Abstract
The southeastern United States harbors one of the most diverse temperate freshwater fish faunas of the world. Unfortunately, due to improper land use practices and habitat degradation, many of the species in this region are imperiled and may become extinct without appropriate conservation efforts. This study examined the population dynamics of an endangered endemic darter of southwest Kentucky, the Relict Darter (Etheostoma chienense) and its sister taxon, the undescribed Clarks Darter (Etheostoma cf. oophylax). Mitochondrial sequence data coupled with SNP data were used to infer population structure, gene flow, genetic variation, and effective population sizes of both species. The results from this study, based on 160 individuals from nine localities, indicate that the endangered Relict Darter possesses limited genetic variation based on mitochondrial DNA haplotypes (N = 4). In addition, SNP data (6.8 k markers) further indicates limited genetic structure (K = 1), as well as a low effective population size (143–918), suggesting that the Relict Darter can be managed as a single, panmictic conservation unit. It is suggested that conservation efforts be taken to protect remaining habitats, augment the system with artificial spawning substrates, and, as a last resort (if needed), supplement the natural population with captive reared individuals. Ethesotoma cf. oophylax showed some genetic variation among distant sites, but more samples are needed throughout the range in order to fully understand the population dynamics of this species.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Understanding the population dynamics of a species is a vital component of wildlife and fisheries conservation (Vrijenhoek 2005; Waples and Do 2010). Elucidating the patterns and processes of genetic structure is a critical step that allows resource managers to make informed decisions for managed and imperiled species (Gido et al. 2016; Scribner et al. 2016). Unfortunately, for many imperiled species, information on genetic structure and gene flow is often lacking making conservation efforts a challenge. Furthermore, it is typically assumed that geographically restricted species possess low levels of genetic diversity as a result of their limited distribution and small population sizes (Frankham 2010). However, some empirical studies have indicated that this is not always the case (Eliades et al. 2011). Given the delicate nature of imperiled species, collecting population genetic data can play a pivotal role in focusing conservation efforts to susceptible areas.
Assessing levels of gene flow, genetic structure, and effective population size are three areas that are particularly useful when assessing the conservation status of imperiled species (Frankham 1996, 2015). This type of information is often used to guide resource managers by allowing them to make necessary management decisions by identifying appropriate management units (Piller et al. 2005; Mee et al. 2015). In particular, knowledge of the magnitude and directionality of gene flow, as well as effective population size and population genetic structure, are key factors in assessing the genetic health of endangered species. This type of information has provided useful data that has allowed resource managers to make scientifically informed management decisions regarding commercial and endangered species (Westemeier et al. 1998; Vrijenhoek 2005; Waples and Do 2010; Russello et al. 2015).
The southeastern United States harbors one of the most diverse temperate freshwater fish faunas in the world. Unfortunately, the region also contains a large number of imperiled fishes (Warren et al. 1994, 2000). One of these groups, the darters (Teleostomi: Percidae), are an ecologically diverse group of more than 200 species and multiple genera (Page 1983; Near et al. 2011). Etheostoma is the most diverse genus in the family and includes species that have restricted distributions and high habitat stringencies. All species are benthic and have life-histories that are intimately associated with the benthos. To date, all species of Etheostoma that have been studied lack a swimbladder (Evans and Page 2003), and many species, for which there is data, lack a pelagic larval drift phase and are assumed to possess minimal migratory abilities (Douglas et al. 2013).
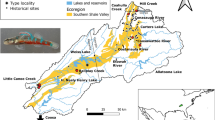
This comparative study examines genetic structure and gene flow of two closely related darter species. First, the Relict Darter, Etheostoma chienense, an imperiled species is endemic to the Bayou Du Chien (BDC), a small (554 km2) direct tributary of the Mississippi River in southwest Kentucky (Page et al. 1992) (Fig. 1). Etheostoma chienense is listed as a federally endangered species due to its restricted distribution, the paucity of spawning habitat, and habitat fragmentation caused by poor land use practices in the area (Piller and Burr 1998, 1999). In a recent USFWS conservation status review (Floyd 2018) it was noted that no information was known about genetic variation, gene flow and effective population size of E. chienense. The Clarks Darter, E. cf. oophylax, is the other focal species of this study. Previous studies have noted the distinctiveness of this lineage, but to date, it has not yet received a formal species description (Near et al. 2011; Harrington et al. 2013; Harrington and Near 2015). This lineage is restricted to the nearby Clarks River (878 km2), a tributary to the lower Tennessee River, in southwest Kentucky and northern central Tennessee. Both taxa possess a reproductive strategy known as egg-clustering in which eggs are deposited on the underside of firm substrates and then guarded by a male (Page 1983; Page et al. 1992; Piller and Burr 1999). Etheostoma chienense is opportunistic in its choice of spawning substrates and utilizes both natural and man-made materials (Piller and Burr 1999). Nothing is known about the spawning substrate preferences of E. cf. oophylax, but it presumably uses natural materials, as they are abundant in the system. Relative to E. chienense, populations of E. cf. oophylax are larger and more widespread, spawning habitat is more readily available, and it does not receive any level of state or federal protection. Presumably, E. cf. oophylax possesses similar life-history traits as E. chienense, but little information is known about larval drift, migratory abilities, age and growth, and diet for either species (Table 1).
The size and dendrology of a stream system, as well as the availability of spawning habitat, life-history influences, and population size play major roles in the connectivity of aquatic populations (Frankham 2010; Thomaz et al. 2016; Camak and Piller 2018). Therefore, this study employed a comparative population genetic approach to investigate genetic structure, levels of gene flow, and effective population sizes of two closely related congeners that occupy similar stream habitats in different drainage basins and possess similar life-history traits. Therefore, we undertook a comparative study to investigate these characteristics for the Relict Darter, E. chienense, and a closely related taxon, the Clarks Darter, E. cf. oophylax. It is assumed that any differences in gene flow between species are most likely attributable to differences in the abundance of each species, conservation status, and/or availability of spawning habitat (Camak and Piller 2018). This comparative approach offers the best opportunity to understand the genetic diversity of these closely related, ecologically similar taxa, one of which is imperiled and one that is conservationally secure.
The specific objectives of this study are as follows. First, to assess levels of intrabasin genetic structure and intraspecific genetic variation of two sister taxa (Near et al. 2011; Harrington et al. 2013; Harrington and Near 2015), Etheostoma chienense and Etheostoma cf. oophylax. We hypothesize that the levels of gene flow between subpopulations are expected to be similar for each taxon due to what is assumed to be similar migratory capabilities, but that the levels of genetic variation within each species are expected to be greater for E. cf. oophylax due to its larger population size and its occupancy of a larger sub-basin. Second, to estimate the effective population size (NE) of each species. We hypothesized that NE will be higher for E. cf. oophylax in comparison to E. chienense for the same reasons mentioned above. To address these questions, mitochondrial DNA sequences -cytochrome b- and double digest restriction site associated DNA -ddRADseq- (Peterson et al. 2012) data were gathered and analyzed. The genetic diversity observed within cytochrome b provides information regarding the maternal genetic diversity of the two species, while the ddRADseq data provided a much more robust multilocus dataset to examine the population dynamics of both E. chienense and E. cf. oophylax.
Materials and methods
Tissue collection
Due to the endangered status of E. chienense, extreme caution was taken for collecting fin clips of this species. Seine nets and a backpack electrofisher were used to collect the darters from the Bayou du Chien (BDC) in early October of 2017. The upper lobe of the caudal fin of each specimen was clipped and placed in 1.5 mL tubes containing 95% ethanol for storage and transport. All darters were released back to the stream after tissues were taken. With the assistance of the United States Fish and Wildlife Service and the Kentucky Department of Fish and Game, tissue samples from 174 Relict Darters from nine localities were collected from the Bayou du Chien system. The distance from each locality to the next was, on average, approximately 5 km of river distance (Fig. 1). The number of darters collected from site to site varied drastically due to its limited habitat availability throughout the stream. The tissues collected for this study covered most of the known range of the species (Table 2).
Etheostoma cf. oophylax also was collected using seine nets and a backpack electrofisher. However, given this species is not imperiled, whole specimens were fixed in 10% formalin after fin clips were taken and put into 1.5 mL tubes containing 95% ethanol. Voucher specimens were archived in the Southeastern Louisiana University Vertebrate Museum. All E. cf. oophylax specimens were collected from headwater streams of the East Fork Clarks River in the days immediately following the E. chienense collection in October of 2017. Localities for E. cf. oophylax were chosen to best mimic the between-locality river distances utilized for E. chienense collections in order to make direct comparisons in downstream analyses between the two species. The collection area comprised approximately 14% of the known range of E. cf. oophylax (Fig. 1). One-hundred eighty-six individuals of E. cf. oophylax were collected from 8 different sites. A number of sites in the Clarks River basin were mostly dry with only isolated pools available for sampling. Six individuals, collected in 2009, from the West Fork of the Clarks River were obtained from the Peabody Museum of Natural History at Yale University (YPM ICH 023032 A-F) and were included in the ddRAD portion of data analysis.
DNA extraction and PCR
Extraction protocols for genomic DNA followed the procedure for the Promega Wizard SV 96 Genomic DNA Purification System and/or the Qiagen DNeasy Blood and Tissue Kit. The cytochrome b gene of the mitochondrial genome was amplified via PCR using the primers from Schmidt and Gold (1993) with the following protocol: initial denaturation 96 °C for 2:00 min, 30 cycles of 93° for 30 s, 47° for 30 s, 72° for 2:00 min, with a final extension of 72° for 10 min. Each PCR reaction contained each of the following: 0.75 μL MgCl, 2.5 μL 10 × Buffer, 0.5 μL dNTP’s, 0.25 μL Taq polymerase, 1 μL of extracted DNA, 19 μL of nuclease free water, and 0.5 μL of both forward and reverse primers (10 ng/uL). PCR amplicons were sent to Genewiz for Sanger Sequencing.
Mitochondrial DNA analyses
The recovered haploid sequences were aligned and trimmed in Geneious using MAFFT to create a final data matrix used for downstream analyses. Haplotype networks were created independently for each species in the program PopArt v1.7. Statistical tests included: Tajima’s D for neutrality, Fu’s F statistic for neutrality, and haplotype diversity which were calculated in the program DnaSP (5.10) (Librado and Rozas 2009).
ddRAD library prep and sequencing
Four ddRAD libraries were prepped at Southeastern Louisiana University. Library preparation roughly followed the protocol from Peterson et al. (2012). Briefly, the DNA was purified, digested with restriction enzymes, ligated to common and unique oligos, PCR amplified, cleaned up, multiplexed, and size selected for 350–600 bp fragment lengths using a Blue Pippen. Libraries were run on a Bioanalyzer for a quality check before sequencing. A total of 174 E. chienense specimens and 186 E. cf. oophylax specimens were prepared for ddRADseq. Libraries were sent to the University of Oregon’s Genomics & Cell Characterization Core Facility for Illumina Sequencing on the Hiseq 4000 for 100 bp single end reads. The six samples received from Yale University had previously been prepped and sequenced using the same protocol. The raw fastq data files for each sample were added into the ipyrad pipeline to create the data files used for downstream analysis.
ddRAD assembly
Raw datafile files were run through the program FastQC to check the overall quality of the reads and the Illumina run. Ipyrad v0.7.30 (Eaton and Overcast 2020) was used to assemble the datasets for both species from the raw sequence reads. Individuals with fewer than 100,000 total raw reads were removed from the dataset. Reads that contained more than 5 bases with a low quality phred score (< 33) were excluded. Reads were then clustered based on a 90% similarity threshold and reads with fewer than 6X coverage were filtered out. A maximum of 5 ambiguous base calls and 8 heterozygous sites per read were allowed during filtering. The dataset for each species was aligned using a min80 threshold; which means that loci not retained in at least 80 individuals at the end of the assembly were filtered out. After a full assembly run, individuals with fewer than 1000 remaining loci were removed, and the assembly pipeline was re-run with the same parameters. Additional filtering was done using vcftools (Danecek et al. 2011). In vcftools we identified the percentage of missing data in each individual and manually removed individuals with greater than 80% missing data for the E. chienense and E. cf. oophylax East Fork datasets. Using the same method, we excluded individuals with greater than 40% missing data for the E. cf. oophylax East Fork and West Fork dataset. Vcftools was also used to exclude loci (SNPs) with a 70% call rate or lower. The R package PCAdapt was used to filter loci under selection with an expected false discovery rate lower than 10% (α = 0.1). Three datasets were assembled for downstream analysis: E. chienense; E. cf. oophylax from the East Fork of the Clarks River; and E. cf. oophylax with 35 of the highest quality samples from the East Fork and six samples from the West Fork.
Population structure
To estimate population structure of each species, the program STRUCTURE (Pritchard et al. 2000) was used to estimate the ancestry of each individual to a given number of populations (K). For E. chienense, K values 1–9 were run and for E. cf. oophylax K values 1–8, one K for each sampling locality of each species. Finally, using the samples of E. cf. oophylax from the West Fork (n = 6) with a subset of samples from each site of the East Fork of the Clarks River (n = 29), K values 1–2 were tested to determine if there is any genetic structure between the two forks of the Clarks River. A burnin period of 10,000 iterations were run initially and an additional 50,000 iterations were run post burnin, where the posterior probabilities of each individual to a cluster were drawn. STRUCTURE was run 10 times for each estimated K value to ensure convergence of the results. STRauto (Chhatre and Emerson 2017) was used to run STRUCTURE in parallel to optimize the efficiency of the runs. Results of each STRUCTURE run were summarized using STRUCTUREHarvester (Earl 2012) and average Ln(P) of each K was plotted in order to visualize the K value with the highest log likelihood for each set of runs. That K value was then used to examine the population structure of each species. After the optimal K value was determined for each species, the results were summarized and graphically visualized using the programs CLUMPP (Jakobsson and Rosenberg 2007) and DISTRUCT (Rosenberg 2004).
Discriminant Analysis of Principal Components (Jombart et al. 2010) was run for each dataset as a secondary test of population structure. The R packages adegenet (Jombart and Ahmed 2011) and poppr were used to perform each DAPC and plot the results of each analysis with 95% confidence ellipses of each site for each species.
Weighted FST method developed by Weir and Cockerham (1984) was used to calculate a proxy for pairwise genetic distance between sites calculated in hierfstat (Goudet 2005). The R package hierfstat was also used to assess the significance of the pairwise FST by determining which values were significantly different from zero. Geographic distances were calculated using ArcGIS using river distances (km) rather than straight line distances. Isolation by distance (IBD) was examined with a Mantel Test implemented in R with the package ‘ape’ (Paradis et al. 2004) using the pairwise FST and geographic river distances for E. chienense and E. cf. oophylax (East Fork only). Due to limited sampling of the range, IBD was not tested with the samples from West Fork of the Clarks River, as this would not be an effective test of genetic structure for the dataset given the large sampling gaps between sites.
Population statistics
Given the results of the population structure tests, separate collection localities were no longer used in the assessment of each taxon, and all individuals of each taxon were treated as one panmictic population. Expected and observed heterozygosities, pairwise site FST, inbreeding coefficients, and Tajima’s D were each estimated using vcftools. Confidence values for pairwise FST were estimated using the r package hierfstat. Heterozygosity and inbreeding coefficients were estimated for each individual given the loci available for that individual. Effective population size estimations of each species were performed using the program NEEstimator v2.1 (Do et al. 2014) with lowest allele frequency values of 0.05, 0.02, and 0.01.
Results
Mitochondrial DNA
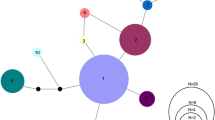
The alignment used for E. chienense (n = 151) was 669 nucleotides in length with a total of three haplotypes. For E. cf. oophylax- EF (n = 168), the alignment was 589 nucleotides in length with a total of nine unique haplotypes. These alignments are freely available on GenBank (BankIt2350217: MT539389—MT539556 and BankIt2350222: MT539557—MT539707). Haplotype networks displayed different levels of genetic variation within each species. Etheostoma chienense possesses only three haplotypes (Fig. 2a) throughout its known range for this cytochrome b fragment, while nine haplotypes were recovered for E. cf. oophylax (Fig. 2b) from a limited portion of its range.
Haplotype networks of a E. chienense (n = 151) and b E. cf. oophylax (n = 168). Each circle represents a unique haplotype, and the size of the circle is proportional to the number of individuals with that haplotype. Colors represent the nine different sampling sites and each tick mark represents the number of nucleotide differences between haplotypes
Tajima’s D and Fu’s F tests for neutrality were not significant for every subset of collection localities of both species; however, both Tajima’s D (− 1.855, p = 0.002) and Fu’s F (− 6.725, p = 0.001) were negative and significant for the E. cf. oophylax when analyzed as a single population. These were the only significant statistical tests with the mitochondrial data. The overall haplotype diversity (H) within E. cf. oophylax (0.251 as a whole, range 0.077–1 from each site) is much lower than the haplotype diversity within E. chienense (0.545 as a whole, range 0.242–0.833 from each site). Even though there are three times as many haplotypes for E. cf. oophylax, 141 (86.5%) individuals share a single haplotype. While in E. chienense, the most common haplotype is shared by only 80 individuals (52.9%) (Fig. 2).
Illumina sequencing
All four raw data files passed the quality check in FastQC with over 1.4 billion reads received among the raw Illumina data files which are available via NCBI SRA (biosample accession numbers: SAMN15018085-SAMN15018179; SAMN15075015-SAMN15075067; SAMN15079195-SAMN15079276; SAMN15086545-SAMN15086608). After SNP filtering in both ipyrad and vcftools, the final datasets used for downstream analyses for E. chienense and E. cf. oophylax had 6871 SNPs for 90 individuals (average 37.4% missing data per individual) and 9876 SNPs for 108 individuals (average 20.7% missing data per individual), respectively. These alignments and ipyrad parameters are available via Dryad (https://doi.org/10.5061/dryad.fqz612jqb). The dataset containing samples from both forks of the Clarks River included 35 individuals with 10,709 SNPs (average 12.0% missing data per individual). Three separate datasets mentioned above were used for downstream analyses.
Population structure
Each of the three datasets were run in STRUCTURE. The log probability of E. chienense was highest for K = 1 (Fig. 3a). Etheostoma cf. oophylax also had the highest log probability of K = 1 (Fig. 3b). As for the dataset with samples from the East and West forks of the Clarks River, K = 2 had the highest log probability (Fig. 3c). STRUCTURE plot shows admixture between forks of the Clarks River (Fig. 4). Discriminant Analysis of Principal Components (DAPC) shows number of clusters of E. chienense and E. cf. oophylax (East Fork) to be K = 1 (Fig. 5a, b). All sites for both species have undifferentiable overlap. Etheostoma cf. oophylax shows strong clustering between the East and West Forks of the Clarks River (K = 2), with there being no overlap between samples from opposite forks (Fig. 5c).
F ST values and isolation by distance
Pairwise Weir and Cockerham (1984) weighted FST values were generated for both species. For E. chienense, pairwise FST site comparisons ranged from − 0.0539 to 0.0819 (Table 3), and E. cf. oophylax (East Fork) ranged from − 0.1707 to 0.0434 (Table 4). Pairwise FST of E. cf. oophylax from the East and West Forks was 0.1941 with 95% confidence between 0.1232 and 0.2650.
The Isolation by Distance (IBD) comparisons using a Mantel test of E. chienense showed a positive relationship between genetic and geographic distance, but it was not statistically significant (p = 0.13). The same IBD analysis for E. cf. oophylax (East Fork) also yielded a non-significant (p = 0.32) positive relationship between geographic and genetic distance.
General genetic diversity
As shown earlier, population structure analysis for E. chienense and E. cf. oophylax both resulted in K = 1 from the STRUCTURE and DAPC analyses, and both tested as not significant for IBD. Therefore, heterozygosity and inbreeding coefficients and Tajima’s D were estimated for all of the collection localities together, not on a site to site basis. Expected and observed heterozygosity among all individuals analyzed for E. chienense is He = 0.0702 and Ho = 0.0625, respectively (Table 5). Expected and observed heterozygosity among all individuals analyzed for E. cf. oophylax (East Fork) is He = 0.0646 and Ho = 0.0547. Inbreeding coefficient for E. chienense throughout its range and E. cf. oophylax throughout the sampled localities of the East Fork Clarks River is FIS = 0.1089 and FIS = 0.1378, respectively (Table 5). Tajimas D tested as insignificant (p > 0.1) for all loci for E. chienense, and all but 7 of 9,876 loci for E. cf. oophylax.
Effective population size
Results of the effective population size estimates using NE Estimator were different for each dataset but confidence ranges overlapped. The estimated effective population sizes of E. chienense range from 221.5 to 337.9, depending on the lowest allele frequency threshold incorporated in the analysis (Table 6). 95% parametric confidence intervals range from 143.4 to 918.6. As for E. cf. oophylax from the East Fork, the estimated effective population size is between 19.6 and 244.6 with 95% non-parametric confidence of 9.2–517.3 (Table 6). Effective population size estimation was not carried out for E. cf. oophylax from both forks of the Clarks River due to low sample size and a likely biased estimation.
Discussion
This study incorporated data from both the mitochondrial and nuclear genomes to assess genetic diversity and population structure of E. chienense, a federally protected species, endemic to the Bayou du Chien system of western Kentucky. The results from this study demonstrate that E. chienense can be managed as a single, panmictic conservation unit. While the initial goal of this study was to use E. cf. oophylax as a baseline for comparison, it was discovered that the population level dynamics of this taxon are much more complex than expected. Given that only a small portion of its range was sampled in hopes to reproduce the geographic distances between sites of the E. chienense collections, it appears that much of the genetic diversity of this species may exist in other portions of the range that were not sampled heavily, or at all. Therefore, it is concluded that more proficient sampling throughout the range of E. cf. oophylax is needed to appropriately assess this species, but the results from the analysis of E. chienense can still be beneficial to conservation officials. With knowledge of the genetic diversity, population structure, gene flow and effective population size, conservation officials can make more informed decisions regarding the management and protection of E. chienense.
Genetic structure of E. chienense
Based on multiple analyses, the number of estimated genetic clusters within E. chienense is estimated to be one. The Bayou du Chien is a small stream system and one might expect that a species inhabiting a small stream would be panmictic. A multitude of other studies, however, suggest that life-history features, the amount of connectivity, and habitat preferences can play substantial roles in the degree of population genetic structure and gene flow of aquatic species (Thomaz et al. 2016; Pilger et al. 2017; Camak and Piller 2018). Based on data from closely related congeners (Evans and Page 2003; Douglas et al 2013), it is assumed that E. chienense has minimal dispersal capabilities given its small size (< 80 mm SL) and the lack of a swimbladder. Therefore, it is somewhat surprising that there are no genetic clusters in the Bayou du Chien.
For many species inhabiting lotic environments, larval drift plays a major role in dispersal. A study conducted by Douglas et al (2013) showed that there is significant variation in larval drift phases of darters. They included eleven species of Etheostoma and five of these were shown to exhibit pelagic larval drift for at least 28 days. Three species in their study, E. percnurum, E. marmorpinnum, E. sitikuense, are closely related to E. chienense, all members of the subgenus Catonotus, and occupy similar types of low-gradient streams, and possess similar life-history characteristics. None of these species possess a pelagic larval drift phase. While E. chienense was not included in their study, it is possible that pelagic larval drift could be a contributing factor to the panmixia of E. chienense, but it cannot be known for sure until more comprehensive life history data is collected for this species. Alternatively, in terms of flooding event intensity and frequency, the Bayou du Chien is an unpredictable system that is subjected to increased flow events following periods of heavy rainfall. The intensity of these types of events are exacerbated in areas that have been channelized (Piller and Burr 1998). These types of stochastic events could facilitate downstream dispersal of larvae, juveniles, and adults.
Genetic structure of E. cf. oophylax
Samples of E. cf. oophylax were obtained from the upper reaches of the East Fork of the Clarks River in an effort to mimic the sampling design used for E. chienense in the Bayou du Chien system and allow for direct comparisons. Unfortunately, the sampling design incorporated limits our ability to discuss the complex nature of genetic diversity and isolation in this taxon. The Clarks River is divided into two relatively large forks (Fig. 1). When individuals from both forks are included in the analyses, E. cf. oophylax shows weak population structure in the STRUCTURE analysis but strong structure in the DAPC analysis between individuals of these two forks (K = 2). There are a few plausible explanations as to why this type of genetic structure was recovered. First, the two forks could each harbor their own genetic clusters, suggesting that dispersal from one fork to the other is less common than dispersal within each fork. In other words, the split of the two forks could be acting as a barrier to migration. This type of structure has been shown in other freshwater organisms (Perkin and Gido 2012; Huey et al 2017), and appears to be the leading cause for this population structure.
A second hypothesis is isolation by distance. It is unlikely that IBD is the mechanism underlying the observed diversity between forks, given that the Clarks river in its entirety is only about 107 km in length. However, IBD should not be ruled out until it is tested with appropriate sampling throughout the range of this species.
Next, since a smaller number of individuals were included in this analysis, including six from the West Fork and 29 from the East Fork of the Clarks River, the filtering approach employed in the study resulted in the inclusion of slightly more loci (i.e. 10,709 loci) than the other analyses. As a result, the statistical power to detect fine-scale genetic structure increases dramatically as more loci are analyzed (Rafiei et al. 2018). In this case, detection of two clusters may simply be an artifact of including more loci rather than the existence of fine-scale structure. Additional sampling throughout both forks of the Clarks River would be needed to address this issue.
Finally, it should be mentioned that during the collection of the E. cf. oophylax from the East Fork, there were several collection sites that were nearly dried up during the fall sampling period, leaving only isolated pools where darters were collected. It is assumed that these pools reconnect seasonally throughout the spring and summer months when more rainfall is present. This type of seasonal environmental pressure can cause serious fluctuations regarding the population dynamics of a species (Faulks et al. 2010). While there was no significant genetic variation between or among sites of the East Fork, more data regarding the seasonal fluctuations of isolated pools in the Clarks River as well as more complete sampling of the species range is needed to make claims regarding the potential for these pools to drive the fixation of alleles in subpopulations.
Genetic diversity
Etheostoma chienense is endangered, therefore it is not unexpected that extremely low levels of genetic diversity were observed in this species. For the cytochrome b haplotype data, only three haplotypes were recovered and when coupled with the extremely low heterozygosity observed from the SNP data, this suggests that this species may be a victim of genetic drift or inbreeding depression. When E. chienense was first discovered as a unique lineage, census population estimates were dangerously low (Page et al. 1992; Piller and Burr 1998). It is entirely possible that this species was bottlenecked for many years as a result of the expansion of agricultural land in the basin, and the concomitant reduction of the riparian zone and increased siltation in the Bayou du Chien system. After the listing of this species as endangered, populations of E. chienense may have stabilized as a result of a greater degree of protection in the basin. However, the dangerous effects of a population bottleneck and subsequent genetic drift may have already been rooted within the imperiled species (Nei et al. 1975), leading to low levels of genetic diversity across the range.
Etheostoma cf. oophylax from the East Fork of the Clarks River displayed nine mitochondrial haplotypes from 168 individuals and a dangerously low observed heterozygosity level from the SNP data. However, it is highly unlikely that these metrics accurately portray the genetic diversity within E. cf. oophylax given the samples were only collected from a small portion of the range. Because the pairwise fixation index was four times higher between East and West Forks (FST = 0.194) of the Clarks River than between any pair of East Fork sites, it is evident that there is genetic diversity throughout the range of this species, but not in the sampled localities of the East Fork. This phenomenon of low observed heterozygosity could be the result of a Wahlund effect caused by sampling a subpopulation within a larger metapopulation (Wahlund 1928; Waples 2014; Zhivotovsky 2015). The inclusion of additional samples throughout the entire range of E. cf. oophylax may provide additional support and evidence that the Clarks River population has higher heterozygosities and larger effective population sizes than currently estimated.
There is some disagreement with the mitochondrial data and the SNP data in the test for neutrality with E. cf. oophylax. A star shaped haplotype network containing multiple rare alleles and one common allele dominating the population, coupled with a significant Tajima’s D test and Fu F test can be indicative of a recent population expansion (Liu et al. 2006; van der Heyden 2007). However, the SNP data had only 7 loci -of 9876- that had significant Tajima’s D values. Only a handful of SNPs agreed with the results from the mitochondrial DNA test with the vast majority of tests being non-significant. While it is possible that the female population has undergone a recent population expansion, we believe it is more likely that a single mitochondrial gene, cytochrome b, may not be representative of the population history of the E. cf. oophylax.
Effective population size
The estimated effective population size of E. chienense is critically low with the most conservative upper and lower bounds between 143 and 918 individuals. This is substantially lower than the census size estimates of Piller and Burr (1998), which were estimated between 9553 and 31,293 individuals across the Bayou du Chien system. Differences between census and effective population size estimates are not an uncommon occurrence among vertebrate populations, with effective population size estimates often being orders of magnitude lower than census size estimates (Turner et al. 2002; Belmar‐Lucero et al. 2012). Low effective population size correlates directly with the loss of rare alleles and can contribute to genetic drift (Berthier et al. 2002), which likely has occurred within this species given the low heterozygosity and absence of genetic structure across the range.
The idea of minimum viable population size (MVP) was proposed by Shaffer (1981) and suggests that there are minimum population thresholds that should be maintained in wild populations to sustain species in both the short and long term. Some argue that a minimum effective population size of 50 is needed to avoid inbreeding depression and avoid extinction of a species, whereas others suggested that a value of 500 is required to retain evolutionary potential and to minimize the influence of genetic drift. This idea is known as the 50/500 rule (Soule 1980; Franklin 1980) and is not without debate (Jamieson and Allendorf 2012; Frankham et al 2014; Franklin et al. 2014; Rosenfeld 2014). Regardless of what value or minimum threshold is required, the relatively low effective population size values recovered in this study suggest that the long-term persistence of the Relict Darter may be in jeopardy.
Effective population size estimates of E. cf. oophylax also are also dangerously low with the most conservative lower and upper bounds of 19 and 244 individuals. No published information is known regarding census sizes of this taxon; however, it is also likely that the effective population estimate for E. cf. oophylax is substantially lower than its census size. In addition, it is likely that the effective population size of E. cf. oophylax is severely underestimated here due to the sampling scheme, which is based on a small portion of the known range of this taxon. The primary reason for an underestimated effective population size is likely due to the Wahlund effect (Wahlund 1928). Effective population size can be severely underestimated if only a subpopulation of a metapopulation is sampled. Throughout the collection of the samples for this study, E. cf. oophylax was very common at almost every locality, suggesting that the population size is large, and that the estimated effective population size is underestimated due to sampling design.
Conclusions
This study represents the first comprehensive population genetic review of the endangered Relict Darter with samples collected throughout the range. Genome wide SNP data coupled with a single mitochondrial marker provide reliable results regarding the status of this species. Etheostoma chienense is a federally endangered species and the results of this study indicate very little genetic variation throughout its range and dangerously low effective population size estimates. The effects of a population bottleneck and subsequent genetic drift appear to be intimately associated with this species. Genetic clustering analyses suggest there is only one panmictic population and that the number of management units (MU’s) should be one.
These results suggest that a conservation and management plan needs to be implemented in the Bayou du Chien basin. A greater degree of habitat protection and stream restoration, including the re-development of riparian zones in agriculturally impacted areas, could stabilize stream banks and reduce siltation in heavily impacted areas. Furthermore, E. chienense is an egg-clustering species that attaches its eggs to the undersides of hard substrates. Prior to human modification to the Bayou du Chien system, E. chienense likely utilized natural woody materials as spawning substrates. The elimination of riparian zones throughout large portions of the Bayou du Chien system have reduced the amount of available allochthonous woody material and therefore reduced the availability of spawning substrates in the system. As a result, E. chienense has become opportunistic in its choice of spawning substrates and will readily attach eggs to the undersides of any firm spawning substrate in the system including tires and roofing shingles, in addition to woody debris (Piller and Burr 1999). As suggested by Piller and Burr (1999), the addition of artificial spawning substrates during the spring reproductive season could increase reproductive output and lead to a greater degree of recruitment into the natural population, which in turn, could increase population sizes of E. chienense throughout the Bayou du Chien system. This approach could be particularly effective if this was conducted over several successive years.
This study cannot provide reliable population genetic conclusions regarding E. cf. oophylax due to the sampling design employed. However, it is clear that there is some underlying genetic structure between forks of the Clarks River that will require more research to confidently quantify genetic variation throughout this species. Low effective population size estimates are likely due to the sampling design as well. Therefore, until a more inclusive sampling of this species is performed, the reliability of the effective population size estimates of E. cf. oophylax should be considered tentative.
Overall, this study highlights the effectiveness of SNP data to quantify many population level metrics of endangered species including gene flow and population structure, overall genetic variation, and effective population size. While SNP data is very effective at quantifying these population dynamic metrics, effective sampling across a species’ range is still necessary when evaluating the genetic health of a species. The results reported here can be used to assist management officials in providing additional support to the endangered Relict Darter.
References
Belmar-Lucero S, Wood JL, Scott S, Harbicht AB, Hutchings JA, Fraser DJ (2012) Concurrent habitat and life history influences on effective/census population size ratios in stream-dwelling trout. Ecol Evol 2:562–573
Berthier P, Beaumont MA, Cornuet JM, Luikart G, Anderson EC, Williamson EG, Wright S (2002) Likelihood-based estimation of the effective population size using temporal changes in allele frequencies: a genealogical approach. Genetics 160:741–751
Camak DT, Piller KR (2018) Going with the flow: testing the role of habitat isolation among three ecologically divergent darter species. Copeia 106:375–387
Chhatre VE, Emerson KJ (2017) StrAuto: automation and parallelization of STRUCTURE analysis. BMC Bioinform 18:192
Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, McVean G (2011) The variant call format and VCFtools. Bioinformatics 27:2156–2158
Do C, Waples RS, Peel D, Macbeth GM, Tillett BJ, Ovenden JR (2014) NeEstimator v2: re-implementation of software for the estimation of contemporary effective population size (Ne) from genetic data. Mol Ecol Resour 14:209–214
Douglas M, Keck BP, Ruble C, Petty M, Shute JR, Rakes P, Hulsey CD (2013) Pelagic larval duration predicts extinction risk in a freshwater fish clade. Biol Lett 9:20130672
Earl DA (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361
Eaton DA, Overcast I (2020) ipyrad: interactive assembly and analysis of RADseq datasets. Bioinformatics 36:2592–2594
Eliades NGH, Gailing O, Leinemann L, Fady B, Finkeldey R (2011) High genetic diversity and significant population structure in Cedrus brevifolia Henry, a narrow endemic Mediterranean tree from Cyprus. Plant Syst Evol 294:185–198
Evans JD, Page LM (2003) Distribution and relative size of the swim bladder in Percina, with comparisons to Etheostoma, Crystallaria, and Ammocrypta (Teleostei: Percidae). Environ Biol Fishes 66:61–65
Faulks LK, Gilligan DM, Beheregaray LB (2010) Islands of water in a sea of dryland: hydrological regime predicts genetic diversity and dispersal in a widespread fish from Australia’s arid zone, the Golden Perch (Macquaria ambigua). Mol Ecol 19:4723–4737
Floyd M (2018) Five year review: Relict Darter (Etheostoma chienense). Kentucky Ecological Services Field, Frankfurt Kentucky
Frankham R (1996) Relationship of genetic variation to population size in wildlife. Conserv Biol 10:1500–1508
Frankham R (2010) Challenges and opportunities of genetic approaches to biological conservation. Biol Conserv 143:1919–1927
Frankham R, Bradshaw CJA, Barry BW (2014) Genetics in conservation management: revised recommendations for the 50/500 rules, Red List criteria and population viability analyses. Biol Conserv 170:56–63
Frankham R (2015) Genetic rescue of small inbred populations: meta-analysis reveals large and consistent benefits of gene flow. Mol Ecol 24:2610–2618
Franklin R (1980) Evolutionary change in small populations. In: Soule ME, Wilcox BA (eds) Conservation biology: an evolutionary-ecological perspective. Sinauer Associates, Sunderland, pp 135–140
Franklin IR, Allendorf FW, Jamieson IG (2014) The 50/500 rule is still valid-reply to Frankham et al. Biol Conserv 176:284–285
Gido KB, Whitney JE, Perkin JS, Turner TF (2016) Fragmentation, connectivity and fish species persistence in freshwater ecosystems. In: Krkosek M, Olden JD, Closs GP (eds) Conservation of Freshwater Fishes. Cambridge University Press, Cambridge, pp 292–323
Goudet J (2005) Hierfstat, a package for R to compute and test hierarchical F-statistics. Mol Ecol Notes 5(1):184–186
Harrington RC, Benavides E, Near TJ (2013) Phylogenetic inference of nuptial trait evolution in the context of asymmetrical introgression in North American darters (Teleostei). Evolution 67:388–402
Harrington RC, Near TJ (2015) Phylogenetic relationships of Goneaperca and the evolution of parental care in darters (Teleostei:Percidae). Mol Phylogenet Evol 84:158–165
Huey JA, Balcombe SR, Real KM, Sternberg D, Hughes JM (2017) Genetic structure and effective population size of the most northern population of the Australian River Blackfish, Gadopsis marmoratus (Richardson 1848): implications for long-term population viability. Freshw Sci 36:113–123
Jakobsson M, Rosenberg NA (2007) CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23:1801–1806
Jamieson IG, Allendorf FW (2012) How does the 50/500 rule apply to MVPs? Trends Ecol Evol 27:578–584
Jombart T, Ahmed I (2011) adegenet 1.3-1: new tools for the analysis of genome-wide SNP data. Bioinformatics 27:3070–3071
Jombart T, Devillard S, Balloux F (2010) Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet 11:94
Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452
Liu JX, Gao TX, Zhuang ZM, Jin XS, Yokogawa K, Zhang YP (2006) Late Pleistocene divergence and subsequent population expansion of two closely related fish species, Japanese anchovy (Engraulis japonicus) and Australian Anchovy (Engraulis australis). Mol Phylogenet Evol 40:712–723
Mee JA, Bernatchez L, Reist JD, Rogers SM, Taylor EB (2015) Identifying designatable units for intraspecific conservation prioritization: a hierarchical approach applied to the Lake Whitefish species complex (Coregonus spp.). Evol Appl 8:423–441
Near TJ, Bossu CM, Bradburd GS, Carlson RL, Harrington RC, Hollingsworth PR Jr, Keck BP, Etnier DA (2011) Phylogeny and temporal diversification of darters (Percidae: Etheostomatinae). Syst Biol 60:565–595
Nei M, Maruyama T, Chakraborty R (1975) The bottleneck effect and genetic variability in populations. Evolution 29:1–10
Page LM (1983) Handbook of Darters. TFH Publications, Neptune City
Page LM, Ceas PA, Swofford DL, Buth DG (1992) Evolutionary relationships within the Etheostoma squamiceps complex (Percidae; subgenus Catonotus) with descriptions of five new species. Copeia 1992:615–646
Paradis E, Claude J, Strimmer K (2004) APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20(2):289–290
Perkin JS, Gido KB (2012) Fragmentation alters stream fish community structure in dendritic ecological networks. Ecol Appl 22:2176–2187
Peterson BK, Weber JN, Kay EH, Fisher HS, Hoekstra HE (2012) Double digest RADseq: an inexpensive method for de novo SNP discovery and genotyping in model and non-model species. PLoS ONE 7:e37135
Pilger TJ, Gido KB, Propst DL, Whitney JE, Turner TF (2017) River network architecture, genetic effective size and distributional patterns predict differences in genetic structure across species in a dryland stream fish community. Mol Ecol 26:2687–2697
Piller KR, Burr BM (1998) Distribution and population estimates of the federally endangered Relict Darter, Etheostoma chienense, Bayou du Chien, Kentucky. J Kentucky Acad Sci 59:64–75
Piller KR, Burr BM (1999) Reproductive biology and spawning habitat supplementation of the relict darter, Etheostoma chienense, a federally endangered species. Environ Biol Fishes 55:145–155
Piller KR, Wilson CC, Lee CE, Lyons J (2005) Conservation genetics of inland Lake Trout in the upper Mississippi River basin: stocked or native ancestry? Trans Am Fish Soc 134:789–802
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Rafiei V, Banihashemi J-Díaz RM, Navas-Cortés JA, Landa BB, Jiménez-Gasco MM, Turgeon BG, Milgroom MG (2018) Comparison of genotyping by sequencing and microsatellite markers for unravelling population structure in the clonal fungus Verticillium dahliae. Plant Pathol 67:76–86
Rosenberg NA (2004) DISTRUCT: a program for the graphical display of population structure. Mol Ecol Notes 4:137–138
Rosenfeld JS (2014) 50/500 or 100/1000? Reconciling short- and long-term recovery targets and MVPs. Biol Conserv 176:287–288
Russello MA, Waterhouse MD, Etter PD, Johnson EA (2015) From promise to practice: pairing non-invasive sampling with genomics in conservation. PeerJ 3:1106
Schmidt TR, Gold JR (1993) Complete sequence of the mitochondrial cytochrome b gene in the cherryfin shiner, Lythrurus roseipinnis (Teleostei: Cyprinidae). Copeia 1993 (3):880
Scribner KT, Lowe WH, Landguth E, Luikart G, Infante DM, Whelan GE, Muhlfeld CC (2016) Applications of genetic data to improve management and conservation of river fishes and their habitats. Fisheries 41:174–188
Shaffer ML (1981) Minimum population sizes for species conservation. Bioscience 31:131–134
Soule ME (1980) Thresholds for survival: maintaining fitness and evolutionary potential. In: Soule ME, Wilcox BA (eds) Conservation biology: an evolutionary-ecological perspective. Sinauer Associates, Sunderland, pp 151–169
Thomaz AT, Christie MR, Knowles LL (2016) The architecture of river networks can drive the evolutionary dynamics of aquatic populations. Evolution 70:731–739
Turner TF, Wares JP, Gold JR (2002) Genetic effective size is three orders of magnitude smaller than adult census size in an abundant, estuarine-dependent marine fish (Sciaenops ocellatus). Genetics 162:1329–1339
Von der Heyden S, Lipinski MR, Matthee CA (2007) Mitochondrial DNA analyses of the Cape Hakes reveal an expanding, panmictic population for Merluccius capensis and population structuring for mature fish in Merluccius paradoxus. Mol Phylogenet Evol 42:517–527
Vrijenhoek RC (2005) Conservation genetics of freshwater fish. J Fish Biol 53(Suppl A):394–412
Wahlund S (1928) Zusammensetzung von Populationen und Korrelationserscheinungen vom Standpunkt der Vererbungslehre aus betrachtet. Hereditas 11:65–106
Waples RS, Do C (2010) Linkage disequilibrium estimates of contemporary Ne using highly variable genetic markers: a largely untapped resource for applied conservation and evolution. Evol Appl 3:244–262
Waples RS (2014) Testing for Hardy-Weinberg proportions: have we lost the plot? J Hered 106:1–19
Warren ML, Burr BM, Taylor CA (1994) The Relict Darter, Etheostoma chienense (Percidae): status review of a Kentucky endemic. Trans Kentucky Acad Sci 55:20–27
Warren ML Jr, Burr BM, Walsh SJ, Bart HL Jr, Cashner RC, Etnier DA, Freeman BJ, Kuhajda BR, Mayden RL, Robison HW, Ross ST (2000) Diversity, distribution, and conservation status of the native freshwater fishes of the southern United States. Fisheries 25:7–31
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38 (6):1358
Westemeier RL, Brawn JD, Simpson SA, Esker TL, Jansen RW, Walk JW, Kershner EL, Bouzat JL, Paige KN (1998) Tracking the long-term decline and recovery of an isolated population. Science 282:1695–1698
Zhivotovsky LA (2015) Relationships between Wright’s FST and FIS statistics in a context of Wahlund effect. J Hered 106:306–309
Acknowledgements
We would like to thank Anna Gruszkiewicz, Aaron Krolow, and Arely Ramirez for assistance in the field. We would also like to thank Mike Floyd, Mike Compton, and Matt Thomas for providing tissue samples from several Relict Darter localities. Members of the Near Lab at Yale University, Ava Ghezelayagh, Daemin Kim, Dan MacGuigan, Tom Near, & Rich Harrington provided ddRAD library prep training and assistance with the ipyrad assembly of raw Illumina data. I would also like to thank David Camak for his guidance with analysis of this data. Funding was provided through a grant to KRP from the Kentucky Department of Fish and Wildlife Resources via the Cooperative Endangered Species Conservation Fund (Section 6 of the Endangered Species Act). Additional logistical support was provided by the U.S. Fish and Wildlife Service, Kentucky Ecological Services Field Office. Portions of this research were conducted with high performance computational resources provided by the Louisiana Optical Network Infrastructure.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Kattawar, J., Piller, K.R. Comparative population genetics of the federally endangered Relict Darter, and its sister taxon the Clarks Darter (Teleostei: Percidae). Conserv Genet 21, 957–970 (2020). https://doi.org/10.1007/s10592-020-01300-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-020-01300-7