Abstract
Understanding landscape impacts on gene flow is necessary to plan comprehensive management and conservation strategies of both the species of interest and its habitat. Nevertheless, only a few studies have focused on the landscape genetic connectivity of the European wildcat, an umbrella species whose conservation allows the preservation of numerous other species and habitat types. We applied population and landscape genetics approaches, using genotypes at 30 microsatellites from 232 genetically-identified wildcats to determine if, and how, landscape impacted gene flow throughout France. Analyses were performed independently within two population patches: the historical north-eastern patch and the central patch considered as the colonization front. Our results showed that gene flow occurred at large spatial scales but also revealed significant spatial genetic structures within population patches. In both population patches, arable areas, pastures and permanent grasslands and lowly fragmented forested areas were permeable to gene flow, suggesting that shelters and dietary resources are among the most important parameters for French wildcat landscape connectivity, while distance to forest had no detectable effect. Anthropized areas appeared highly resistant in the north-eastern patch but highly permeable in the central patch, suggesting that different behaviours can be observed according to the demographic context in which populations are found. In line with this hypothesis, spatial distribution of genetic variability seemed uneven in the north-eastern patch and more clinal in the central patch. Overall, our results highlighted that European wildcat might be a habitat generalist species and also the importance of performing spatial replication in landscape genetics studies.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The landscape, defined as a heterogeneous area for at least one factor of interest (Turner et al. 2001), in which organisms evolve has many implications for population ecology. For instance, its composition (i.e. which habitat or cover type are present and how much, Turner & Gardner 2015) determines the amount of available habitat for a given species or population, which is, in turn, linked to individual performances, population dynamics and thus distribution and persistence (Fahrig 2003 and references therein). In addition to composition, landscape configuration (i.e. the arrangement of spatial elements within the landscape, Turner & Gardner 2015) also interacts with population ecology since it participates in determining landscape connectivity. Landscape connectivity is defined as ‘‘the degree to which the landscape facilitates or impedes movement among resource patches’’ (Taylor et al. 1993). It refers to both structural and functional connectivity, the former relying on physical characteristics and the latter integrating behavioural responses of organisms to this physical landscape structure (Taylor et al. 2006). By affecting individual movements, landscape also impacts natal and breeding dispersal and thus gene flow and population genetic structures. Gene flow contributes to maintaining genetic diversity, therefore preventing inbreeding depression (Keller & Waller 2002) and fixation of deleterious alleles (Lynch et al. 1995), preserving adaptive potential of populations (Frankham et al. 2004) and avoiding extinction vortices (Crooks & Sanjayan 2006). Studying the impact of landscape on gene flow and describing landscape genetic connectivity is thus of primary importance to plan comprehensive management and conservation strategies.
Landscape genetics, combining population genetics, landscape ecology and spatial statistics, allows for the studying of landscape genetic connectivity and identifying landscape features that are permeable (i.e. favouring) or resistant (i.e. impeding) to gene flow (Manel et al. 2003). Several landscape characteristics have thus been shown to impact gene flow in numerous carnivore and herbivore taxa (e.g. natural and anthropogenic linear landscape features, Riley et al. 2006, Cullingham et al. 2009, Portanier et al. 2018; type of available habitats, Perez-Espona et al. 2008; or the degree of habitat fragmentation, Wan et al. 2018). In addition to structural landscape characteristics, genetic connectivity in landscapes can be impacted by other ecologically relevant factors like prey density (e.g. Yumnam et al. 2014), human disturbances (landscape of fear, Laundré et al. 2001, see e.g. Larroque et al. 2016a) or climate change (e.g. Wasserman et al. 2012). Furthermore, since landscape connectivity depends in part on the behaviour of individuals, it can vary according to the species and to the classes of individuals within species (e.g. sexes, Portanier et al. 2018; ages, Larroque et al. 2016a).
Within species, differences might also be observed between populations as a result of variations in ecological factors (e.g. mountainous and non-mountainous areas in the Pine marten Martes martes, Larroque et al. 2016b) and ecological context (e.g. behavioural differences of dispersers in the Iberian lynx Lynx pardinus, Ferreras 2001). Differences in dispersal behaviour between individuals and their consequences for spatial genetic structure have also been reported between subpopulations characterized by different demographic histories. This is for instance the case in invasive species, between already-colonized sites and spreading invasion fronts (Pizzatto et al. 2017) or between historical and recolonizing populations of European wildcats Felis silvestris silvestris (Hartmann et al. 2013; Würstlin et al. 2016). Studying landscape genetic connectivity in different populations of the same species is therefore of paramount importance to better understand the effects of the landscape on population processes (Short-Bull et al. 2011). It is especially relevant in the current context of global changes impacting the composition and configuration of landscapes (e.g. human activities, climate change; Foley et al. 2005; Turner & Gardner 2015) and animal behaviour (Marchand et al. 2014; Tarjuelo et al. 2015). Despite an increasing amount of research done in this field in recent decades, the impacts of landscape on populations of numerous species are still unknown, especially in understudied small Felidae (Anile & Devillard 2015).
Although being classified as ‘Least Concern’ by the International Union for the Conservation of Nature, European wildcat populations are declining (Yamaguchi et al. 2015) and it is considered an endangered taxa across most of its geographical range, being legally protected by European laws (Appendix II of CITES, Annex IV of the EU Habitats Directive 92/43/EEC, Appendix II of the Bern Convention) and other national legislation (Lozano & Malo 2012, Breitenmoser et al. 2019, Gil-Sanchez et al. 2020). In addition, like many carnivores, European wildcats are considered an umbrella species. Their conservation thus allows preserving numerous other species and habitat types (e.g. Eurasian lynx, Lynx lynx and mustelids, Noss et al. 1996, Virgos et al. 2002, Jerosch et al. 2009, 2018). Several threats to wildcat populations have been identified, such as human persecution and activities, habitat loss and hybridization (Yamaguchi et al. 2015, Breitenmoser et al. 2019). However, to date, while numerous studies have focused on hybridization with domestic cats (Felis silvestris catus, e.g. Beaumont et al. 2001, Oliveira et al. 2008, Hertwig et al. 2009, Devillard et al. 2014, Steyer et al. 2018, Quilodrán et al. 2019, Beugin et al. 2016, 2020, Tiesmeyer et al. 2020), only a few have focused on genetic structures and landscape genetic connectivity (Daniels et al. 2001; Hartmann et al. 2013; Mattucci et al. 2013, 2016; Steyer et al. 2016; Würstlin et al. 2016; Westekemper et al. 2021) and almost none have reported on the French wildcat despite being one of the largest populations in Europe (but see Say et al. 2012).
Spatial ecology studies nevertheless showed strong relationships between several habitat types, ecological factors and wildcat presence or habitat selection, suggesting that landscape might influence gene flow. The presence of forested areas, providing shelters (i.e. both features that allow moving without being seen by humans, predators or preys and that constitutes resting/nursing sites) have been shown to favour wildcat presence in several populations (e.g. Klar et al. 2008, Jerosch et al. 2009, Beugin et al. 2016, Anile et al. 2019). However, open areas (scrublands, meadows, pastures) providing preys are also important habitat categories for this species (Klar et al. 2008; Lozano et al. 2003; Lozano 2010). These variations in habitat preferences suggest that that the presence of both shelters and preys are more crucial for habitat suitability than the habitat type itself (Monterroso et al. 2009; Lozano 2010; Silva et al. 2013; Jerosch et al. 2018; Anile et al. 2019). Shelters might indeed be found in the dense vegetation of forested areas, but also in scrubs or tall herbaceous vegetation of open areas (Klar et al. 2008, Jerosch et al. 2009, 2018, Anile et al. 2019). Forest fragmentation seem to be another important landscape feature which has been shown to negatively impact wildcat presence (Anile et al. 2019). Similarly, human infrastructures and presence appear to be avoided (Klar et al. 2008; Oliveira et al. 2018). In the present study, we were therefore interested in determining if these diverse landscape features also impact gene flow in European wildcats, contributing valuable information for their conservation and management.
Using genotypes at 30 microsatellites from 232 individuals sampled in the French wildcat populations (excluding the Pyrenean population), we applied population and landscape genetics approaches to decipher the genetic structure of populations and to investigate which landscape elements determine gene flow. We selected landscape features based on previous knowledge on European wildcat habitat preferences. Given the importance of shelters and preys (small mammals and rabbits, Malo et al. 2004; Apostolico et al. 2015) for habitat suitability, we expected habitat categories providing such resources to be permeable to gene flow. Opposingly, habitat categories not providing such resources or being human-dominated were expected to have neutral or negative impacts on gene flow. We also accounted for forest fragmentation and expected a higher permeability of less fragmented forested patches as compared to fragmented ones. Finally, since the French population has previously been shown to be divided in two subpopulations genetically differentiated separated by the Paris-Lyon highway (Say et al. 2012), all genetic analyses were performed within subpopulations. The north-eastern population patch is thought to be the historical endemic patch, having been settled for longer than the central patch, thought to represent the colonization front of the population and thus being in expansion (Say et al. 2012). Since different dispersal behaviours and spatial genetic structures can be observed in settled or colonizing populations (see Hartman et al. 2013, Würstlin et al. 2016, Pizzato et al. 2017), differences in the permeability of landscape features to gene flow between the two population patches could also be expected.
Materials and methods
Study population and data collection
Samples mostly came from the long-term monitoring of road-killed wildcats in France (1999–2016, Say et al. 2012; Devillard et al. 2014) covering the distribution of the European wildcat as established in 2012 (Say et al. 2012), and a broad peripheral zone representing the putative expansion area. Surveys were conducted by officers of the French Office for Biodiversity (OFB), professionals of various hunting associations and by naturalists specifically trained to recognize the wildcat coat pattern. Road-killed animals were collected if they exhibited all typical wildcat pelage characteristics: (i) a tail with a large, rounded, black tip and at least two black bands that completely encircled the tail; (ii) one thin, straight, dorsal stripe, interrupted at the root of the tail; (iii) lateral stripes that are not pronounced and not linked to the back stripe; (iv) light-tawny or grey-coloured fur. A tissue sample of the ear or hairs (with bulbs) was collected for each specimen. The sample set was enhanced by local hair-trapping protocols. Overall, n = 413 samples were available for the present study (346 road-killed animals, 33 dead animals from unknown causes, 34 hair-trapped animals).
DNA extraction and microsatellite genotyping
DNA extraction was conducted at the Antagene Laboratory (La-Tour-de-Salvagny, France, http://www.antagene.com/) using Nucleospin 96 Tissue kit (Macherey–Nagel, Düren, Germany), in sterile conditions (i.e. a designated extraction room free of DNA, using disposable sterile tools, cleaning bench with bleach) and in the presence of negative and positive extraction controls. The samples were lysed overnight at 56 °C (according to manufacturer) and DNA was purified and isolated using purification columns and vacuum filtration. DNA was eluted with 100 µL of elution buffer to obtain final concentrations between 20–100 ng/µL. Extracts were stored in labelled 96-strip tube plates in a − 20 °C freezer. For each DNA sample, 32 microsatellites were amplified by three multiplexed PCRs (polymerase chain reaction, see Supplementary data 1, Table S1 and Menotti-Raymond et al. 1999; Pilgrim et al. 2005; Beugin et al. 2016, 2020) and analysed in three runs (one for each multiplex) with an automated sequencer. PCR reactions were prepared step-by-step according to a unidirectional workflow starting in a clean room with positive air pressure to prepare sensitive reagents (enzymes and DNA primers) and continuing in a pre-PCR room for combining DNA and reagents using filtered tips.
Three negative and positive controls were included per PCR reaction. PCR amplifications were then performed in a dedicated post-PCR area in 96-well microplates with a final volume of 10 µl containing 5 µl of mastermix Taq Polymerase (Type-It Microsatellite PCR Kit, Qiagen, Hilden, Germany), and either 1.66 µL of a first pool of 15 pairs of primers, 0.91 µl of a second pool of 9 pairs of primers or 0.88 µl of a third pool of 11 pairs of primers at a concentration from 0.12 to 1.00 µM each, and a mean of 30 ng of genomic DNA. Each pair of primers was coupled with fluorescent dye. Our PCR thermal protocol consisted of 5 min at 95 °C, followed by 35 cycles: 95 °C for 30 s, 56.8 °C, 57 °C or 59 °C for 90 s, and 72 °C for 60 s; and ended by an extension step at 60 °C for 30 min. PCR products were resolved on an ABI PRISM 3130 XL capillary sequencer (ThermoFisher Scientific, Waltham, Massachusetts) under denaturing conditions (Hi-DiTM Formamide, ThermoFisher Scientific, Waltham, Massachusetts) with an internal size marker prepared once and dispatched equally in all sample wells of each multiplex run. The electropherograms were analyzed independently by two analysts using GENEMAPPER 4.1 (ThermoFisher Scientific, Waltham, Massachusetts) to determine the allele sizes for each marker of each individual. When the genotypes determined by each analyst was not congruent, the electropherograms were read again, reading errors were resolved, and in case of persistent disagreement, ambiguous results were considered as missing data.
Molecular identification of wildcats
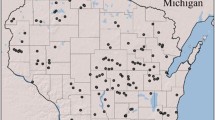
While morphological identification of wildcats can be challenging, molecular identification has been shown to have greater accuracy (Nussberger et al. 2013; Devillard et al. 2014). To reliably identify wildcats among the 413 individuals genotyped in the present study, we followed O’Brien et al. (2009), Say et al. (2012) and Devillard et al. (2014) methodology, using STRUCTURE software (Pritchard et al. 2000). As detailed in Supplementary data 2, this approach, of which the accuracy was also assessed (see Supplementary data 2), allowed us to reliably identify 272 “pure” wildcats among the 413 collected samples. Molecular identification was performed on all the available samples, including samples from the Pyrenees (south-western France). However, only a restricted number of “pure” wildcats (n = 40) were available for this part of France precluding a within-population landscape genetics analysis in the Pyrenees. No wildcats have been observed or detected in areas between the Pyrenees and the central part of France (Fig. 1). Genetic connectivity was thus unexpected between these two areas and Pyrenean samples were not considered in subsequent landscape genetics analyses, which included 232 individuals continuously distributed over central-east France.
Spatial distribution of the wildcats sampled in the north-eastern (green), central (blue), and Pyrenean (orange) population patches. In grey, the national distribution of the species (10 × 10 km grid from the European Environmental agency) from OFB data collected between 2000 and 2017 (validation from morphological and/or anatomical characters, see Devillard et al. 2014 for details). The red line indicates the Paris-Lyon highway (see Say et al. 2012). The main border countries are indicated (BE: Belgium, DE: Germany, CH: Switzerland, IT: Italy, ES: Spain)
Genotyping errors
In the wildcat dataset, two microsatellites among the 32 amplified and genotyped (F37 and FCA577) had a high number of unamplified individuals (58.8% and 51.1%, respectively) and were excluded from further analyses. We performed a correspondence analysis using the Genetix v4.05.2 software (Belkhir et al. 2004) for the 30 remaining loci in order to detect and exclude outliers. We then used MICROCHECKER v.2.2.3 (Van Oosterhout et al., 2004) to search for the presence of null alleles. Finally, three identical genotypes were identified using the matching option in GenAlex v.6.501 (Peakall & Smouse, 2006, 2012). These three genotypes were merged in a consensual genotype that was considered in downstream analyses.
Population genetics analyses
In a previous study on spatial genetic structure of French wildcats, Say et al. (2012) evidenced a clear subdivision of the continuously distributed wildcats from the central-east part of France. Two subpopulations were identified: the north-eastern and the central patches. Based on these previous results and after having verified that the spatial structure is still present in our dataset (Supplementary data 3), we divided it in two subsets for subsequent analyses. The subdivision was done based on the Paris-Lyon axis (Fig. 1), supposed to be responsible for the genetic differentiation observed between the two patches (Say et al. 2012, Supplementary data 3). A total of 118 (38 females, 58 males and 22 with undetermined sex) and 114 (40 females, 62 males and 12 with undetermined sex) individuals were assigned to the central and north-eastern patches, respectively. We determined classic genetic diversity indices (see Table 1) and tested for Hardy–Weinberg equilibrium departure and the presence of linkage disequilibrium between pairs of loci using FSTAT v.2.9.3.2 software (Goudet 1995, 2001), while observed heterozygosity was determined using the hierfstat package (Goudet 2005) in R- 3.6.0 (R Core Team 2019) for the two patches separately.
The spatial genetic structure within each patch was investigated by testing for isolation by distance (IBD) and by a sPCA (Spatial Principal Component Analysis, Jombart et al. 2008; Montano & Jombart 2017). Patterns of IBD were tested using Mantel tests to measure the correlation between pairwise genetic distances (Rousset’s âr, Rousset 2000) and the log-transformed pairwise geographic Euclidean distance (EuD) between individuals, as recommended by Rousset (2000). We calculated genetic distances using SPAGeDI v.1.5 (Hardy & Vekemans 2002) and Mantel tests were performed using the ecodist package for R (Goslee & Urban 2007) with 9999 permutations. In the sPCA, we set the connection network using the inverse of pairwise Euclidean distances between individuals since the spatial extents of study areas were much larger than wildcats home range sizes (Germain et al. 2008; Beugin et al. 2016). The significance of local and global spatial structures was tested performing eigenvalue tests (n = 9999, Montano & Jombart 2017).
Landscape genetics analyses
We used a mixed linear models selection framework within each of the central and north-eastern patches to assess if landscape impacted gene flow in French wildcats. We determined if the geographic distance (isolation by distance (IBD) model), the landscape (isolation by resistance (IBR) models) or the combination of both (IBR + IBD models) better explained between-individual genetic distances. IBR models were based either on a habitat map (HAB) or on a combination of habitat map and distance to forest patches (HAB + Dforest). While in IBD models, the Euclidean geographic distance (i.e. in straight line) is supposed to drive genetic differentiation, in the IBR models, ecological distances (i.e. accounting for the landscape resistance to movements, McRae 2006), are expected to determine genetic differentiation between individuals or populations.
Here, we used least cost distances (LCD) as ecological distances and calculated the cumulative cost along the least-cost path, as recommended by Etherington & Holland (2013) using the costDistance function in the gdistance R package (van Etten 2017). Resistance surfaces used to calculate LCD accounted for nine landcover categories (Fig. 2, Table 2, Supplementary data 9, Figure S10) expected to be more or less resistant to wildcat movements (Table 2). Since we hypothesized that unfragmented forest patches would be more favourable than fragmented ones, forested areas were categorized in four different sub-classes to account for the level of fragmentation. For each forest patch, the area was divided by the perimeter to calculate a fragmentation index (Fi, ranging from 0.85 to 202.09 m/km2, mean = 9.49 ± 4.19 SD). The four quartiles of the fragmentation index (q1 < 7.29 m/km2; 7.29 ≤ q2 < 9.40; 9.40 ≤ q3 < 11.56 and q4 ≥ 11.56) were used to classify each pixel in one of the four sub-classes describing low, medium low, medium high and high degrees of fragmentation.
Habitat map and landscape variables were derived from the European landscape database CORINE Land Cover 2012 level 3 (European Environment Agency, 2015). Land cover maps were rasterized using QGIS v2.12.1 (Open Source Geospatial Foundation Project http://qgis.osgeo.org) to a grid cell size of 3160 × 3160 m (i.e. spatial resolution of 10 km2) and the habitat category assigned to each pixel was the one of the pixel centroid. This spatial resolution of 10 km2 was chosen to encompass a wildcat home range while accounting for its between-population variability (Germain et al. 2008; Anile et al. 2017, Beguin et al. 2016) in order to have a biological meaningful spatial resolution. To define the limits of the study area, we put a buffer of 100 km around the most extreme wildcat spatial locations to avoid artificial map boundary effects and subsequent bias in the least-cost algorithm and resistance values estimations (Koen et al. 2010). In both population patches, arable areas, pastures and permanent grasslands, and lowly fragmented forested areas were the more represented habitat classes (Supplementary data 4, Table S2).
Based on previous knowledge of habitat suitability studies, habitat categories that may provide shelters and preys were expected to be permeable to gene flow (forested areas, pastures and permanent grasslands and crops) while other habitat categories, not providing such resources or being human-dominated (and thus in which a high level of disturbance can occur), were expected to have neutral or negative impacts on gene flow (arable areas, anthropized areas and water bodies, see Table 2 and Supplementary data 9 Figure S10). Although we had expectations about the permeability of these landscape elements (Table 2), we applied an approach without a priori assumptions, since it was shown that habitat selection and suitability results are not always representative of landscape resistance to gene flow (e.g. Mateo-Sanchez et al. 2015; Roffler et al. 2016; Portanier et al. 2018; Westekemper et al. 2021). We thus alternatively assigned five different resistance values to the nine landscape variables, leading to 1,953,125 different resistance surfaces (scenarios hereafter). Resistance values varied from 1 (totally permeable) to 100 (totally resistant) by an increment of 25, so that all landscape elements have been tested for resistance values of 1, 25, 50, 75 and 100 (see Portanier et al. 2018 for a similar approach). In models considering distances to forest patches, the Euclidean distance between the centres of each pixel to the nearest forest patch was calculated. Since forested areas were expected to be permeable to gene flow, this distance was considered as an additional cost to the habitat raster costs (i.e. rasters were added). Distances to forest patches were beforehand rescaled to between 1 and 100 to be in the same order of magnitude as habitat resistance values.
To identify the best scenarios, we used linear mixed models with the maximum-likelihood population effect parameterization (MLPE, Clarke et al. 2002; van Strien et al. 2012), as implemented in mlpe.rga function of the ResistanceGA R package (Peterman 2018). MLPE models allow accounting for the non-independence of values in pairwise distance matrices (Clarke et al. 2002; van Strien et al. 2012). For each population patch, four sets of models were created, fitting genetic distances (G) as the response variable and (i) the log-transformed LCD calculated on the habitat map as an explanatory variable (models G ~ log(LCDHAB)i for the ith scenario; i.e. HAB models), (ii) the log-transformed LCD calculated on the habitat map and Euclidean distance as explanatory variables (models G ~ log(LCDHAB)i + log(EuD); i.e. HAB + IBD models), (iii) the log-transformed LCD calculated on the raster combining habitat map and distance to forest patches as an explanatory variable (models G ~ log(LCDHAB+Dforest)i; i.e. HAB + Dforest models) and (iv) the log-transformed LCD calculated on the raster combining habitat map and distance to forest patches and Euclidean distance as explanatory variables (models G ~ log(LCDHAB+Dforest)i + log(EuD); i.e. HAB + Dforest + IBD models). These models were fitted using LCD calculated under each of the 1,953,125 scenarios in both the central and the north-eastern patches. Within each model set, all models were compared with each other and to the null IBD model (G ~ log(EuD)) using the Akaike information criterion (with correction for finite sample size, AICc). It allowed the determining of which scenario was the best descriptor of genetic differentiation. Using AICc values obtained with this best scenario, we compared the different model sets with each other, to determine if the inclusion of IBR significantly improved model fit. We considered that models had equivalent support if ΔAICc < 2 and that an improvement was significant when ΔAICc > 2. When several scenarios were equivalent, we averaged resistance values of landscape elements to interpret the landscape effects on gene flow.
For landscape genetics analyses, when two individuals were located in the same raster cell, we randomly selected one individual in order to have a single individual per cell. Accordingly, a total of 106 (33 females, 51 males and 22 with undetermined sex) and 107 (38 females, 58 males and 11 with undetermined sex) individuals were considered in landscape genetics analyses from the central and north-eastern patches, respectively. We performed all landscape genetics analyses using R-3.5.2 (R Core Team 2018).
Results
Population genetic structure
Neither outliers nor null alleles (all f < 0.08, Van Oosterhout et al.’s estimator, Van Oosterhout et al. 2004) were detected in the north-eastern and central patches. Slight departures from Hardy–Weinberg equilibrium were observed in each population patches (p-values of Fis were equal to Bonferroni adjusted nominal level: 0.00167 and 95% confidence interval did not include zero, Table 1) and no linkage disequilibrium was detected among pairs of loci (nominal level: 0.00012). Genetic diversity appeared to be relatively high (Table 1 and Supplementary data 5, Table S3 for locus specific results) in both patches. The Mantel test between genetic distances and Euclidean geographic distances revealed a significant pattern of isolation by distance in the central patch (r = 0.13, p = 0.002) while not in the north-eastern patch, albeit the signal was not far from the significance threshold (r = 0.07, p = 0.08). The sPCA revealed a significant global spatial structure in both patches (global test, p = 0.0001 for the north-eastern and the central patch), while no local structure was detected (local test, p = 0.95 and p = 0.94 for the north-eastern and the central patch, respectively).
In the north-eastern patch, we kept the first positive axis and no negative axis of sPCA (see Supplementary data 6, Figure S7A, C). The first positive eigenvalue was 0.14 while the second was 0.12 and all the others were lower than 0.09. This first sPCA axis distinguished individuals from the north-east and the east, although genetic differentiation seemed to be moderate (Fig. 3A). In the central patch, we kept the first positive axis and no negative axis (see Supplementary data 6, Figure S7B, D). This first positive eigenvalue was 0.14 while the next were lower than 0.11. This axis revealed no clear spatial pattern suggesting genetic exchanges or dispersal events occurring at the scale of the population in this patch (Fig. 3B).
Landscape impacts on gene flow
In both the north-eastern and central wildcat population patches, the models accounting for both IBD and IBR had the lowest AICc (Table 3). More precisely, across scenarios, 44% and 49% of the models G ~ log(LCD)i + log(EuD) had a better support (lower AICc) than the G ~ log(EuD) model for the north-eastern and the central patch, respectively, indicating an effect of landscape on genetic distances. Accounting for distances to forest patches did not increase the fit of models since in both population patches, models including Dforest had higher AICc than the models considering only habitat effects (Table 3). In the following, only results regarding HAB + IBD models will be reported. In the north-eastern patch, 1399 scenarios received equivalent support (ΔAICc < 2) while in the central patch 625 scenarios were equivalent (Table 4), and were used to average resistance values of habitat types.
In the north-eastern area, the historical and endemic deme, the most permeable elements were arable areas, pastures and permanent grasslands and lowly fragmented forested areas (Table 4). In this population patch, all other forest categories (from medium low to highly fragmented, q2, q3, q4) had an intermediate level of resistance as well as permanent crops and water bodies. Finally, anthropized areas were highly resistant to gene flow. Arable areas, pastures and permanent grasslands, low and medium low forested areas and anthropized areas had, in addition, highly conserved values across the equivalent scenarios (small differences between resistance values in the best scenario and the averaged resistance values over the equivalent scenarios, see Supplementary data 7, Figure S8) suggesting that these landscape elements are among the most important in determining landscape impacts on gene flow in the north-eastern population patch.
Similarly, in the central population patch, corresponding to the colonization front, arable areas, pastures and permanent grasslands and lowly fragmented forested areas were highly permeable to gene flow (Table 4) as in the north-eastern area. In this population patch however, medium low fragmented forested areas (q2) and, surprisingly, anthropized areas were also highly permeable to gene flow. All other considered habitat categories (fragmented forested areas (q3, q4), permanent crops and water bodies) showed intermediate levels of resistance. Based on the variation of resistance values between the first top scenario and resistance values obtained after averaging across 625 scenarios, arable areas, pastures and permanent grasslands, lowly fragmented forested areas and anthropized areas appeared to be among the most important landscape feature determining gene flow in the colonization front (Supplementary data 7, Figure S8).
Discussion
The aim of this study was to determine if, and how, landscape features impacted gene flow in European wildcats in France. We therefore investigated population genetic diversity and structures in two French population patches well-separated by the Paris-Lyon axis. These two populations do not have the same demographic status: the north-eastern population is the endemic historical population while the central one is more recent and expands toward the south-west of the country. We detected high genetic diversity and the presence of a spatial genetic structure in both patches. In the historical north-eastern patch, the spatial distribution of genetic variability suggested the presence of two wildcat groups, with moderate genetic differentiation and thus high levels of gene flow. The spatial genetic structure of the expanding central deme was more diffuse and followed an isolation by distance pattern. In both population patches, considered landscape features had an impact on gene flow but not the distance to forested areas. Arable areas, pastures and permanent grasslands and lowly fragmented forested areas were permeable to gene flow and were among the most important drivers of gene flow in both populations, confirming the importance of resources and shelters for European wildcats. Anthropized areas also had a strong impact on gene flow in both population patches but with an opposite effect as they were highly resistant in the north-eastern patch and highly permeable in the central patch. Between populations differences in genetic structures and landscape genetic connectivity suggested that different behaviours might be observed according to the demographic context of the populations.
Genetic diversity and spatial genetic structure of French wildcat populations
Genetic diversity was relatively high in both population patches, as previously observed in both France (Say et al. 2012) and other countries (e.g. Germany, Hartmann et al. 2013, Würstlin et al. 2016, Steyer et al. 2016; Sardinia, Mattucci et al. 2013). In both population patches, a significant global structure was revealed by sPCA but spatial patterns of genetic differentiation varied. In the north-eastern patch, two genetic groups might be distinguished and isolation by distance was absent or weak. More precisely, wildcats living near the German and the Swiss borders seemed to be slightly differentiated from each other. It is noteworthy that this genetic structure was weak since it was not detected by the clustering approach (see Supplementary data 8, Figure S9). In the central patch, no clear spatial interpretation could be found when looking at the sPCA scores, the spatial genetic signal appeared more clinal and followed an isolation by distance pattern.
Such genetic patterns agreed with our expectations. Indeed, the north-eastern patch is considered as the historical French wildcat population deme while the central patch is thought to be the colonization front of the population. Having been settled for a longer time, individuals in the north-eastern patch might be expected to be less mobile, performing less exploratory movements and to be more faithful to their home range. Indeed, alternative dispersal strategies and enhanced dispersal capacities can be observed in the colonization front as compared to core species distribution (Simmons & Thomas 2004). This phenomenon has been particularly studied in invasive species (Lindström et al. 2013; Courant et al. 2019). Different dispersal behaviours might thus explain the uneven distribution of genetic variability in the north-eastern patch and the more clinal one in the colonization front.
Landscape elements determining gene flow in European wildcats
In both French wildcat population patches, landscape had an impact on gene flow. Indeed, models accounting for isolation by resistance received better support than models accounting only for isolation by distance. Since distance to forested areas has been previously identified as an important landscape feature for wildcat habitat suitability (Klar et al. 2008), we expected this landscape characteristic to also impact gene flow. Surprisingly, models including habitat characteristics and distance to forests received less support than models including habitat only, suggesting that distance to forest patches had no impact on gene flow and that the wildcat is not as forest-specialist as previously thought for its dispersal.
In the habitat-only models, forested areas with low levels of fragmentation were nevertheless among the most important drivers of gene flow in both population patches and were permeable to gene flow, especially in the central patch. In accordance with the negative effect of forest fragmentation on the presence of felids (European wildcat, Anile et al. 2019; Eurasian lynx, Niedzialkowska et al. 2006), forest patches with a higher degree of fragmentation (namely, q3 and q4) were more resistant to gene flow. It is nevertheless noteworthy that, in the present study, fragmented forests represented only a limited proportion of available forested areas (see Supplementary data 4, Table S2). Accordingly, evidencing an impact of forest fragmentation in an area in which forests are mostly unfragmented might be challenging (Short-Bull et al. 2011). It might explain why fragmented forested areas had a high variability among equally supported scenarios, suggesting that these land-cover categories were less determining for wildcat gene flow (see Supplementary data 7, Figure S8). Without focusing on the fragmentation degree, forested areas have been shown to be positively selected by the European wildcat in spatial ecology studies (e.g. Sarmento et al. 2006, Beugin et al. 2016, Oliveira et al. 2018, Kilshaw et al. 2016, but see Silva et al. 2013). This preference was associated with the need for individuals to include shelters in their home range (e.g. vegetation cover, dead wood or dense structures in forested areas), and more particularly, for breeding females (Beugin et al. 2016; Jerosch et al. 2018; Oliveira et al. 2018). Permeability of forested areas observed in the present study thus revealed that the presence of shelters is also one of the main parameters determining European wildcat gene flow.
Another crucial point for wildcat habitat suitability is prey availability (Klar et al. 2008, Monterosso et al. 2009, Silva et al. 2013). We therefore expected habitats providing small mammals and rabbits (wildcat prey, Malo et al. 2004; Apostolico et al. 2015) as well as shelters to be permeable to gene flow. However, while permanent crops can provide both (e.g. rabbits in vineyards, Barrio et al. 2010; and shelters in trees and shrubs in orchards, Jerosch et al. 2018), relatively high resistance values were observed for this landscape variable which had, in addition, highly variable values across equivalent scenarios (Supplementary data 7, Figure S8). Permanent crops included essentially vineyards, but also fruit trees and berry plantations (Table 2, Supplementary data 9, Figure S10) where human presence might be high, explaining the higher-than-expected resistance to gene flow. Indeed, wildcats were strongly persecuted in the twentieth century in Europe and in France (see Lozano & Malo 2012; Von Thaden et al. 2021 and references therein) and are known to be wary and elusive animals. As a consequence, human presence and infrastructures have been repeatedly shown to be avoided by wildcats (e.g. Klar et al. 2008; Oliveira et al. 2018). This landscape variable was nevertheless, as fragmented forested areas, only sparsely present in the study areas (Supplementary data 4, Table S2). Concluding about its effects on gene flow is thus challenging.
In contrast, despite probable high human presence, arable areas and pastures and permanent grasslands were among the most important drivers of gene flow (Supplementary data 7, Figure S8) and were permeable to gene flow in both study areas. These two landscape variables can host rabbits and small mammals (Lombardi et al. 2007; de la Peña et al. 2003). However, while we expected pastures and permanent grasslands to also provide shelters (e.g. hedgerows, scattered trees) and thus to be permeable to gene flow, arable areas were expected to be more resistant, providing prey but no, or few, shelters. Indeed, this habitat category included mostly non-irrigated arable land which represent annually harvested non-permanent crops, often under a crop rotation system (see Table 2 and Supplementary data 9, Figure S10). In Mediterranean areas (Lozano et al. 2003; Lozano 2010) and in Germany (Jerosch et al. 2009, 2017, 2018) shrubs, tall herbaceous vegetation and cereal/rapeseed fields have nevertheless been shown to represent a high enough degree of shelter to be favourable to wildcat presence. The same can occur in France, crops cultivated in arable lands might be dense or tall enough to provide shelters. In addition, notwithstanding the large agricultural re-parcelling of rural areas that occurred in France at the end of the twentieth century and which dramatically reduced the number of hedgerows, such linear landscape features might still be present at a non-negligible rate and provide additional shelters and covered pathways for dispersal. Positive effects of agricultural areas for wildcat gene flow have also recently been evidenced in Germany (Westekemper et al. 2021).
An a priori counter-intuitive result was the intermediate level of resistance observed for water bodies in both population patches, since we expected them to provide neither resources nor shelters. Water bodies have previously been identified as either limiting (Hartmann et al. 2013) or not (Würstlin et al. 2016) European wildcat gene flow. These discrepancies can be linked to river characteristics, such as river width, water level and flow velocity but also to the different histories of populations, one being the result of recent colonization events and the other being settled since numerous decades (Würstlin et al. 2016). Similar contrasting results can be observed in spatial ecology studies, with watercourses being negatively correlated to wildcat presence in Scotland (Silva et al. 2013) and riparian areas being positively selected in Germany (Klar et al. 2008). The positive influence of riparian habitats was associated with the presence of high diversity and abundance of prey in conjunction with shelters provided by riparian vegetation (Virgos 2001; Sullivan & Sullivan 2006; Klar et al. 2008). In the present study, due to rasterization in spatial units of 10 km2, pixels assigned to the water bodies category in resistance surfaces are likely the largest parts of water bodies and courses, expected to be the most resistant to gene flow (Würstlin et al. 2016). The intermediate resistance values we observed might suggest that these areas in France were not providing particularly ideal amounts of resources or shelters but still enough to not be completely avoided. Alternatively, the occurrence of both water bodies with suitable riparian vegetation and areas without vegetation in this landscape variable (Table 2) might have resulted in this intermediate resistance level. It is noteworthy that resistance values nevertheless varied between the equivalent scenarios (Supplementary data 7, Figure S8), suggesting that this habitat category was not among the main drivers of gene flow and that conclusions about it should be considered with caution.
Finally, since human presence and infrastructures (e.g. roads) have been shown to be avoided by European wildcats in several populations (e.g. Klar et al. 2008, Monterosso et al. 2009, Oliveira et al. 2018, Anile et al. 2019, Westekemper et al. 2021, but see Mueller et al. 2020), anthropized areas were expected to be resistant to gene flow. This avoidance has been suggested to be a result of disturbances (noise, light, walkers, dogs and feral cats, Klar et al. 2008) and has also been reported in other felid species (e.g. Eurasian lynx, Sunde et al. 1998, female jaguars, Panthera onca, Conde et al. 2010). Disturbances might thus translate into gene flow limitations since very high resistance values were assigned to anthropized areas in the north-eastern population patch. Interestingly, a completely different effect was observed in the central patch since anthropized areas had a very high permeability to gene flow. It is noteworthy that, in both population patches, anthropized areas were key landscape features determining gene flow (Supplementary data 7, Figure S8). While wildlife avoidance of anthropized areas can be easily understood, permeability is more difficult to understand.
A high density in human population has, for instance, been suggested as an explanation of a higher number of visits made by wolves (Canis lupus) in house-yards (Kojola et al. 2006). In the central patch, more pixels were classified as anthropized than in the north-eastern patch, which might explain a higher propensity to use human-dominated habitats. In addition, even if felid species are often described as avoiding human presence, they can be found in urban areas (e.g. bobcats, Tigas et al. 2002), distances between resting sites and human presence or roads can be low (e.g. for bobcats, Sunde et al. 1998; European wildcat, Jerosch et al. 2009) and some individuals might be observed very close to human infrastructures (e.g. males jaguar, Conde et al. 2010). European wildcats have, furthermore, been suggested to habituate to human presence as long as other favourable habitats were available in close proximity (Jerosch et al. 2009). Westekemper et al. (2021) also showed that human settlements were resistant to gene flow but particularly when they were in synergy with other landscape features such as roads or absence of suitable habitats. All these elements might participate in increasing the permeability of anthropized areas to gene flow in the central patch. Finally, the central patch is considered the colonization front of the population and dispersal behaviour might be different (for instance, less shy) in this demographic context, thus decreasing anthropized areas’ resistance to gene flow. In line with this hypothesis, in this area, none of the habitat categories considered had a resistance value higher than 50 and four imposed no resistance to gene flow. In addition, the spatial distribution of genetic variability was less patchy than in the north-eastern patch. Dispersal of European wildcats across human-dominated landscape and roads has been recently reported in a context of spatial recolonization (Mueller et al. 2020).
Such increased frequentation of human-dominated landscape might have important conservation implications regarding introgression. Expansion of wildcat populations in areas already occupied by domestic counterparts might indeed result in introgression from domestic to wild cats (Nussberger et al. 2018). This was suggested to be a result of negative density-dependent dispersal (most probably of male wildcats): dispersing individuals avoiding areas occupied by conspecifics being thus more prone to encounter domestic individuals (Quilodrán et al. 2019). The higher use of anthropized areas (hypothesized to host higher domestic cat densities) for movements observed in the expanding population here might be proposed as an explanation for hybridization rate and demographic status correlation observed elsewhere. Hybridization with domestic cats is one of the most severe threats to European wildcats (Hertwig et al. 2009; Yamaguchi et al. 2015) and, while expansion is a positive fact for this endangered species, special attention should be given in the future regarding the introgression it might result in.
Conclusion and perspectives
While historically considered as a forest specialist, evidence is accumulating to describe the European wildcat as a habitat generalist species for which the co-occurrence of shelters, resources and landscape heterogeneity are highly important (Klar et al. 2008, Monterroso et al. 2009, Lozano et al. 2003, Silva et al. 2013, Kilshaw et al. 2016, Jerosch et al. 2017, 2018). Similar importance of shelters and prey for habitat suitability has been shown in other felids, also highlighting the importance of shrubs and not only forested areas for these species (e.g. Iberian lynx, Palomares 2001; Fernandez et al. 2003). Impacts of habitat categories on gene flow described in both French population patches in the present study were in line with these results, revealing a higher than expected permeability to gene flow of open and agricultural habitats. Our study also suggested that differences in demographic context in which population patches are found might lead to differences in the permeability of anthropized areas, which can, in turn, have implications in terms of introgression from domestic cats.
Altogether these results highlighted the importance of performing spatial replication in conservation genetics studies to accurately assess the effects of landscape characteristics on population structure, genetic diversity and persistence probability (Short-Bull et al. 2011; Habel et al. 2013). Additional spatial replications in other more or less anthropized ecological contexts (e.g. Mediterranean areas, other European countries) would represent an interesting pursuit to the present study and help assess the spatial variability in the risk of introgression in other expanding populations provided that habitat variables are standardized across studies. In addition, recent detection of European wildcats in areas between the Pyrenees and the French Central massif have been reported (S. Ruette, pers. comm.). It is, for now, unknown if these individuals reached these areas from the Pyrenees or from the central patch studied here. Further genetic analyses would allow the answering of such questions and determine if similar landscape features are permeable to gene flow.
Future landscape genetics studies in European wildcats should be performed sex-specifically, an improvement that sample sizes of the present study did not allow. Indeed, recent spatial ecology studies highlighted that females and males use space differently and might exhibit different dispersal abilities and behaviours (Würstlin et al. 2016; Beugin et al. 2016; Oliveira et al. 2018; Jerosch et al. 2017, 2018). These differences can translate into differences in landscape connectivity (see e.g. Portanier et al. 2018), the description of which would help in designing appropriate conservation strategies. Similar studies should also be performed on hybrid individuals living in the wild. Hybrids and wild individuals seem to have different spatial behaviour and habitat requirement: hybrids being more flexible in their habitat choices than wildcats (Germain 2007; Germain et al. 2008). These differences might have important implications for hybridization and introgression. A better knowledge about dispersal and landscape connectivity for hybrid individuals would help designing conservation plans preventing hybridization when needed. Finally, it would also be relevant to account explicitly for linear landscape features. Indeed, roads and rivers might play a crucial role in determining wildcat gene flow (Hartmann et al. 2013; Westekemper et al. 2021). While no strong barrier effect was detected in the present study, since no strong within-patch genetic structure was detected, gaining better knowledge about which characteristics of linear features might be resistant to gene flow (e.g. river and roads width, water velocity, presence of border/riparian vegetation) would also be useful for wildcat conservation.
Data Availability
Microsatellite data are available from the figshare repository at 10.6084/m9.figshare.19144643. Precise spatial coordinates for wild individuals are available upon reasonable demand from the corresponding and the last author.
Code availability
Not applicable.
References
Anile S, Devillard S (2015) Study design and body mass influence RAIs from camera trap studies: evidence from the Felidae. Anim Conserv 19:35–45
Anile S, Bizzarri L, Lacrimini M, Sforzi A, Ragni B, Devillard S (2017) Home-range size of the European wildcat (Felis silvestris silvestris): a report from two areas in central Italy. Mammalia 82:1–11
Anile S, Devillard S, Bernardino R, Rovero F, Mattucci F, Lo Valvo M (2019) Habitat fragmentation and anthropogenic factors affect wildcat Felis silvestris silvestris occupancy and detectability on Mt Etna. Wildl Biol. https://doi.org/10.2981/wlb.00561
Apostolico F, Vercillo F, La Porta G, Ragni B (2015) Long-term changes in diet and trophic niche of the European wildcat (Felis silvestris silvestris) in Italy. Mamm Res 61:109–119
Barrio IC, Bueno CG, Tortosa FS (2010) Alternative food and rabbit damage in vineyards of southern Spain. Agr Ecosyst Environ 138:51–54
Beaumont M, Barrat EM, Gottelli D, Kitchener AC, Daniels MJ, Pritchard JK, Brudford MW (2001) Genetic diversity and introgression in the Scottish wildcat. Mol Ecol 10:319–336
Belkhir K, Borsa P, Chikhi L, Raufaste N, Bonhomme F (2004) GENETIX 4.05, logiciel sous Windows TM pour la génétique des populations. Laboratoire Génome, Populations, Interactions, CNRS UMR 5000, Université de Montpellier II, Montpellier (France)
Beugin MP, Leblanc G, Queney G, Natoli E, Pontier D (2016) Female in the inside, male in the outside: insights into the spatial organization of a European wildcat population. Conserv Genet 17:1405–1415
Beugin MP, Salvador O, Leblanc G, Queney G, Natoli E, Pontier D (2020) Hybridization between Felis silvestris silvestris and Felis silvestris catus in two contrasted environments in France. Ecol Evol 10:263–276
Breitenmoser U, Lanz T, Breitenmoser-Würsten C (2019) Conservation of the wildcat (Felis silvestris) in Scotland: review of the conservation status and assessment of conservation activities. Scottish wildcat action
Clarke RT, Rothery P, Raybould AF (2002) Confidence limits for regression relationships between distance matrices: Estimating gene flow with distance. J Agric Biol Environ Stat 7:361–372
Conde DA, Colchero F, Zarza H, Christensen NL, Sexton JO, Manterola C, Chavez C, Rivera A, Azuara D, Ceballos G (2010) Sex matters: modeling male and female habitat differences for jaguar conservation. Biol Cons 143:1980–1988
Courant J, Secondi J, Guillemet L, Vollette E, Herrel A (2019) Rapid changes in dispersal on a small spatial scale at the range edge of an expanding population. Evol Ecol 33:599–612
Crooks KR, Sanjayan M (2006) Connectivity conservation. Cambridge University Press, Cambridge
Cullingham CI, Kyle CJ, Pond BA, Rees EE, White BN (2009) Differential permeability of rivers to raccoon gene flow corresponds to rabies incidence in Ontario, Canada. Mol Ecol 18:43–53
Daniels MJ, Beaumont M, Johnson PJ, Balkharry D, MacDonald DW, Barratt E (2001) Ecology and genetics of wild-living cats in the north-east of Scotland and the implications for the conservation of the wildcat. J Appl Ecol 38:146–161
de la Peña NM, Butet A, Delettre Y, Paillat G, Morant P, Le Du L, Burel F (2003) Response of the small mammal community to changes in western French agricultural landscapes. Landscape Ecol 18:265–278
Devillard S, Jombart T, Léger F, Pontier D, Say L, Ruette S (2014) How reliable are morphological and anatomical characters to distinguish European wildcats, domestic cats and their hybrids in France? J Zool Syst Evol Res 52:154–162
Etherington TR, Penelope Holland E (2013) Least-cost path length versus accumulated-cost as connectivity measures. Landscape Ecol 28:1223–1229
Fahrig L (2003) Effects of habitat fragmentation on biodiversity. Annu Rev Ecol Evol Syst 34:487–515
Fernandez N, Delibes M, Palomares F, Mladenoff DJ (2003) Identifying breeding habitat for the Iberian lynx: inferences from a fine-scale spatial analysis. Ecol Appl 13:1310–1324
Ferreras P (2001) Landscape structure and asymmetrical inter-patch connectivity in a metapopulation of the endangered Iberian lynx. Biol Cons 100:125–136
Foley JA, DeFries R, Asner GP, Barford C, Bonan G, Carpenter SR, Chapin FS, Coe MT, Daily GC, Gibbs HK, Helkowski JH, Holloway T, Howard EA, Kucharik CJ, Monfreda C, Patz JA, Prentice IC, Ramankutty N, Snyder PK (2005) Global consequences of land use. Science 309:570–574
Frankham R, Ballou JD, Briscoe DA (2004) A primer of conservation genetics. Cambridge University Press, New York
Germain E, Benhamou S, Poulle ML (2008) Spatio-temporal sharing between the European wildcat, the domestic cat and their hybrids. J Zool 276:195–203
Germain E (2007) Approche éco-éthologique de l’hybridation entre le Chat forestier d’Europe (Felis silvestris silvestris Schreber 1777) et le Chat domestique (Felis catus L.). PhD thesis, Université Reims Champagne-Ardenne
Gil-Sanchez JM, Barea-Azcon JM, Jaramillo J, Herrera-Sanchez FJ, Jimenez J, Virgos E (2020) Fragmentation and low density as major conservation challenges for the southernmost populations of the European wildcat. PLoS ONE 15:e0227708
Goslee SC, Urban DL (2007) The ecodist package for dissimilarity-based analysis of ecological data. J Stat Softw 22:1–19
Goudet J (1995) FSTAT: a computer program to calculate F-statistics. J Hered 86:485–486
Goudet J (2001) FSTAT, a program to estimate and test gene diversities and fixation indices (version 2.9.3). http://www2.unil.ch/popgen/softwares/fstat.htm. Updated from Goudet (1995)
Goudet J (2005) HIERFSTAT, a package for R to compute and test hierarchical F-statistics. Mol Ecol Notes 5:184–186
Habel JC, Husemann M, Finger A, Danley PD, Zachos FE (2013) The relevance of time series in molecular ecology and conservation biology. Biol Rev 89:484–492
Hardy OJ, Vekemans X (2002) SPAGeDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol Ecol Notes 2:618–620
Hartmann SA, Steyer K, Kraus RHS, Segelbacher G, Nowak C (2013) Potential barriers to gene flow in the endangered European wildcat (Felis silvestris). Conserv Genet 14:413–426
Hertwig ST, Schweizer M, Stepanow S, Jungnickel A, Bohle UR, Fischer MS (2009) Regionally high rates of hybridization and introgression in German wildcat populations (Felis silvestris, Carnivora, Felidae). J Zool Syst Evol Res 47:283–297
Jerosch S, Gotz M, Klar N, Roth M (2009) Characteristics of diurnal resting sites of the endangered European wildcat (Felis silvestris silvestris): implications for its conservation. J Nat Conserv 18:45–54
Jerosch S, Gotz M, Roth M (2017) Spatial organisation of European wildcats (Felis silvestris silvestris) in an agriculturally dominated landscape in central Europe. Mamm Biol 82:8–16
Jerosch S, Kramer-Schadt S, Gotz M, Klar N, Roth M (2018) The importance of small-scale structures in an agriculturally dominated landscape for the European wildcat (Felis silvestris silvestris) in central Europe and implications for its conservation. J Nat Conserv 41:88–96
Jombart T, Devillard S, Dufour AB, Pontier D (2008) Revealing cryptic spatial patterns in genetic variability by a new multivariate method. Heredity 101:92–103
Keller LF, Waller DM (2002) Inbreeding effects in wild populations. Trends Ecol Evol 17:230–241
Kilshaw K, Montgomery RA, Campbell RD, Hetherington DA, Johnson PJ, Kitchener AC, Macdonald DW, Millspaugh JJ (2016) Mapping the spatial configuration of hybridization risk for an endangered population of the European wildcat (Felis silvestris silvestris) in Scotland. Mamm Res 61:1–11
Klar N, Fernández N, Kramer-Schadt S, Herrmann M, Trinzen M, Büttnerf I, Niemitzb C (2008) Habitat selection models for European wildcat conservation. Biol Cons 141:308–319
Koen EL, Garroway CJ, Wilson PJ, Bowman J (2010) The effect of map boundary on estimates of landscape resistance to animal movement. PLoS ONE 5:1–8
Kojola I, Hallikainen V, Mikkola K, Gurarie E, Heikkinen S, Kaartinen S, Nikula A, Nivala V (2006) Wolf visitations close to human residences in Finland: the role of age, residence density, and time of day. Biol Cons 198:9–14
Larroque J, Ruette S, Vandel JM, Queney G, Devillard S (2016a) Age and sex-dependent effects of landscape cover and trapping on the spatial genetic structure of the stone marten (Martes foina). Conserv Genet 17:1293–1306
Larroque J, Ruette S, Vandel J-M, Devillard S (2016b) Divergent landscape effects on genetic differentiation in two populations of the European pine marten (Martes martes). Landscape Ecol 31:517–531
Laundré JW, Hernandez L, Altendorf KB (2001) Wolves, elk, and bison: reestablishing the ‘“landscape of fear”’ in Yellowstone National Park, USA. Can J Zool 79:1401–1409
Lindström T, Brown GP, Sisson SA, Phillips BL, Shine R (2013) Rapid shifts in dispersal behavior on an expanding range edge. PNAS 110:13452–13456
Lombardi L, Fernandez N, Moreno S (2007) Habitat use and spatial behaviour in the European rabbit in three mediterranean environments. Basic Appl Ecol 8:453–463
Lozano J (2010) Habitat use by European wildcats (Felis silvestris) in central Spain: what is the relative importance of forest variables? Anim Biodivers Conserv 33:143–150
Lozano J, Malo A (2012) Conservation of the European wildcat (Felis silvestris) in mediterranean environments: a reassessment of current threats. In: William GS (ed) Mediterranean Ecosystems. Nova Science Publishers, NewYork, pp 1–31
Lozano J, Virgós E, Malo AF, Huertas DL, Casanovas JG (2003) Importance of scrub—pastureland mosaics for wild- living cats occurrence in a Mediterranean area: Implications for the conservation of the wildcat (Felis silvestris). Biodivers Conserv 12:921–935
Lynch M, Conery J, Burger R (1995) Mutation accumulation and the extinction of small populations. Am Nat 146:489–518
Malo AF, Lozano J, Huertas DL, Virgós E (2004) A change of diet from rodents to rabbits (Oryctolagus cuniculus). Is the wildcat (Felis silvestris) a specialist predator? J Zool 263:401–407
Manel S, Schwartz MK, Luikart G, Taberlet P (2003) Landscape genetics: combining landscape ecology and population genetics. Trends Ecol Evol 18:189–197
Marchand P, Garel M, Bourgoin G, Dubray D, Maillard D, Loison A (2014) Impacts of tourism and hunting on a large herbivore’s spatio-temporal behavior in and around a French protected area. Biol Cons 177:1–11
Mateo-Sanchez MC, Balkenhol N, Cushman SA, Perez T, Dominguez A, Saura S (2015) A comparative framework to infer landscape effects on population genetic structure: are habitat suitability models effective in explaining gene flow? Landscape Ecol 30:1405–1420
Mattucci F, Oliveira R, Bizzarri L, Vercillo F, Anile S, Ragni B, Lapini L, Sforzi A, Alves PC, Lyons LA, Randi E (2013) Genetic structure of wildcat (Felis silvestris) populations in Italy. Ecol Evol 3:2443–2458
Mattucci F, Oliveira R, Lyons LA, Alves PC, Randi E (2016) European wildcat populations are subdivided into five main biogeographic groups: Consequences of Pleistocene climate changes or recent anthropogenic fragmentation? Ecol Evol 6:3–22
McRae BH (2006) Isolation by resistance. Evolution 60:1551–1561
Menotti-Raymond M, David VA, Lyons LA, Schäffer AA, TomlinJF HMK, O’Brien SJ (1999) A Genetic Linkage Map of Microsatellites in the Domestic Cat (Felis catus). Genomics 57:9–23
Montano V, Jombart T (2017) An Eigenvalue test for spatial principal component analysis. BMC Bioinformatics 18:562
Monterroso P, Brito JC, Ferreras P, Alves PC (2009) Spatial ecology of the European wildcat in a Mediterranean ecosystem: dealing with small radio-tracking datasets in species conservation. J Zool 279:27–35
Mueller S, Reiners TE, Steyer K, von Thaden A, Tiesmeyer A, Nowak C (2020) Revealing the origin of wildcat reappearance after presumed long-term absence. Eur J Wildl Res 66:94
Niedzialkowska M, Jedrzejewski W, Mysłajek RW, Nowak S, Jedrzejewska B, Schmidt K (2006) Environmental correlates of Eurasian lynx occurrence in Poland – Large scale census and GIS mapping. Biol Cons 133:63–69
Noss RF, Quigley HB, Hornocker MG, Merrill T, Paquet PC (1996) Conservation biology and carnivore conservation in the Rocky mountains. Conserv Biol 10:949–963
Nussberger B, Greminger MP, Grossen C, Keller LF, Wandeler P (2013) Development of SNP markers identifying European wildcats, domestic cats, and their admixed progeny. Mol Ecol Resour 13:447–460
Nussberger B, Currat M, Quilodrán CS, Ponta N, Keller LF (2018) Range expansion as an explanation for introgression in European wildcats. Biol Cons 218:49–56
O’Brien J, Devillard S, Say L, Vanthomme H, Leger F, Ruette S, Pontier D (2009) Preserving genetic integrity in a hybridising world: are European wildcats (Felis silvestris silvestris) in eastern France distinct from sympatric feral domestic cats? Biodivers Conserv 18:2351–2360
Oliveira R, Godinho R, Randi E, Alves PC (2008) Hybridization versus conservation: are domestic cats threatening the genetic integrity of wildcats (Felis silvestris silvestris) in Iberian Peninsula? Philos Trans R Soc Lond B Biol Sci 363:2953–61
Oliveira T, Urra F, López-Martín JM, Ballesteros-Duperón E, Barea-Azcón JM, Moléon M, Gil-Sanchez JM, Alves PC, Diaz-Ruiz F, Ferreras P, Monterroso P (2018) Females know better: Sex-biased habitat selection by the European wildcat. Ecol Evol 8:9464–9477
Palomares F (2001) Vegetation structure and prey abundance requirements of the Iberian lynx: implications for the design of reserves and corridors. J Appl Ecol 38:9–18
Peakall R, Smouse PE (2006) GENALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Peakall R, Smouse PE (2012) GenALEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 28:2537–2539
Perez-Espona S, Perez-Barberia FJ, Mcleod JE, Jiggins CD, Gordon IJ, Pemberton JM (2008) Landscape features affect gene flow of Scottish Highland red deer (Cervus elaphus). Mol Ecol 17:981–996
Peterman WE (2018) ResistanceGA: An R package for the optimization of resistance surfaces using genetic algorithms. Methods Ecol Evol 9:1638–1647
Pilgrim KL, McKelvey KS, Riddle AE, Schwartz MK (2005) Felid sex identification based on noninvasive genetic samples. Mol Ecol Notes 5:60–61
Pizzatto L, Both C, Brown G, Shine R (2017) The accelerating invasion: dispersal rates of cane toads at an invasion front compared to an already-colonized location. Evol Ecol 31:533–545
Portanier E, Larroque J, Garel M, Marchand P, Maillard D, Bourgoin G, Devillard S (2018) Landscape genetics matches with behavioral ecology and brings new insight on the functional connectivity in Mediterranean mouflon. Landscape Ecol 33:1069–1085
Pritchard J, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Quilodrán CS, Nussberger B, Montoya-Burgos JI, Currat M (2019) Hybridization and introgression during density-dependent range expansion: European wildcats as a case study. Evolution 73:750–761
R Core Team (2018) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna
Riley SPD, Pollinger JP, Sauvajot RM, York EC, Bromley C, Fuller TK, Wayne RK (2006) A southern California freeway is a physical and social barrier to gene flow in carnivores. Mol Ecol 15:1733–1741
Roffler GH, Schwartz MK, Pilgrim KL, Talbot SL, Sage GK, Adams LG, Luikart G (2016) Identification of landscape features influencing gene flow: how useful are habitat selection models? Evol Appl 9:805–817
Rousset F (2000) Genetic differentiation between individuals. J Evol Biol 13:58–62
Sarmento P, Cruz J, Tarroso P, Fonseca C (2006) Space and habitat selection by female European wild cats (Felis silvestris silvestris). Wild Biol Pract 2: 79–89.
Say L, Devillard S, Léger D, Pontier D, Ruette S (2012) Distribution and spatial genetic structure of European wildcat in France. Anim Conserv 15:18–27
Short-Bull RAS, Cushman SA, Mace R, Chilton T, Kendall KC (2011) Why replication is important in landscape genetics: American black bear in the rocky Mountains. Mol Ecol 20:1092–1107
Silva AP, Kilshaw K, Johnson PJ, MacDonald DW, Rosalino LM (2013) Wildcat occurrence in Scotland: Food really matters. Divers Distrib 19:232–243
Simmons AD, Thomas CD (2004) Changes in dispersal during species’ range expansions. Am Nat 164:378–395.
Steyer K, Kraus RH, Molich T, Anders O, Cocchiararo B, Frosch C, Geib A, Gotz M, Herrman M, Hupe K, Kohnen A, Kruger M, Muller F, Pir JB, Reiners TE, Roch S, Schade U, Schiefenhovel P, Siemmund M, Simon O, Steeb S, Streif S, Streit B, Thein J, Tiesmeyer A, Trinzen M, Vogel B, Nowak C (2016) Large-scale genetic census of an elusive carnivore, the European wildcat (Felis s. silvestris). Conserv Genet 17:1183–1199
Steyer K, Tiesmeyer A, Muñoz-Fuentes V, Nowak C (2018) Low rates of hybridization between European wildcats and domestic cats in a human-dominated landscape. Ecol Evol 8:2290–2304
Sullivan TP, Sullivan SD (2006) Plant and small mammal diversity in orchard versus non-crop habitats. Agr Ecosyst Environ 116:235–243
Sunde P, Stener SO, Kvam T (1998) Tolerance to humans of resting lynxes Lynx lynx in a hunted population. Wildl Biol 4:177–183
Tarjuelo R, Barja I, Morales MB, Traba J, Benítez-López A, Casas F, Arroyo B, Delgado MP, Mougeot F (2015) Effects of human activity on physiological and behavioral responses of an endangered steppe bird. Behav Ecol 26:828–838
Taylor PD, Fahrig L, Henein K, Merriam G (1993) Connectivity is a vital element of landscape structure. Oikos 68:571–573
Taylor P, Fahrig L, With K (2006) Landscape connectivity: a return to the basics. In: Crooks KR, Sanjayan M (eds) Connectivity conservation. Cambridge University Press, Cambridge, pp 29–43
Tiesmeyer A, Ramos L, Manuel Lucas J, Steyer K, Alves PC, Astaras C, Brix M, Cragnolini M, Domokos C, Hegyeli Z, Janssen R, Kitchener AC, Lambinet C, Mestdagh X, Migli Monterosso P, Mulder JL, Schockert V, Youlatos D, Pfenninger M, Nowak C (2020) Range-wide patterns of human-mediated hybridisation in European wildcats. Conserv Genet 21:247–260
Tigas LA, Van Vuren DH, Sauvajot RM (2002) Behavioral responses of bobcats and coyotes to habitat fragmentation and corridors in an urban environment. Biol Cons 108:299–306
Turner MG, Gardner RH, O’Neill RV (2001) Landscape ecology in theory and practice. Springer, New-York
Turner MG, Gardner RH (2015) Landscape ecology in theory and practice, 2nd edn. Springer, New-York
van Etten J (2017) R Package gdistance: distances and routes on geographical grids. J Stat Softw 76:1–21
van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P (2004) MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4:535–538
van Strien MJ, Keller D, Holderegger R (2012) A new analytical approach to landscape genetic modelling: least-cost transect analysis and linear mixed models. Mol Ecol 21:4010–4023
Virgos E (2001) Relative value of riparian woodlands in landscapes with different forest cover for medium-sized Iberian carnivores. Biodivers Conserv 10:1039–1049
Virgos E, Telleria JL, Santos T (2002) A comparison on the response to forest fragmentation by medium-sized Iberian carnivores in central Spain. Biodivers Conserv 11:1063–1079
Von Thaden A, Cocchiararo B, Mueller SA, Reiners TE, Reinert K, Tuchscherer I, Janke A, Nowak C (2021) Informing conservation strategies with museum genomics: long-term effects of past anthropogenic persecution on the elusive European wildcat. Ecol Evol 11:17932–17951
Wan YW, Cushman SA, Ganey JL (2018) Habitat fragmentation reduces genetic diversity and connectivity of the mexican spotted owl: a simulation study using empirical resistance models. Genes 9:403
Wasserman TN, Cushman SA, Shirk AS, Landguth EL, Littell JS (2012) Simulating the effects of climate change on population connectivity of American marten (Martes americana) in the northern Rocky Mountains, USA. Landscape Ecol 27:211–225
Westekemper K, Tiesmeyer A, Steyer K, Nowak C, Signer J, Balkenhol N (2021) Do all roads lead to resistance? State road density is the main impediment to gene flow in a flagship species inhabiting a severely fragmented anthropogenic landscape. Ecol Evol 00:1–14
Würstlin S, Segelbacher G, Streif S, Kohnen A (2016) Crossing the Rhine: a potential barrier to wildcat (Felis silvestris silvestris) movement? Conserv Genet 17:1435–1444
Yamaguchi N, Kitchener A, Driscoll C, Nussberger B (2015) Felis silvestris. IUCN Red List Threat Species 2015:e.T60354712A50652361. https://doi.org/10.2305/IUCN.UK.2015-2.RLTS.T60354712A50652361.en
Yumnam B, Jhala YV, Qureshi Q, Maldonado JE, Gopal R, Saini S, Srinivas Y, Fleischer RC (2014) Prioritizing tiger conservation through landscape genetics and habitat linkages. PLoS ONE 9:e111207
Acknowledgements
We thank all the students, technicians and officers, especially Jean-Luc Wilhem, for their help in the collection of cats and in the laboratory. This study was supported by the French Biodiversity Agency (Office Français de la Biodiversité) and the University of Lyon-CNRS. We also gratefully acknowledge the CC LBBE/PRABI for providing computer resources, and the bioinformatics team of LBBE for their advice on computational optimization of scripts. Finally, we thank Mrs. Elizabeth Kennedy-Overton for English proofreading and one anonymous reviewer for useful comments on the first version of the manuscript.
Funding
This research project and E. Portanier were funded by the Office Français de la Biodiversité, Université Claude Bernard Lyon 1, the Laboratoire de Biométrie et Biologie Évolutive and the Antagene Laboratory.
Author information
Authors and Affiliations
Contributions
EP, SR and SD: conceptualized and designed the research. FL and LH: did the field work. GQ: conducted laboratories steps. EP: conducted data analyses except for molecular identification which were performed by TG and SD. EP: wrote the first draft of the manuscript. All authors contributed in interpreting the results and writing the paper.
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare that they have no conflict of interest.
Ethics Approval
Not applicable.
Consent to participate
Not applicable.
Consent to Publish
Not applicable.
Plant Reproducibility
Not applicable.
Clinical Trials Registration
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Portanier, E., Léger, F., Henry, L. et al. Landscape genetic connectivity in European wildcat (Felis silvestris silvestris): a matter of food, shelters and demographic status of populations. Conserv Genet 23, 653–668 (2022). https://doi.org/10.1007/s10592-022-01443-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-022-01443-9