before they are described? The need for conservation assessments of cryptic species complexes using an amblyopsid cavefish (Amblyopsidae: Typhlichthys) as a case study. Biodivers Conserv 22:1799–1820
Niemiller ML, Zigler KS, Fenolio DB (2013c) Cave Life of TAG. National Speleological Society
Niemiller ML, Zigler KS, Hart PB, Kuhajda BR, Armbruster JW, Ayala BN et al (2016) First definitive record of a stygobiotic fish (Percopsiformes, Amblyopsidae, Typhlichthys) from the Appalachians karst region in the eastern United States. Subterr Biology 20:39–50
Niemiller ML, Taylor SJ, Bichuette ME (2018) Conservation of Cave Fauna, with an Emphasis on Europe and the Americas. In: Cave Ecology, pp. 451–478
Niemiller ML, Taylor SJ, Slay ME, Hobbs HHI (2019) Bidiversity in the United States and Canada. Encyclopedia of caves. Culver, DC, White, WB & Pipan, pp 163–176
Niemiller ML, Helf K, Toomey RS (2021) Mammoth Cave: a hotspot of subterranean biodiversity in the United States. Diversity 13:1–21
Porras-Hurtado L, Ruiz Y, Santos C, Phillips C, Carracedo A, Lareu MV (2013) An overview of STRUCTURE: applications, parameter settings, and supporting software. Front Genet 4:1–13
Poulson TL (1963) Cave adaptation in amblyopsid fishes. Am Midl Nat 70:257–290
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Proudlove GS (2010) Biodiversity and distribution of the subterranean fishes of the World. Biology of Subterranean Fishes. CRC Press Enfield, New Hampshire, pp 41–64. Trajano, E, Bichuette, ME & Kapoor, BG)
Scanlon BR, Faunt CC, Longuevergne L, Reedy RC, Alley WM, McGuire VL et al (2012) Groundwater depletion and sustainability of irrigation in the US High Plains and Central Valley. Proc Natl Acad Sci U S A 109:9320–9325
Sciuto K, Moschin E, Moro I (2017) Cryptic cyanobacterial diversity in the Giant Cave (Trieste, Italy): the New Genus Timaviella (Leptolyngbyaceae), vol 38. Cryptogamie, Algologie, pp 285–323
Seaber PR, Kapinos FP, Knapp GL (1987) In: Survey (ed) Hydrological unit maps. USG) Denver, CO
Slay ME, Niemiller ML, Sutton M, Taylor SJ (2016) Cave Life of the Ozarks. National Speleological Society
Soares D, Niemiller ML (2013) Sensory adaptations of fishes to subterranean environments. BioScience, pp 274–283
Soares D, Niemiller ML (2020) Extreme Adaptation in Caves. Anat Rec (Hoboken) 303:15–23
Starnes WC, Etnier DA (1986) Drainage evolutionn and fish biogeography of the Tennessee and Cumberland Rivers drainage realm. In: The zoogeography of North American Freshwater Fishes (eds. Hocutt, CH & Wiley, EO). Wiley New York, pp. 325–361
Strecker U, Hausdorf B, Wilkens H (2012) Parallel speciation in Astyanax cave fish (Teleostei) in Northern Mexico. Mol Phylogenet Evol 62:62–70
Swofford DL (1982) Genetic variability, population differentiation, and biochemical relatioships in the family Amblyopsidae. Eastern Kentucky University Richmond, Kentucky
Tarsi K, Tuff T (2012) Introduction to Population demographics. Nat Educ Knowl 3:1–3
Trajano E, Gallão JE, Bichuette ME (2016) Spots of high diversity of troglobites in Brazil: the challenge of measuring subterranean diversity. Biodivers Conserv 25:1805–1828
Trontelj P, Douady CJ, Fišcher C, Gibert J, Gorički Å, Lefébure T et al (2009) A molecular test for cryptic diversity in ground water: how large are the ranges of macro-stygobionts? Freshw Biol 54:17
Trontelj P, Blejec A, Fiser C (2012) Ecomorphological convergence of cave communities. Evolution 66:3852–3865
Wada Y, van Beek LPH, Bierkens MFP (2012) Nonsustainable groundwater sustaining irrigation: a global assessment. Water Resour Res 48:1–18
Zakšek V, Sket B, Gottstein S, Franjevic D, Trontelj P (2009) The limits of cryptic diversity in Groundwater: Phylogeography of the Cave shrimp Troglocaris anophthalmus (Crustacea: Decapoda: Atvidae). Mol Ecol 18:931–946
Acknowledgements
We would like to thank our funding sources including the Cave Research Foundation Graduate Research Award (AWD-002400), the National Speleological Society Ralph Stone Fellowship (AWD-002172), the North American Native Fishes Association Conservation Research Grant (AWD-002112), the Sigma Xi Honor Society Grants in Aid of Research (AWD-002221) award to PBH, NSF-1839915 to PC, NSF DEB-1011216 to MLN, NSF DEB-1350474 to MLN, Tennessee Wildlife Resources Agency grant nos. ED-06–02149-00 and ED-08023417–00 to MLN, and Kentucky Department of Fish and Wildlife Resources grant no. PON2-660-1000003354 to MLN. We thank the Missouri Department of Conservation, Tennessee Department of Environment and Conservation, Tennessee Wildlife Resources Agency, National Park Service, U.S. Forest Service, Southeastern Cave Conservancy, Inc., Carroll Cave Conservancy, National Speleological Society, and private landowners for permitting access to caves on their properties. We thank the state agencies of Alabama, Georgia, Kentucky, Missouri, and Tennessee, the National Park Service, and the U.S. Forest Service for scientific collecting permits. Collection of specimens and tissue samples was authorized by Middle Tennessee State University IACUC (protocol no. 04–006), University of Tennessee-Knoxville IACUC (no. 1589 − 0507), and Yale University IACUC (no. 2012–10681). We would like to thank the following for their assistance with this project: F. Alda for assistance in data analysis and bioinformatics. C.D.R. Stephen, B.R. Kuhajda, and T. Fobian for tissue specimen collection assistance. E.D. Burress for discussion. The Chakrabarty Lab including D. Elias and S. Rodriguez-Machado for intellectual support. The following museums for museums for their specimen loans: Alabama Department of Conservation of Natural Resources, Louisiana State University Museum of Natural Sciences, Yale Fish Tissue Collection, North Carolina State Museum, and University of Tennessee Tissue Collection.
Author information
Authors and Affiliations
Contributions
All authors contributed to project conceptualization. Data collection and analysis performed by PBH. Sample collection by MLN, JWA, PBH, and PC. All authors contributed to manuscript writing.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Conflict of interest
The authors do not declare any conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hart, P.B., Niemiller, M.L., Armbruster, J.W. et al. Conservation implications for the world’s most widely distributed cavefish species complex based on population genomics (Typhlichthys, Percopsiformes). Conserv Genet 25, 165–177 (2024). https://doi.org/10.1007/s10592-023-01562-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-023-01562-x