Abstract
The Western Massasauga (Sistrurus tergeminus) is a small North American rattlesnake found west of the Mississippi River. Sistrurus tergeminus has previously been divided into two putative subspecies, Desert (S. t. edwardsii) and Prairie Massasaugas (S. t. tergeminus) based upon qualitative variation in morphology, coloration, and habitat. The Desert Massasauga subspecies has been formally petitioned for federal listing under the U.S. Endangered Species Act. Our overarching goal was to evaluate genetic structure and genomic differentiation between specimens of the two putative subspecies in an effort to inform ongoing conservation assessments. To that end, we generated whole genome sequence data for both putative taxa and then developed nearly 200 genetic markers from different fractions of the genome (~ 50 intergenic and ~ 50 genic markers from each of the two subspecies) to test for population structure across much of the Western Massasauga range. Mean genomic divergence between subspecies was 0.0041 ± 0.0080 (Kimura’s 2-parameter distance) for nuclear sequences and 0.0175 ± 0.0031 for mitochondrial sequences, both exceedingly low values which approach the minimum of zero. Admixture analyses and F-statistics both indicated that regardless of how the markers were partitioned, genetic structure was oriented far more along a geographic axis (isolation-by-distance) than a taxonomic axis (i.e., between putative subspecies). Overall, our analyses provide little support that formal protection of the purported Desert Massasauga is warranted based on the homogeneity of the collective Western Massasauga gene pool.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Rattlesnake populations are subject to a long litany of existential threats including climate change, disease, overexploitation, human persecution, habitat degradation and fragmentation (Clark et al. 2010, 2011; Colley et al. 2017; Fitzgerald and Painter 2000). Many rattlesnake populations are now of conservation concern (at least to biologists, if not to much of the general public), but legal protections generally require biological delineation. For example, the U.S. Endangered Species Act (ESA) requires that listings apply to “distinct population segments.” Ideally, these biotic entities coincide with the formal, accepted Linnean taxonomy, but unfortunately that is not always the case (O’Brien and Mayr 1991).
The taxonomy of rattlesnakes is convoluted and has been controversial at times (Crother et al. 2011, 2013; Holycross et al. 2008; Knight et al. 1993; Murphy et al. 2002). Rattlesnakes are a monophyletic lineage of American pit vipers that are divided into two reciprocally monophyletic genera, Crotalus and Sistrurus, that diverged from one another about 12.5 MYA (Fig. 1; Blair and Sánchez-Ramírez 2016). Genetic analyses have confirmed the evolutionary distinctiveness of the Eastern Massasauga (S. catenatus) and have clarified the evolutionary relationships of the Pygmy Rattlesnake (S. miliarius) and the Western Massasauga (S. tergeminus), but they provide relatively weak support for distinguishing S. tergeminus edwardsii (Desert Massasauga) from S. tergeminus tergeminus (Prairie Massasauga) (Crother et al. 2011; Kubatko et al. 2011). The few available sequence data suggest that genomic differentiation (if any) between these two putative taxa may be limited due to various interacting factors (e.g., gene flow, overlapping ranges, etc.).
Evolutionary tree of the clade in Viperidae containing Sistrurus. The two rattlesnake genera (Sistrurus and Crotalus) diverged from one another about 12.5 MYA. A Newark file was generated using TimeTree.org (Kumar et al. 2017) and visualized with the Interactive Tree of Life v6 (Letunic and Bork 2021). Depiction of the tree is based upon external data and is provided purely for biological context
Western Massasauga subspecies could be in the early stages of speciation (Kubatko et al. 2011). The Desert Massasauga, historically recognized in the American Southwest from northern Mexico to southern Colorado and central Texas, is a small rattlesnake that lives in xeric grasslands feeding largely off centipedes and lizards whereas the Prairie Massasauga, found in central Texas up to central Iowa and Missouri, occupies mesic grasslands and feeds more on small mammals (Holycross and Mackessy 2002). The original distinction between these two subspecies is traced back to surveys by Howard Gloyd based on the Prairie Massasauga’s larger size, darker ventral coloration, higher number of midbody scale rows, and more numerous ventral scales and dorsal blotches (Gloyd 1955) but there is no obvious geographical barrier separating Desert and Prairie Massasaugas (Ryberg et al. 2015; Fig. 2). Since the original distinction (Gloyd 1955), the morphological characters mentioned above have been shown to vary geographically in independent ways, casting doubt on where and if subspecific boundaries exist in this species (e.g., Stebbins 1980; Klauber 1982; Conant and Collins 1991; Mackessy 2005; Kubatko et al. 2011). Genetic diversity has been studied in specific populations (Anderson et al. 2009; Gibbs et al. 2011; McCluskey and Bender 2015), but has not definitively resolved this boundary issue and determined whether the Western Massasauga consists of one or two distinct gene pools (Kubatko et al. 2011; Ryberg et al. 2015) that might comprise a “distinct population segment” as defined by the ESA.
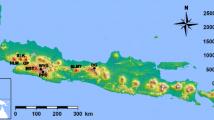
Map of sampling locations. The counties in which genotyped individuals were collected are shaded blue for the Desert Massasauga (S. t. edwardsii) and brown for the Prairie Massasauga (S. t. tergeminus) based on Dixon and Werler (2005). Approximate range of the Western Massasauga is shaded in gray. Note that actual Western Massasauga habitat is highly fragmented throughout much of its range (Anderson et al. 2009). A blue asterisk marks the location of Matagorda Island. Photos of individuals from Presidio County, Texas (Desert Massasauga; left) and Wheeler County, Texas (Prairie Massasauga; right) are two examples of the Western Massasauga. Photo credit: T. Hibbitts
Torstrom et al. (2014) wrote that “Determining the validity of the subspecies rank should no longer be argued since this classification is applied in management and legislation; rather, the focus should be on determining the best and most consistently reliable method to discern subspecies” and we agree with them. We refer the interested reader to Hillis (2020) for a philosophical discussion of the interplay between subspecies taxonomy and geography. The failure of morphological characters to reliably distinguish the two Western Massasauga subspecies is problematic given the petition to formally list the Desert subspecies under the ESA (Wild Earth Guardians 2010). While we generally favor some version of the biological species concept, in which a (sub)species is a group of genetically compatible interbreeding natural populations that are genetically isolated from other such groups, our personal views on subspecies concepts are largely irrelevant because the U.S. Fish and Wildlife Service (USFWS) needs to make comprehensive decisions based on the best available science. We conducted this study to provide USFWS with genetic information across the range of the entire species, including those populations that historically were described as subspecies.
Our purpose herein is to evaluate the contiguity (or lack thereof) in the Western Massasauga gene pool and help guide decision-making regarding formal conservation protection of the Desert Massasauga (S. t. edwardsii). Our first goal was to sequence the genomes of both S. t. tergeminus and S. t. edwardsii and subsequently generate a curated suite of single nucleotide polymorphism (SNP) markers. We did so by developing a marker array designed (a) to best utilize suboptimal DNA obtained from roadkill (our primary source of tissue); (b) to avoid ascertainment biases due to potential subspecific differentiation; and (c) to query two orthogonal aspects of genetic diversity: functional protein-coding genes as well as more neutral variants in intergenic regions far from known genes. Our second goal was to employ the markers and genomic sequences to ascertain genetic and genomic differentiation between the two putative entities across much of their respective geographic ranges in order to inform impending conservation decisions, with particular emphasis on the use of genomic data in subspecies delineation.
If S. t. edwardsii and S. t. tergeminus are distinct enough that they merit different management strategies, then we expect those differences would manifest themselves by sundering the Western Massasauga gene pool. First, we might expect subspecific differences reflected in the genomes themselves (assembly sizes, TE content, GC content, etc.). Second, we would expect sequence differentiation (K2P) in both nuclear and mitochondrial genomes to be significantly greater than zero and consistent with values from pairs of other, well-established reptilian subspecies. Third, we would expect patterns of genetic differentiation (FST) to reveal obvious discontinuities between subspecies, including ascertainment biases where genetic differentiation was exaggerated when assessing one taxon with markers developed in the other taxon. Fourth, we would expect clusters of individuals from each subspecies to group distinctly in a PCA. Fifth, we would expect admixture analyses to reveal sharp departures from k = 1 (i.e., strong support for k = 2). Sixth, we would expect evidence of a Wahlund effect. Finally, we might see subspecific evidence of local adaptation. Herein, we provide data and analyses that critically evaluate these expectations with the aim of informing decisions associated with the conservation and management of the Western Massasauga.
Materials and methods
Sample collection and DNA extraction
Western Massasauga samples were collected opportunistically via driving surveys (Table S1; Fig. 2). Most snakes were found dead on the road (n = 23). For the specimens caught live, we obtained scale clips or shed skins (n = 2). Three samples from Matagorda Island in southeast Texas were recovered as fire mortalities (n = 3). Additional samples were obtained from previous studies (n = 13) or harvested from museum samples (n = 12). Both Fetzner’s technique (1999) for shed skin and a standard phenol–chloroform extraction for other tissues were used to extract DNA for analysis. Extracted DNA was cleaned using a Zymo dsDNA Clean and Concentrate kit (Zymo Research, Irvine, CA) and electrophoresed on agarose gels to confirm DNA quality and quantity. For genome sequencing, one reference sample was chosen from each putative subspecies (TJH 3595 and CSA 1). We chose individuals based on DNA quality and quantity as well as on geographic region of origin. Therefore, out of the highest quality and quantity DNA samples, we chose samples from Hood and Ward Counties as exemplars within the range of each putative subspecies (Fig. 2). We confirmed that both samples were morphologically assigned to the putative subspecies they should represent based on the amount of dark ventral pigment and number of dorsal scale rows at midbody. We did so to increase the likelihood that each sequenced individual was a representative member of each putative subspecies (see Table S1 for sample metadata).
Genome sequencing and assembly
We created independent genomic libraries for both of the representative samples, then generated both paired-end and mate-pair reads using an Illumina NovaSeq (S4 2 × 150) platform; details can be found in Online Resources. Paired-end library insert sizes were ~ 350 bp and mate-pair insert sizes were restricted to 300–1500 bp. Reads were trimmed with Trimmomatic v0.39 (Bolger et al. 2014) to exclude base qualities less than Phred-20 and read lengths < 30 bp, and remaining reads were assessed with FastQC v0.11.7 (Andrews 2010). We estimated genome size with kmergenie v1.6982 (Chikhi and Medvedev 2014) and assembled genomes de novo with AByss v2.1.5 (Jackman et al. 2017) and SOAPdenovo2 v240 (Luo et al. 2012).
We also generated mitochondrial genome assemblies for both nominal subspecies. We employed MITObim v1.8 (Hahn et al. 2013), matching kmers of 21 bp and syncing paired read data to extend from seed sequences obtained from publicly available data (see Table S2).
Genomic divergence between putative S. tergeminus subspecies
To estimate pairwise nuclear genetic distances between putative taxa, we employed Kimura’s 2-parameter (K2P; Kimura 1980) distances using sequences from large, ostensibly orthologous scaffolds. We did so using scaffolds greater than 100 kbp in length from our best assembly of each representative from the two subspecies (Prairie: ABySS, Desert: SOAPdenovo2; both with kmer = 60), the C. viridis reference genome, and an unpublished assembly for the Eastern Massasauga (courtesy of L. Gibbs; see acknowledgements and Online Resources). We aligned all four genome assemblies (C. viridis and Eastern, Desert, and Prairie Massasaugas) pairwise using BLAST+ v2.10.0 (Camacho et al 2009) and calculated K2P distances with the dist.dna function of the ape v5.4 R package (Paradis et al. 2004). In Western Massasaugas, these data included 858 scaffolds totaling 0.12 Gb for the Prairie Massasauga and 147 scaffolds comprised of 0.02 Gb for the Desert Massasauga (Table S3). For each comparison, we plotted the distribution of K2P distances for each aligned sequence with ggplot2 v.3.32 (Wickham 2016). For context, we compiled K2P values from the literature for genetic distance between other (i.e. non-Massasauga) snakes or calculated them ourselves using publicly available genomes and the same methods above.
To estimate pairwise genomic mtDNA distances, we obtained mitochondrial genome assemblies for Crotalus (2 spp.) and Sistrurus (1 sp.) (see Online Resources) for comparison with both our assemblies. Sequences were then rotated via gapless alignment by CSA (Fernandes et al. 2009) and multiply aligned in MEGA-X v10.1.8 (Kumar et al. 2018) using the ClustalW algorithm (Thompson et al. 1994), and we then estimated pairwise genetic distances again using the K2P model (Kimura 1980). Standard errors for each pairwise distance were estimated with 500 bootstrapped replicates. We again compiled values comparing non-Massasauga snakes and non-avian reptiles from available mitochondrial datasets.
SNP identification and genotyping
We developed a SNP genotyping array for use in a Fluidigm microfluidic system that works well with nontraditional sources of tissue (e.g., roadkill) that yield suboptimal DNA (Carroll et al. 2018). To identify variants (i.e., candidate SNP markers), we mapped paired end reads from our two representative samples against the C. viridis reference genome (Schield et al. 2019) because of its chromosome-level assembly, high quality annotation, and the relatively recent divergence time from S. tergeminus of 12.5 MYA (Blair and Sánchez-Ramírez 2016). We mapped reads with BWA v0.7.17 (Li 2013) and used Picard v2.18.2 (Broad Institute 2018) to filter and quality check our alignments. We then called variants for each sample using GATK HaplotypeCaller v3.8.1 (Van der Auwera et al. 2013). We hard-filtered variants to include only those with (a) read depth between 10 × and 100 × ; (b) a strand odds ratio greater than 4.0; (c) quality by depth (an estimate of base quality as a function of allele depth) less than 5.0; and (d) quality scores greater 30.0. We used IGV v2.5.3 to further restrict these variants to SNPs greater than 20 bp from neighboring SNPs and a neighboring GC content less than 65%. Additionally, we chose only SNPs at loci with flanking regions of contiguously mapping reads for at least 10 kbp to reduce chances of linkage disequilibrium (LD) between our markers.
From 7,985,022 filtered variants, we designed 96 putatively neutral markers from intergenic regions and 96 putatively “adaptive” markers from exonic genic regions (as indicated by the annotations of the reference genome). To minimize ascertainment bias, we selected half of each marker type listed above (intergenic versus genic) from each representative subspecies, irrespective of whether SNPs were polymorphic in the other subspecies (Fig. S1). Using all 192 loci and the Fluidigm microfluidics platform, we genotyped 88 samples (Table S1) that represented snakes collected from across the Western Massasauga range (Fig. 2). We also included three technical replicates each for one Desert and one Prairie sample to estimate genotyping error rates. Samples were genotyped using the Fluidigm Biomark HD platform.
Subspecies affiliations were designated entirely on the basis of geography. We collected four samples (3 from Matagorda Island, TX and 1 from Coleman County, TX) from near disputed subspecies boundaries in north-central Texas and south Texas. Our samples from Matagorda Island are more morphologically similar to S. t. tergeminus (e.g., darker ventral coloration, higher midbody scale row counts). However, the nearest mainland populations are in south Texas and morphologically/geographically classify as S. t. edwardsii. Our sample from Coleman County was assigned to S. t. edwardsii due to habitat continuity to S. t. edwardsii range in the south and west and disjunction from S. t. tergeminus habitat to the north and east.
Population genetic analyses
We used the Fluidigm analysis software to call genotypes, pruning data by removing loci that failed to produce distinct genotype clusters in greater than 20% of individuals and by removing individuals that were successfully genotyped at fewer than 80% of remaining loci. We estimated the genotyping error rate (\(e\)) from our three technical replicates as described in Doyle et al. (2016) using the equation \(e=\frac{m}{ds}\), where \(m\) is the number of pairwise mismatches between replicates of the majority consensus including miscalls and amplification failures, \(d\) is the number loci in each replicate, and \(s\) is the total number of replicates. We tested for LD between pairs of loci using GENEPOP v4.7.3 (Rousset 2008) and subsequently removed one member of each linked pair from subsequent analyses.
We computed indices of genetic diversity using GENEPOP. We calculated minor allele frequency (MAF), observed heterozygosity (HO), tested for deviations from Hardy–Weinberg Equilibrium (HWE) at each locus, and estimated FST using various subsets of the 192 markers (ALL—all markers, HWE—markers in HWE, GENIC—markers in exonic regions, INTERGENIC—markers in intergenic regions, STT—markers identified from Prairie representative, and STE—markers identified from Desert representative) to partition variances in allele frequencies both functionally and taxonomically. Putative subspecies were evaluated as separate datasets and artificially split in half to produce random “populations” for comparison. FST was then calculated for each of 500 randomization trials using all markers and compared against the Desert–Prairie FST to discern if the inter-subspecific was greater than the intra-subspecific FST. We then calculated the FST, FIS, and R2 (as a measure of LD) for each locus or pair and the means for all Western Massasauga samples together. Additionally, we adjusted the p-values for deviations from HWE with the sequential Bonferroni method (Waples and Allendorf 2015) across all loci in order to identify locus-specific deviations. To test for the Wahlund effect (overall heterozygote deficiency due to population structure), we plotted the relationships of FIS and FST for the entire dataset (all individuals of both presumptive subspecies) and calculated a linear regression. If population structure is causing deviations from HWE, we would expect a positive mean FIS and a positive linear relationship with slope = 1 for FIS/FST (Waples and Allendorf 2015).
We evaluated population structure by first conducting a PCA with the 139 HWE loci (i.e., those in HWE) in adegenet v2.1.2 (Jombart 2008). Second, we conducted sNMF Bayesian admixture analyses using the program LEA v2.0 (Frichot et al. 2014; Frichot and François 2015). For the admixture analyses of all data subsets, we included loci regardless of whether they were in HWE and used LEA’s minimal cross-entropy approach to determine optimum α (a regularization parameter) and k (the number of theoretical ancestral populations). Additionally, we ran k = 2 with α = 5 to directly test for signatures of two distinct subspecies.
Results
Genome sequencing and assembly
We sequenced about 100 Gb per subspecies, averaging approximately 50 × coverage for the Desert Massasauga and 46 × coverage for the Prairie Massasauga (Tables S4, S5 for summary statistics). Species in the family Viperidae have a mean genome size of 2.05 ± 0.49 Gb (Gregory 2020) similar to our assembly size estimates for Western Massasauga (1.897 Gb for Desert and 1.937 Gb for Prairie). GC content was very similar for Desert (40.0%) and Prairie (39.75%) Massasaugas. Mitochondrial assemblies were 17,416 bp and 17,396 bp for the Desert and Prairie individuals respectively (Fig. S3). From a sample of n = 2 individuals, we cannot say whether the length variation (20 bp) is partitioned among subspecies or individuals but we strongly suspect there is individual variation based on Ryberg et al. (2015).
Genomic divergence between putative S. tergeminus subspecies
Our categorical pairwise alignments of large nuclear scaffolds for K2P estimates covered up to 10% of the S. tergeminus genome and thus should reasonably reflect the overall genomic divergence between presumptive taxa. Genome assembly quality (i.e., contiguity) contributed to the observed K2P variance in our dataset, with higher continuity resulting in longer sequence alignments and increased likelihood of identifying those regions with higher divergence. For example, the observed variance in K2P values positively correlated with the degree of genome continuity when comparing C. viridis with the Eastern (N50 = 1 Gbp), Prairie (N50 = 23 kbp) and Desert (N50 = 8 kbp) genome assemblies (Table S3). Across many scaffolds, K2P distances averaged only 0.0041 ± 0.0080 for intraspecific comparisons between Desert and Prairie Massasaugas (Fig. 3f). As expected, interspecific comparisons between either Western Massasauga assembly and the Eastern Massasauga were larger (specifically, ~ 2 ×) at 0.008 ± 0.0040 (Fig. 3d, e). Finally, intergeneric K2P distances between Crotalus and Sistrurus species averaged 0.021 ± 0.0103 (Fig. 3a–c), roughly 5X larger than the infraspecific comparisons. Overall, these K2P data are consistent with the idea that genomic divergence increases concomitantly with taxonomic divergence. All estimates for genetic distances between nuclear genome assemblies are depicted in Fig. 3 and reported in Table 1. Alignment statistics are reported in Table S3.
Distribution of pairwise genetic distance estimates (K2P; Kimura 1980) between ≥ 100 kb scaffolds in nuclear genome assemblies: a–c represents K2P estimates derived from local alignments of ≥ 100 kb scaffolds to the C. viridis chromosome-level assembly, d, e represents K2P estimates from local alignments of ≥ 100 kb S. tergeminus scaffolds to the S. catenatus scaffold-level assembly, and f K2P estimates derived from local alignments of ≥ 100 kb scaffolds between putative S. tergeminus sub-species. Horizontal dashed lines in each panel identify the mean K2P value for each alignment. These plots illustrate relative (not absolute) divergence among taxa and neither Western Massasauga subspecies departs substantively from the other
The K2P distance for mitochondrial DNA between Desert and Prairie Massasaugas was 0.0103 ± 0.0008. Interspecific distances between Eastern and Western Massasauga mtDNA genomes were about nine-fold higher, 0.0940 ± 0.0027 and 0.0907 ± 0.0026 when comparing the Eastern Massasauga to the Desert and Prairie Massasauga sequences respectively. Intergenic distances between Sistrurus and Crotalus spp. ranged from 0.1451 ± 0.0034 (Prairie Massasauga—Eastern Diamondback) to 0.1567 ± 0.0034 (Desert Massasauga—Timber Rattlesnake).
SNP identification and genotyping
We ultimately genotyped 184 SNPs in 78 individuals (i.e., only 8 SNPs and 10 samples consistently failed to amplify). We subsequently removed 13 markers due to gametic phase disequilibrium. Our final set consisted of 171 markers, including 83 intergenic and 88 genic, 78 of which were from the Desert Massasauga and 93 from the Prairie Massasauga. As expected given our sampling regime, we found no unknown replicate samples and no first or second-degree relatives among our samples. Our genotyping error rate (\(e\)) was low, averaging 0.0082 (Desert: 0.0072, Prairie: 0.0091), and is heretofore ignored in population-level analyses.
Our genic markers include those within genes related to metabolism, venom production, and immune function among others (Table S6). In contrast, our intergenic markers reside at least 10 kb from known protein-coding genes. We partitioned our markers in this fashion to parse these two aspects of genomic variation if needed (e.g., due to signals of local adaptation). Similarly, half of the markers were developed from S. t. tergeminus reads and half from S. t. edwardsii reads to reduce taxon-specific ascertainment biases. Overall, however, our analyses revealed no substantive differences among the data partitions (i.e., ALL, HWE, GENIC, INTERGENIC, STT, and STE; Table 2) and thus for simplicity below we refer to results for ALL markers. For example, HO was 0.300 ± 0.250 for all markers and did not differ between genic and intergenic markers (0.331 ± 0.245 and 0.267 ± 0.253; Fisher’s exact test of 0.849, p > 0.05). Deviations from HWE were no more common in the genic markers (Fisher’s exact test of 0.480, p > 0.05) than in the intergenic markers. We tested for the Wahlund effect for all subsets of data and found no such evidence given the mean FIS for all markers was not significantly different than zero (FIS = 0.073 ± 0.395) and the slope of the relationship was greater than 1 (slope = 1.995 ± 0.583, adjusted R2 = 0.0659).
Population genetic analyses
We genotyped 78 individual snakes that were retained through filtering. Our Western Massasauga samples were collected across seven US states (Arizona, Colorado, Kansas, Missouri, New Mexico, Oklahoma, Texas) and one Mexican state (Coahuila). For metadata of genotyped samples, see Table S1. After Bonferroni correction, we found 32 out of 171 loci to be out of expected HWE proportions, 15 in heterozygote excess and 17 in a heterozygote deficit (Table S6). For our population statistics, we found an overall mean FST across all markers of 0.0264 ± 0.0525 and 0.0308 ± 0.0548 for markers in HWE. Mean FST between putative subspecies was 0.0318 ± 0.0612 for genic SNPs and 0.0198 ± 0.0414 for intergenic SNPs. FST also was calculated separately for SNPs selected from the reads of each subspecies (Desert mean = 0.0358 ± 0.0588; Prairie mean = 0.0178 ± 0.0458). In case we might have misassigned the subspecies affiliation of the four samples from ambiguous geographies, we redesignated subspecies assignments for the three samples from Matagorda Island and the one from Coleman County, then recalculated FST for all SNP markers. When we did so, we found qualitatively the same results (FST = 0.0365 ± 0.0644 after redesignation as S. t. tergeminus; FST = 0.0264 ± 0.0525 with original designation as S. t. edwardsii). Table 3 reports FST calculations for all data subsets.
We retained 16 of the axes from our PCA, explaining a cumulative 51% of the variation. The samples did not cluster according to subspecies status along any axis, though samples did show some separation according to geography on the primary two principal axes (Fig. 4). Samples are plotted according to their position on the first two primary axes (11.3% of variation cumulatively).
PCA of genotyped Western Massasauga samples. Desert (blue–grey diamonds) and Prairie (brown circles) samples are generally clustered in the first two axes of eigenspace. Cumulatively, PCs1 and 2 account for 11.3% of the variance in the data. Consistent with an isolation-by-distance model, AZ and NM samples (Desert) fall further to the left of PC1 and CO (Desert) samples tend towards the top of PC2. All three southeastern TX samples from Matagorda Island (Desert) are distinctly in the lower right of the plot (circled), suggesting that these data have the capacity to identify isolated populations. As an example, we have circled the sole MO sample (Prairie) that is both farthest east and furthest on PC1 from the AZ (also circled) and NM samples of any Prairie Massasaugas. Overall, these data provide little if any support for subspecific designations in the Western Massasauga
The Bayesian admixture analysis conducted in LEA determined the optimum parameters that best fit the data were α = 5 and k = 4. Admixture results for 40 different scenarios, including k = 4 and α = 5 as well as for various subsets of the SNP data (genic, intergenic, desert, prairie), can be found in Fig. S2. Our interpretation of these 40 scenarios is that the LEA results never convincingly partition the samples taxonomically. Accordingly, we do not discuss these alternative scenarios at length because they do not directly relate to our core research question which was to quantify the genetic differentiation between the two nominal subspecies. Most critically, we note there was no clear distinction between the two subspecies at the optimal k = 4 (Fig. S2) nor was there a convincing distinction between the two putative subspecies when k = 2 (Fig. 5), especially when considering the possibility of isolation by distance (IBD, see below).
Bayesian admixture analysis for k = 2, α = 5 from LEA using all SNP markers. Each column represents a single individual labelled with sample name and state, with nominal Desert and Prairie Massasaugas split left to right by the dark line, with samples for each putative subspecies sorted separately by admixture coefficient. The results shown here are most consistent with a single gene pool for Western Massasauga; if each nominal subspecies represented a distinct population segment, we would expect to see a genetic discontinuity that corresponded with the taxonomic designations. For other admixture plots, see Fig. S1
Discussion
Herein, we generated new discrete character (DNA sequences) and frequency-based marker datasets (SNP genotypes) to help provide conservation context to pending ESA decisions regarding the Desert Massasauga, S. t. edwardsii. Like all pending ESA decisions (because of the language in the Act), the question is whether a “distinct population segment,” or DPS, exists as claimed. There is a convoluted history associated with this language, and we refer the interested reader to the personal summary provided by Waples (2020). In this particular case, the Desert subspecies is being considered as a DPS. Accordingly, our analyses have focused on whether the two subspecies are distinct from one another.
The petition filed by Wild Earth Guardians (2010) cites habitat degradation and loss (native habitat conversion, overgrazing, urbanization, desertification, water resource depletion, habitat fragmentation and isolation, and road-related mortality), increased death (intentional culls, vehicle-related deaths, and predation), and other factors (disease, naturally low survivability and fecundity, prey loss, drought and climate change, and pet trade collection) that contribute to population declines. Our data do not directly speak to many of these issues, but they do address whether the taxonomic entity S. t. edwardsii actually exists as a genetically distinct population segment as discussed in Waples (2020).
Snakes demonstrate a remarkable diversity in genome size and structure that challenge traditional notions of genome evolution (Pasquesi et al. 2018), and thus we might expect differences in basic genome statistics between two different subspecies. The assembly statistics in Table S4 indicate quite similar genome compositions between both putative subspecies of Western Massasauga. The similarity in assembly size and GC content are indicators that Desert and Prairie Massasauga genomes are, at a gross level, very similar in structure. Clearly this is weak evidence to suggest that the Western Massasauga is not composed of two distinct population segments, but the genomic similarities are certainly consistent with the stronger evidence below.
K2P distance between any two individuals should, in principle, be zero only for monozygotic twins or other clones (barring mutation). Thus, any two randomly chosen individuals from a population should have a K2P value that exceeds 0. Using nuclear DNA sequences, we estimated a K2P value of 0.0041 ± 0.0080 between individual S. t. tergeminus and S. t. edwardsii, a value near the expected lower bound of zero. Clearly, this value suggests very little genomic differentiation between our representatives of each putative subspecies. There is currently no “standard” level of genomic differentiation that corresponds to any level of taxonomic hierarchy, but it seems quite possible that one day this quantitative approach will be useful in the delineation of subspecies and higher taxa. Our survey of comparative K2P values from the literature and publicly available nuclear gene sequences averaged 0.004 ± 0.005, quite similar to our estimates in Western Massasauga. We note that our literature-based K2P estimates are biased by the small number of genes considered (i.e., most studies were not “genomic”), so further research will be required to firmly document the degree of genomic differentiation between putative subspecies. In our context, K2P estimates are relative (not absolute) measures that may be biased in some way, but such biases should be similar with respect to either Western Massasauga taxon. Overall, the genomic K2P evidence supports the idea of exceedingly low differentiation between Western Massasaugas and provides limited or no support for subspecific delineation.
Snake mitochondrial sequence data are far more publicly available than snake nuclear sequence data, and thus more taxonomic context is available. Our mitochondrial K2P estimate between putative Desert and Prairie Massasauga was 0.0103 ± 0.0008 (Fig. 3; Table 1), far below estimates of subspecies-level distances in Macroprotodon spp. (0.093 ± 0.049) (Carranza et al. 2004) and Psammophis spp. (0.067 ± 0.033) (Kelly et al. 2008). Instead, the estimate between the Desert and Prairie mitochondrion corresponds to the range of mean inter-population mtDNA distances for different populations of Psammophis (0.016 ± 0.016; Kelly et al. 2008) and between populations of Natrix maura (0.042 ± 0.008; Guicking et al. 2008). Furthermore, the mtDNA genome sequences, assemblies, and annotations are nearly identical between Desert and Prairie Massasauga (Fig. S3). Overall, our sequence data indicate that the mtDNA divergence between putative subspecies of Western Massasauga is far less than what might be expected of distinct subspecies but falls well within the range of population-level divergence. In other words, our mtDNA data are generally consistent with the idea of a single Western Massasauga taxon that contains modest levels of nucleotide variability but generally inconsistent with data from comparisons between established snake subspecies.
Our overall FST value for all nuclear SNP markers between the two subspecies was low (FST = 0.0264 ± 0.0525), but similar to an independent subspecific comparison of snake taxa (FST = 0.02–0.08 for Micrurus diastema ssp.; Reyes-Velasco et al. 2020). Furthermore, the genetic differentiation between putative Western Massasauga subspecies was greater at genic loci (FST = 0.0318) than at intergenic loci (FST = 0.0198), suggesting that natural selection contributes to the patterns of differentiation observed in our dataset (Table 3). Our study was designed to evaluate the impacts of both drift and selection by employing both genic and intergenic markers; the intent was to capture candidate functional variants that might underlie pronounced genetic differentiation between the two subspecies (e.g., due to environmental or ecological differences). However, the FST data do not capture strong differentiation between the two subspecies, so we are cautious in our interpretation of the differences in genic and intergenic SNP differentiation for several reasons. First, subspecies differentiation was low with both subsets of markers (genic FST = 0.032 vs. intergenic FST = 0.020). Second, we used a small number of markers (N = 171) and specimens (N = 78). Third, our geographic sampling was broad (e.g., our two most distant samples were separated by 1300 km) but sparse. Fourth, the two putative subspecies have mostly disparate geographic ranges (i.e., different environments) and the differentiation we observed in FST values better reflects IBD (see below and Online Resources) than subspecific delineation. We think the most parsimonious explanation for the increased FST at genic loci is the more pronounced influence of natural selection relative to the intergenic loci (which are more influenced by drift), but because of the reasons listed above we did not evaluate potential genotype by environment associations. Overall, we think these SNP data are important not because of potential signs of selection, but because it is clear from these FST results that a very small proportion of the genetic variation within Western Massasaugas differentiates putative subspecies.
The results of our multivariate analyses via PCA also reveal little differentiation between putative subspecies (Fig. 4). If there were genetic differentiation between the two subspecies in question, we would see samples from each subspecies clustering separately on at least one axis. However, Desert and Prairie Massasaugas were largely coincident along all axes. Samples ordinated slightly with geography along axes 1 and 2, with those from populations at the range limits (i.e. AZ, NM, CO, MO, southeast TX—Matagorda Island) tending to ordinate further along axes 1 and 2 with sympatric samples. An exception to this is the lone sample from Mexico, which did not separate from the main grouping of samples. Overall, we think the most parsimonious interpretation of the genetic structure results is simple IBD, and that the PCA data do not provide convincing support for the genetic distinctiveness of formal taxonomic subspecies. This should not be too surprising given the low motility and dispersal capacity of Massasaugas (Mackessy et al. 2005), the large geographic range sampled herein, and the fact that IBD is the de facto null hypothesis in population genetics.
The results of our admixture analyses provide no support for genetically distinct subspecies. LEA results reveal no evidence of strong population structure within Western Massasaugas and, instead, are more consistent with genetic homogeneity, albeit with some IBD, across the sampled range. If the two putative subspecies were genetically differentiated, one would expect that genetic assignment tests could reliable identify a Western Massasauga of unknown origin to one or the other subspecies, but Fig. 5 illustrates how assignment probabilities would be virtually identical. Additionally, if each nominal subspecies represented a distinct population segment, we would expect to see a genetic discontinuity that corresponded with the taxonomic designations in Fig. 5, but that is not the case. Evaluation of admixture scenarios where k > 2 (Fig. S2) would require much more intensive sampling across the range, but we think such scenarios are unlikely because if two or more genetically structured populations are artificially grouped, even if the subpopulations are in HWE, we would expect a Wahlund effect. For example, if the two subspecies were genetically differentiated but we analyzed them as a single unit, we should find an overall deficit of heterozygotes, but we found no such evidence in our analyses (Table S7). This is an admittedly weak test for “distinct populations”, as it is in effect an absence of evidence, but it is consistent with our interpretation of our other analyses.
Finally, we see little evidence for subspecific designation based on the possibility of local adaptation because there is no evidence of strong differential selection on our genic markers; they are no more likely to deviate from HWE than the putatively neutral intergenic markers. We again explicitly acknowledge this is a weak test of population distinction, but there is no obvious taxonomic signal of local adaptation in our dataset.
These data represent the most complete genetic survey of Western Massasaugas available, but our study of course has limitations. We have no ecological (e.g., diet), behavioral (e.g., mate choice), or physiological (e.g., thermal tolerance) data. Furthermore, our genetic data are not ideal as most of our samples were collected via driving surveys and found dead on the road, therefore yielding fragmented DNA unsuitable for many assays (e.g., RadSeq). The Fluidigm SNPtype assay works remarkable well with poor quality DNA (Carroll et al. 2018; von Thaden et al. 2020), but in the end it surveys relatively few markers across the genome. Despite significant effort, we were likewise limited in the number of biological samples we surveyed across a very wide geographic range. All of these limitations add noise to our dataset, but the overall consistency among our various analyses (e.g., genetic/genomic, and frequency/categorical) speak to the significant biological signal that nevertheless remains.
Conclusions
Our data on the Western Massasauga are generally uniform across different data types and different analyses, revealing no obvious genetic discontinuities that yield a distinct population segment. These genetic and genomic data do not support the idea of either Desert or Prairie Massasauga; instead, our data suggest that Western Massasaugas consist of a single, relatively diverse gene pool. Previous evidence sharply distinguished the Western Massasauga from its Eastern sister species (Kubatko et al. 2011; Ryberg et al. 2015). Those studies cast initial doubt on the traditional division of Desert and Prairie Massasaugas as separate subspecies but were lacking power due to restricted sampling. This study, with greater geographic, genetic, and genomic resolution, buttresses previous studies of population structure in Massasaugas (Kubatko et al. 2011; Ryberg et al. 2015). Therefore, based on the genetic and genomic data previously published and herein, we recommend that Western Massasaugas, S. tergeminus, not be considered as two genetically differentiated subspecies but as a genetically unified species. Our data also illustrate the ability of genetics and genomics to help delineate taxa, in this case unifying artificial taxonomic constructs that do not reflect biological realities.
Data availability
Sequencing reads and assemblies have been submitted to NCBI under BioProject accession PRJNA668351.
Code availability
Analyses were conducted in R or Unix environments and scripts are available in the Online Resources.
References
Anderson CD, Gibbs HL, Douglas ME, Holycross AT (2009) Conservation genetics of the Desert Massasauga rattlesnake (Sistrurus catenatus edwardsii). Copeia 2009:740–747. https://doi.org/10.1643/cg-08-152
Andrews S (2010) FastQC: a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc
Blair C, Sánchez-Ramírez S (2016) Diversity-dependent cladogenesis throughout western Mexico: evolutionary biogeography of rattlesnakes (Viperidae: Crotalinae: Crotalus and Sistrurus). Mol Phylogenet Evol 97:145–154. https://doi.org/10.1016/j.ympev.2015.12.020
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. https://doi.org/10.1093/bioinformatics/btu170
Broad Institute (2018) Picard Tools version 2.18.2. GitHub repository. Broad Institute. http://broadinstitute.github.io/picard/
Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL (2009) BLAST+: architecture and applications. BMC Bioinform 10:1–9. https://doi.org/10.1186/1471-2105-10-421
Carranza S, Arnold EN, Wade E, Fahd S (2004) Phylogeography of the false smooth snakes, Macroprotodon (Serpentes, Colubridae): mitochondrial DNA sequences show European populations arrived recently from northwest Africa. Mol Phylogenet Evol 33:523–532. https://doi.org/10.1016/j.ympev.2004.07.009
Carroll EL, Bruford MW, DeWoody JA, Leroy G, Strand A, Waits L, Wang J (2018) Genetic and genomic monitoring with minimally invasive sampling methods. Evol Appl 11:1094–1119. https://doi.org/10.1111/eva.12600
Chikhi R, Medvedev P (2014) Informed and automated k-mer size selection for genome assembly. Bioinformatics 30(1):31–37. https://doi.org/10.1093/bioinformatics/btt310
Clark RW, Brown WS, Stechert R, Zamudio KR (2010) Roads, interrupted dispersal, and genetic diversity in timber rattlesnakes. Conserv Biol 24:1059–1069. https://doi.org/10.1111/j.1523-1739.2009.01439.x
Clark RW, Marchand MN, Clifford BJ, Stechert R, Stephens S (2011) Decline of an isolated timber rattlesnake (Crotalus horridus) population: interactions between climate change, disease, and loss of genetic diversity. Biol Conserv 144:886–891. https://doi.org/10.1016/j.biocon.2010.12.001
Colley M, Lougheed SC, Otterbein K, Litzgus JD (2017) Mitigation reduces road mortality of a threatened rattlesnake. Wildl Res 44:48–59. https://doi.org/10.1071/WR16130
Conant R, Collins JT (1991) A field guide to reptiles and amphibians. Eastern and central North America. Houghton Mifflin Co., Boston
Crother BI, Savage JM, Holycross AT (2011) Case 3571 Crotalinus catenatus Rafinesque, 1818 (currently Sistrurus catenatus) and Crotalus tergeminus Say in James, 1822 (currently Sistrurus tergeminus; Reptilia, Serpentes): proposed conservation of usage by designation of neotypes for both species. Bull Zool Nomencl 68:271–274. https://doi.org/10.21805/bzn.v68i4.a3
Crother BI, Savage JM, Holycross AT (2013) Crotalinus catenatus Rafinesque, 1818 (currently Sistrurus catenatus) and Crotalus tergeminus Say in James, 1822 (currently Sistrurus tergeminus; Reptilia, Serpentes): usage conserved by designation of neotypes for both species. Bull Zool Nomencl 70:271–274. https://doi.org/10.21805/bzn.v68i4.a3
Dixon JR, Werler JE (2005) Texas snakes: a field guide. University of Texas Press, Austin
Doyle JM, Katzner TE, Roemer GW, Cain JW III, Millsap BA, McIntyre CL, Sonsthagen SA, Fernandez NB, Wheeler M, Bulut Z, Bloom PH, DeWoody JA (2016) Genetic structure and viability selection in the golden eagle (Aquila chrysaetos), a vagile raptor with a Holarctic distribution. Conserv Genet 17:1307–1322. https://doi.org/10.1007/s10592-016-0863-0
Fernandes F, Pereira L, Freitas AT (2009) CSA: an efficient algorithm to improve circular DNA multiple alignment. BMC Bioinform 10:230. https://doi.org/10.1186/1471-2105-10-230
Fetzner JW Jr (1999) Extracting high-quality DNA from shed reptile skins: a simplified method. Biotechniques 26:1052–1054. https://doi.org/10.2144/99266bm09
Fitzgerald LA, Painter CW (2000) Rattlesnake commercialization: long-term trends, issues, and implications for conservation. Wildl Soc Bull 28:235–253
Frichot E, François O (2015) LEA: an R package for landscape and ecological association studies. Methods Ecol Evol 6(8):925–929. https://doi.org/10.1111/2041-210X.12382
Frichot E, Mathieu F, Trouillon T, Bouchard G, François O (2014) Fast and efficient estimation of individual ancestry coefficients. Genetics 196(4):973–983. https://doi.org/10.1534/genetics.113.160572
Gibbs HL, Murphy M, Chiucchi JE (2011) Genetic identity of endangered Massasauga rattlesnakes (Sistrurus sp.) in Missouri. Conserv Genet 12:433–439. https://doi.org/10.1007/s10592-010-0151-3
Gloyd H (1955) A review of the Massasauga, Sistrurus catenatus, of the southwestern United States (Serpentes: Crotalidae). Bull Chin Acad Sci 10:83–98
Gregory TR (2020) Animal genome size database. http://www.genomesize.com. Accessed 22 Sep 2020
Guicking D, Joger U, Wink M (2008) Molecular phylogeography of the viperine snake Natrix maura (Serpentes: Colubridae): evidence for strong intraspecific differentiation. Org Divers Evol 8:130–145. https://doi.org/10.1016/j.ode.2007.05.001
Hahn C, Bachmann L, Chevreux B (2013) Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—a baiting and iterative mapping approach. Nucleic Acids Res 41(13):129. https://doi.org/10.1093/nar/gkt371
Hillis DM (2020) The detection and naming of geographic variation within species. Herpetol Rev 51:52–56
Holycross AT, Mackessy SP (2002) Variation in the diet of Sistrurus catenatus (Massasauga), with emphasis on Sistrurus catenatus edwardsii (Desert Massasauga). J Herpetol 36:454–464. https://doi.org/10.1670/0022-1511(2002)036[0454:VITDOS]2.0.CO;2
Holycross AT, Anton TG, Douglas ME, Frost DR (2008) The type localities of Sistrurus catenatus and Crotalus viridis (Serpentes: Viperidae), with the unraveling of a most unfortunate tangle of names. Copeia 2008:421–424. https://doi.org/10.1643/ch-07-095
Jackman SD, Vandervalk BP, Mohamadi H, Chu J, Yeo S, Hammond SA, Jahesh G, Khan H, Coombe L, Warren RL, Birol I (2017) ABySS 2.0: resource-efficient assembly of large genomes using a Bloom filter. Genome Res 27(5):768–777. https://doi.org/10.1101/gr.214346.116
Jombart T (2008) Adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24:1403–1405. https://doi.org/10.1093/bioinformatics/btn129
Letunic I, Bork P (2021) Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res 49(W1):W293–W296
Kelly CMR, Barker NP, Villet MH, Broadley DG, Branch WR (2008) The snake family Psammophiidae (Reptilia: Serpentes): phylogenetics and species delimitation in the African sand snakes (Psammophis Boie, 1825) and allied genera. Mol Phylogenet Evol 47:1045–1060. https://doi.org/10.1016/j.ympev.2008.03.025
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120. https://doi.org/10.1007/BF01731581
Klauber LM (1982) Rattlesnakes: their habits, life histories, and influence on mankind. University of California Press, Berkeley
Knight A, Styer D, Campbell JA, Pelikan S, Densmore LD, Mindell DP (1993) Choosing among hypotheses of rattlesnake phylogeny a best fit rate test for DNA sequence data. Syst Biol 42:356–367. https://doi.org/10.1093/sysbio/42.3.356
Kubatko LS, Gibbs HL, Bloomquist EW (2011) Inferring species-level phylogenies and taxonomic distinctiveness using multilocus data in Sistrurus rattlesnakes. Syst Biol 60:393–409. https://doi.org/10.1093/sysbio/syr011
Kumar S, Stecher G, Suleski M, Hedges SB (2017) TimeTree: a resource for timelines, timetrees, and divergence times. Mol Biol Evol 34:1812–1819. https://doi.org/10.1093/molbev/msx116
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. https://doi.org/10.1093/molbev/msy096
Li H (2013) Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. ArXiv. http://arxiv.org/abs/1303.3997
Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y, Tang J, Wu G, Zhang H, Shi Y, Liu Y, Yu C, Wang B, Lu Y, Han C, Cheung DW, Yiu SM, Peng S, Xiaoqian Z, Liu G, Liao X, Li Y, Yang H, Wang J, Lam TW, Wang J (2012) SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaScience 1(1):2047-217X-1–18. https://doi.org/10.1186/2047-217X-1-18
Mackessy SP, Keinath DA, Mcgee M, Mcdonald D, Ise T (2005) Desert Massasauga rattlesnake (Sistrurus catenatus edwardsii): a technical conservation assessment. https://www.fs.usda.gov/Internet/FSE_DOCUMENTS/stelprdb5182073.pdf. Accessed 22 Sep 2020
McCluskey EM, Bender D (2015) Genetic structure of Western Massasauga rattlesnakes (Sistrurus catenatus tergeminus). J Herpetol 49:343–348. https://doi.org/10.1670/14-016
Murphy RW, Fu J, Lathrop A, Feltham JV, Kovac V (2002) Phylogeny of the rattlesnakes (Crotalus and Sistrurus) inferred from sequences of five mitochondrial DNA genes. In: Schuett GW, Hoggren M, Douglas ME, Greene HW (eds) Biology of the vipers. Eagle Mountain Publishing LC, Eagle Mountain, pp 66–92
O’Brien SJ, Mayr E (1991) Bureaucratic mischief: recognizing endangered species and subspecies. Science 251:1187–1188. https://doi.org/10.1126/science.251.4998.1187
Paradis E, Claude J, Strimmer K (2004) APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20:289–290. https://doi.org/10.1093/bioinformatics/btg412
Pasquesi GIM, Adams RH, Card DC, Schield DR, Corbin AB, Perry BW, Reyes-Velasco J, Ruggiero RP, Vandewege MW, Shortt JA, Castoe TA (2018) Squamate reptiles challenge paradigms of genomic repeat element evolution set by birds and mammals. Nat Commun 9:2774. https://doi.org/10.1038/s41467-018-05279-1
Reyes-Velasco J, Adams RH, Boissinot S, Parkinson CL, Campbell JA, Castoe TA, Smith EN (2020) Genome-wide SNPs clarify lineage diversity confused by coloration in coralsnakes of the Micrurus diastema species complex (Serpentes: Elapidae). Mol Phylogenet Evol 147:106770. https://doi.org/10.1016/j.ympev.2020.106770
Rousset F (2008) GENEPOP’007: a complete re-implementation of the GENEPOP software for Windows and Linux. Mol Ecol Resour 8:103–106. https://doi.org/10.1111/j.1471-8286.2007.01931.x
Ryberg WA, Harvey JA, Blick A, Hibbitts TJ, Voelker G (2015) Genetic structure is inconsistent with subspecies designations in the Western Massasauga Sistrurus tergeminus. J Fish Wildl Manag 6:350–359. https://doi.org/10.3996/122014-JFWM-093
Schield DR, Card DC, Hales NR, Perry BW, Pasquesi GM, Blackmon H, Adams RH, Corbin AB, Smith CF, Ramesh B, Demuth JP, Bertrán E, Tollis M, Meik JM, Mackessy SP, Castoe TA (2019) The origins and evolution of chromosomes, dosage compensation, and mechanisms underlying venom regulation in snakes. Genome Res 29:590–601. https://doi.org/10.1101/gr.240952.118
Stebbins RC (1980) A field guide to western reptiles and amphibians. Houghton Mifflin Company, Boston
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Torstrom SM, Pangle KL, Swanson BJ (2014) Shedding subspecies: the influence of genetics on reptile subspecies taxonomy. Mol Phylogenet Evol 76:134–143
Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, del Angel G, Levy-Moonshine A, Jordan T, Shakir K, Roazen D, Thibault J, Banks E, Garimella KV, Altshuler D, Gabriel S, DePristo MA (2013) From FastQ data to high-confidence variant calls: the genome analysis toolkit best practices pipeline. Curr Protoc Bioinform 43:11.10.1-11.10.33. https://doi.org/10.1002/0471250953.bi1110s43
von Thaden A, Nowak C, Tiesmeyer A, Reiners TE, Alves PC, Lyons LA, Mattucci F, Randi E, Cragnolini M, Galián J, Hegylei Z, Kitchener AC, Lambinet C, Lucas JM, Mölich T, Ramos L, Schockert V, Cocchiararo B (2020) Applying genomic data in wildlife monitoring: development guidelines for genotyping degraded samples with reduced single nucleotide polymorphism (SNP) panels. Mol Ecol Resour 20:662–680. https://doi.org/10.1111/1755-0998.13136
Waples RS (2020) Serendipity and me. ICES J Mar Sci 77:1658–1665
Waples RS, Allendorf F (2015) Testing for Hardy-Weinberg proportions: have we lost the plot? J Hered 106:1–19. https://doi.org/10.1093/jhered/esu062
Wickham H (2016) ggplot2: elegant graphics for data analysis. Springer, New York
Wild Earth Guardians (2010) Petition to list the Desert Massasauga (Sistrurus catenatus edwardsii) under the U.S. Endangered Species Act. https://pdf.wildearthguardians.org/site/DocServer/listing_petition_desert_massasauga.pdf?docID=685&AddInterest=1059. Accessed 22 Sep 2020
Acknowledgements
We thank Andrew Holycross, Lisle Gibbs, Stephen Mackessy, Paul Crump, Jason Jones, Doug Burkett, Ian Latella, Sean Graham, Nathan Rains, and Dan Walker for providing samples of Sistrurus tergeminus and Sistrurus catenatus. We also acquired tissue samples from the Texas Natural History Collection (Travis LaDuc), Gladys Porter Zoo (Thomas deMaar), Museum of Southwestern Biology (Tom Giermakowski), Sternberg Museum of Natural History (Curtis Schmidt), Sam Noble Oklahoma Museum of Natural History (Jessa Watters), and Texas State University (Michael R. J. Forstner). We thank Lisle Gibbs, Alex Ochoa, and Scott Martin for their generosity in sharing the Sistrurus catenatus nuclear and mitochondrial genome assemblies. We thank Drew R. Schield for his help in obtaining and navigating the Crotalus viridis genome assembly. We also thank Drs. Lisle Gibbs, Anna Brüniche-Olsen, Phillip San Miguel, Janna Willoughby, Alex Ochoa and members of the DeWoody Lab for their invaluable feedback and advice at many different parts of the process. This research was supported in part by the National Institute for Food and Agriculture, National Fish and Wildlife Foundation, U.S. Fish and Wildlife Service, and Texas Parks and Wildlife Department. Field work was conducted under the authority of Permits issued by the Texas Parks and Wildlife Department (SPR-0506-662) and the New Mexico Department of Game and Fish (NMDGF Sci# 3742) to Toby J. Hibbitts. This study was approved by the Texas A&M University Institutional Animal Care and Use Committee (AUP 2016-178). This is publication number 1657 of the Biodiversity Research and Teaching Collections at Texas A&M University.
Funding
This research was supported in part by the National Institute for Food and Agriculture, National Fish and Wildlife Foundation, U.S. Fish and Wildlife Service, and Texas Parks and Wildlife Department.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors claim no competing interests.
Ethical approval
Field work was conducted under the authority of Permits issued by the Texas Parks and Wildlife Department (SPR-0506-662) and the New Mexico Department of Game and Fish (NMDGF Sci# 3742) to Toby J. Hibbitts. This study was approved by the Texas A&M University Institutional Animal Care and Use Committee (AUP 2016-178).
Informed consent
Not applicable.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Bylsma, R., Walkup, D.K., Hibbitts, T.J. et al. Population genetic and genomic analyses of Western Massasauga (Sistrurus tergeminus ssp.): implications for subspecies delimitation and conservation. Conserv Genet 23, 271–283 (2022). https://doi.org/10.1007/s10592-021-01420-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-021-01420-8