Abstract
Candy Darters (Etheostoma osburni) and Variegate Darters (E. variatum) are both native to West Virginia and Virginia. The geographic ranges of these two species were historically separated by Kanawha Falls, a natural barrier to fish dispersal located at Glen Ferris, WV. In the early 1980s, Variegate Darters or putative hybrids (E. osburni × E. variatum) were first collected at locations upstream of Kanawha Falls, and have since undergone range expansion. Hybridization with the Variegate Darter was one of the threats that led to the Candy Darter being listed as Endangered under the U.S. Endangered Species Act in 2018. Genetic and morphologic data were examined for individuals from the New, Gauley, and Greenbrier river drainages. Individuals were genotyped using a suite of 5 diagnostic microsatellite loci to investigate potential hybridization. Widespread hybridization was found throughout populations of Candy Darters, with the geographic range of hybridization expanding from 2004 to 2014. A hybrid zone was observed, with the highest levels of Variegate Darter introgression representing the kernel within this zone and the locations of first-generation (F1) hybrids at the periphery. F1 hybrids were morphologically intermediate within and across characters for parental species. Introgressive hybridization threatens the genetic integrity of the Candy Darter, and may lead to population extirpation or extinction.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Interspecific hybridization can lead to population extirpation or species extinction (Rhymer and Simberloff 1996), and has been documented widely within a variety of fish families (Morizot et al. 1991; Seehausen et al. 1997; Taylor et al. 2006; Muhlfeld et al. 2009; Almodóvar et al. 2012; Halas and Simons 2014; Matthews et al. 2016). Of particular conservation concern is introgressive hybridization between a native and a non-native species. Specifically, asymmetric introgressive hybridization or genetic swamping imperils a rare native species. A recent example of this phenomenon involves hybridization between the narrow endemic Candy Darter (Etheostoma osburni; Hubbs and Trautman 1932) of the lower New River drainage, USA, and the widespread Variegate Darter (E. variatum; Kirtland 1840), a non-native close relative.
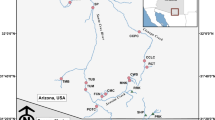
The geographic ranges of Candy and Variegate darters were historically separated by Kanawha Falls, a natural barrier to fish dispersal located at Glen Ferris, WV (Hocutt and Wiley 1986; Stauffer et al. 1995; Page and Burr 2011). The Candy Darter is a narrow endemic of the New River drainage upstream of Kanawha Falls. Its current range includes the Gauley and Greenbrier river drainages of West Virginia and the New River drainage within Virginia (Jenkins and Burkhead 1994; Stauffer et al. 1995). Within West Virginia, historic distribution records have documented this species in the Bluestone River and Indian Creek drainages, where it appears extirpated. The aforementioned streams are components of the New River drainage (Fig. 1). In 2008, due to limited distribution and population reduction, the Candy Darter was listed as vulnerable in West Virginia and Virginia by the American Fisheries Society (Jelks et al. 2008). In 2018, the species was listed as Endangered under the U.S. Endangered Species Act. The Variegate Darter has a wide distribution in the Ohio River drainage, including the Kanawha River drainage below Kanawha Falls and excluding the Wabash and Tennessee River systems; within this range, it occurs in Indiana, Kentucky, New York, Ohio, Pennsylvania, and West Virginia (Hubbs and Black 1940; Stauffer et al. 1995).
Collection sites for materials used in this study. Open circles (towns), lines (dams), and asterisks (waterfalls) represent important landmarks referred to in the text. Major drainages and lakes are labeled. Enumerated site labels correspond with Table 1
The Variegate Darter has been introduced upstream of Kanawha Falls, and now overlaps the range of the Candy Darter. It is currently widespread within the New River drainage of West Virginia downstream of Bluestone dam, the Greenbrier drainage, and the Gauley River drainage downstream of Summersville dam (Wellman 2004; Burns 2007; Cincotta, unpublished data). The introduction of the Variegate Darter may have resulted from one or more bait-bucket releases. Kirtland (1840), in the species description of the Variegate Darter, stated that this darter was “frequently taken by fishermen for bait, and preferred to the common minnows.” The Variegate Darter was first collected upstream of Kanawha Falls by D. Cincotta (West Virginia Division of Natural Resources) in 1982 from the New River at Sandstone Falls (West Virginia Wildlife Resources (WVWR) #437). During 1991 through 1996, the National Park Service collected five specimens (Jesse Purvis, personal communication) from the New River at Sandstone Falls that possessed characters intermediate between Candy and Variegate darters (1991, n = 2; 1995, n = 1; 1996 n = 2; confirmed by Cincotta). In 1993, Cincotta (personal observation) collected one specimen identified as a Variegate Darter from Anthony Creek (a tributary to the Greenbrier River). In 1995 and 1999, Cincotta observed what appeared to be both parental species present in Anthony Creek (WVWR #784). Variegate Darters and/or hybrids were collected in 1997 from the New River at Stone Cliff Bridge (Cincotta et al. 1999) and two years later, a substantial population of Variegate Darters was documented from the Greenbrier River at Ronceverte (Jason Morgan, WV Department of Environmental Protection, personal communication; confirmed by Cincotta; WVWR #1278). Subsequently, in the early 2000s, Wellman (2004) collected Variegate Darters and hybrids from tributaries widely in the New River gorge, but Cincotta collected what appeared to be Candy Darters in the mainstem Gauley River near Swiss within the same time period (i.e., 2002). During 2004 and 2014, Candy Darters, Variegate Darters, and putative hybrids (E. osburni × E. variatum) were collected from the New and Gauley river drainages, and finclips from those specimens were used in the current genetic study.
Based on morphology, hybridization between Variegate and Candy darters was first observed by Cincotta in 1993 from specimens collected in Anthony Creek. Specimens collected prior to then were visually identified as Variegate Darters. The Candy Darter can be morphologically differentiated from the Variegate Darter with higher lateral line scale, dorsolateral saddle, and orange lateral bar counts (or, conversely, dark lateral bar counts); the Candy Darter head generally lacks scales but occasionally has a few scales on the upper portion of the opercle, whereas the Variegate Darter typically possesses scales on the opercle; the Candy Darter breast usually lacks scales whereas the Variegate Darter breast is usually scaled (Hubbs and Trautman 1932; Page 1983; Page and Burr 2011). A genetic analysis using the mitochondrial cytochrome b gene documented hybridization between Variegate and Candy darters in two of 10 specimens collected in 2001 from Glade Creek, a tributary of the New River within the New River gorge, West Virginia (Switzer 2004). Because hybridization and possible introgression could lead to the extirpation or extinction of the Candy Darter, additional genetic research was warranted. In order to better inform management actions under the U.S. Endangered Species Act, it is critical to assess the geographic extent of hybridization between the two species and to identify any remaining pure Candy Darter populations. Therefore, our first objective was to synthesize and map distributional data for Variegate Darter introgression in the Gauley and Greenbrier populations of Candy Darter during two different time-periods (2004 and 2014) using genetic data. Ultimately, it would be useful for managers to identify individuals of hybrid origin based on visual inspection. Therefore, our second objective was to describe morphological characters for the identification of hybrids following genetic confirmation of the selected individuals’ hybrid status.
Materials and methods
Collection and preservation
Specimens were collected using a backpack electrofishing unit and a seine net. A finclip from the right pelvic fin of each specimen from 2004 (n = 148) and 2014 (n = 335) was collected from 11 and 22 sampling locations, respectively (Table 1; Fig. 1). Also, morphological data were obtained from 45 of the 335 specimens from 2014. Fin clips were placed in 99% ethyl alcohol, and then the entire fish was placed in 10% formalin. Specimens then were washed with water and stored in 45% isopropyl alcohol.
Genetic characterization
Switzer et al. (2008) developed a set of 15 polymorphic microsatellite DNA markers for examining genetic variation within populations and assessing potential hybridization between Candy and Variegate darters. Five of these microsatellite loci (Eos-C2, Eos-C3, Eos-C6, Eos-C112, and Eos-C117) were found to be diagnostic for differentiation between the two species. This set of five diagnostic loci was instrumental in detecting introgression and for identifying pure and hybrid individuals.
For samples collected in 2004, total genomic DNA was extracted from each sample using the PUREGENE DNA extraction kit (Gentra Systems, Inc., Minneapolis, MN) and resuspended in TE (10 mM Tris–HCl, pH 8.0, 1 mM EDTA). A total of 148 individuals from 11 sites within the New River drainage were collected (Table 1; Fig. 1). Conditions for microsatellite PCR consisted of 100–300 ng of genomic DNA, 1× PCR buffer, 2 mM MgCl2, 0.2 mM dNTPs, 0.5 µM of each primer (forward primers were fluorescently labeled with FAM, HEX or NED), 1.0 unit of Taq DNA polymerase (Promega) and deionized water to a final volume of 10 µl. The PCR for all loci consisted of an initial denaturation at 94 °C for 2 min; 35 cycles of 94 °C denaturation for 45 s, 58 °C annealing for 45 s, and 72 °C extension for 1 min 30 s; and a 5 min extension at 72 °C. Prior to electrophoresis, 1 µl of PCR product was diluted 1:10 with deionized water and mixed. One µl of the diluted PCR product was added to 12 µl of deionized formamide and 0.5 µl of internal size standard (GENESCAN-500; Applied Biosystems). This mixture was denatured at 95 °C for 3 min, placed on ice for 5 min, and then subjected to capillary electrophoresis on an ABI 3100 automated genetic analyzer. Fluorescent DNA fragments were analyzed, and genotype data were generated using GENESCAN software (Applied Biosystems). Genotyper version 2.0 (Applied Biosystems) DNA fragment analysis software was used to score, bin, and output allelic and genotypic data for each individual. Laboratory processing of these samples occurred at the U.S. Geological Survey Leetown Science Center (WV).
For samples collected in 2014, total genomic DNA was extracted with the Promega SV 96 Genomic DNA Purification System following a modified version of the “Animal Tissues” protocol (Promega Technical Bulletin Part #TB303). Approximately 4 µg (± 2 µg; or approximately 1 mm × 2 mm) of pelvic fin tissue per specimen was prepared in each well of a 96-well plate for a 16-h digestion at 55 °C. The elution process consisted of two 50-µl elutions (for a total elution volume of 100 µl) and was performed with a combination of vacuum and centrifugation. Concentrations of extracted DNA were quantified with a NanoDrop Lite spectrophotometer. Conditions of microsatellite PCR consisted of 20 ng of genomic DNA, 0.4× QIAGEN Multiplex PCR Master Mix, and 0.2 µM of each primer (with the forward primer fluorescently labeled) in a 10-µl final reaction volume. A C1000 Touch (BioRad) thermal cycler was used to conduct the PCR reaction that consisted of an initial denaturation at 95 °C for 15 min; 25 cycles of 94 °C denaturation for 30 s, 57 °C annealing for 90 s, and 72 °C extension for 60 s; and a 60 °C final extension for 30 min. Capillary electrophoresis of PCR product (2 µl per reaction) was conducted on a GenomeLab GeXP Genetic Analysis System (Beckman Coulter) with 400-bp size standard (0.5 µl per reaction) and sample loading solution (27.5 µl per reaction). The associated software, GenomeLab GeXP Series Software Suite, was used to visualize analyzed fragments and manually score alleles to generate genotype data for each individual. ALLELOGRAM v2.2 (Manaster 2009) was used to normalize and bin alleles utilizing a positive control common to all capillary electrophoresis runs. Laboratory processing of these samples occurred at West Virginia University.
Error of laboratory practices was evaluated. Ten percent of the tissue samples were selected with a random number generator. Each of these samples was extracted, amplified, visualized, and scored again according to the aforementioned practices. These data were compared to those of the original series to identify inconsistencies between allele scoring. Error rates were summarized as the number of inconsistently scored alleles divided by the total number of alleles. Error of allele scoring was also evaluated by randomly selecting 14% of the original capillary electrophoresis results and having the results cross-read by another researcher.
The potential for null alleles was investigated with MICROCHECKER v2.2.3 (Van Oosterhout et al. 2003). Default settings were used with a maximum expected allele size of 500 bp, a 95% confidence interval, and 1000 randomizations. The software GENEPOP v4.2 (Raymond and Rousset 1995) was used to assess the conformance of genotype frequencies to Hardy–Weinberg equilibrium (HWE) for all sampling sites. A probability test for each locus in the population was conducted to estimate exact P values with the Markov chain method. The following Markov chain parameters were used: Dememorization number = 1000; 100 batches; and 1000 iterations per batch. The significance level was set at α = 0.05. Sequential Bonferroni adjustments were made for multiple tests (Rice 1989). For all analyses, samples from 2004 to 2014 were run separately.
Hybrids were first identified through a Bayesian analysis of the genetic data, using the software STRUCTURE v2.3.4 (Pritchard et al. 2000). Ten independent runs for K = 2 (representing the two species) with 100,000 Markov Chain Monte Carlo (MCMC) iterations and a burn-in period of 100,000 generations were performed. The default settings, which included correlated allele frequencies, assumed admixture, and no prior information, were used. The composition of each population was determined by assigning individuals to populations based on affinity to the cluster representing hybrids. If an individual had a membership coefficient q ≥ 0.994 for the cluster representing Candy Darters or Variegate Darters, then it was assigned as a pure-species individual, respectively; otherwise, it was assigned hybrid status. This membership coefficient was selected because it was found, upon manual inspection of the diagnostic loci, that individuals with lower coefficients (q < 0.994) possessed a mixture of alleles from the two species.
A second approach to refine hybrid classifications also was implemented. NewHybrids v1.1 Beta 3 (Anderson and Thompson 2002) was used to estimate the posterior probability of the classification of genotyped individuals. Classifications were defined as Candy Darter, Variegate Darter, F1 hybrid, and advanced-generation (F2 + or backcross) hybrid. An individual was required to have a posterior probability of classification of P ≥ 0.998 for Candy Darter; otherwise, it was assigned to the classification with the next-highest posterior probability. This value was used because a natural break in the continuity of posterior probabilities occurs at P = 0.998 and because every individual with a probability P < 0.998 for Candy Darter classification was discovered to have at least one Variegate Darter allele upon manual inspection of the genotype. Individuals that assigned to F1 hybrid were manually inspected to verify that they indeed possessed an F1 genotype (i.e., one Candy Darter allele and one Variegate Darter allele at each locus). If the genotype was not indicative of an F1 hybrid, then it was not considered to be an F1 hybrid. While some individuals seemed to be backcrosses with Candy Darter and other individuals seemed to be the result of multigenerational backcrossing with Candy Darters, the small number of diagnostic loci makes it difficult to reliably distinguish the two. Therefore, this group was collectively referred to as advanced-generation hybrids. This method was designed by corroboration with manual inspection of genotypes, follows natural breaks in the continuity of posterior probabilities, and imposes the strictest rules for assignment of pure parental species, F1 hybrids, and advanced-generation hybrids.
Morphology
Morphological data from F1 hybrids were compared with those of pure male Candy Darters and Variegate Darters (Table 2). The F1 specimens represented one male and one female each from the Gauley and Greenbrier river drainages. The Candy Darters (n = 41) were collected from 12 sites in the Gauley and Greenbrier river drainages (Table 1). Data for Variegate Darters are from Page (1983) and Page and Burr (2011) and specimens (n = 30) collected from Dunkard Creek, Monongalia County, West Virginia. Sex was determined by examining genital papillae and supported by male nuptial characters such as thickened fins, enlarged scale margins, and concentrations of pigment. Data for 14 meristic variables were recorded: dorsal-fin spines, dorsal-fin rays, pectoral-fin rays, pelvic-fin rays, pelvic-fin spines, anal-fin rays, anal-fin spines, scales above lateral line, scales below lateral line, scales along lateral line (left), scales along lateral line (right), circumpeduncle scales, dark lateral bars, and dorsolateral saddles. Presence-absence data were recorded for four variables: breast squamation (embedded), breast squamation (exposed), opercle squamation, and supraopercular squamation. Supraopercular squamation may occur in the dorsolateral region of the head above the opercle (Fig. 2). The minimum, maximum, and modal values for meristic variables are reported.
Results
Genetic characterization
The MICROCHECKER analysis found no evidence for scoring error due to stutter, no evidence for large allele dropout, and no evidence of null alleles at any of the loci in the Gauley and Greenbrier populations of pure Candy Darters. In both 2004 and 2014, collections from all sample sites were in HWE.
During 2004, several sites with only pure Candy Darter populations remained in the Gauley and Greenbrier river drainages (Figs. 3a, 4a). The remaining population of pure Candy Darters in the Gauley River drainage was found upstream of Summersville Dam. This included sites 2, 3, and 5. Remaining clusters of pure Candy Darters in the Greenbrier River drainage were found in locations upstream of site 15 (Anthony Creek) at sites 16, 17, and 19. No Variegate Darter alleles were detected at these locations, which indicated site 15 as the upstream extent of hybridization in the Greenbrier River drainage at this time (Table 1; Fig. 5a). In contrast, populations at downstream locations, such as sites 10, 11, 12, and 13, consisted solely of Variegate Darters or hybrids. A small percentage of Candy Darter alleles were detected at these locations, including two advanced-generation hybrids at site 10. There are no historic records of Candy Darters in this region and so these data indicate that Candy Darter alleles likely were transported to these locations by colonizing hybrids. Hybrid individuals were observed at site 15 along with Variegate Darters; no pure Candy Darters were detected. At site 15, one hybrid individual appeared to be an F1 hybrid (p = 0.562), while the remaining individuals appeared to be advanced-generation hybrids (Fig. 4a). The lack of pure Candy Darters at site 15 indicates that genetic swamping by Variegate Darters has occurred here.
STRUCTURE diagram depicting posterior probability of assignment (vertical axis) at K = 2 clusters for all individuals collected in a 2004 and b 2014. Each individual is represented by a single vertical bar that is partitioned into one to two segments. The length of the segments is proportional to the coefficient of membership (q-value) for each species. Dark and light gray represent the Candy and Variegate darter species, respectively. Site numbers are indicated in brackets. The relative position of Summersville Dam is indicated with a dashed line
Posterior probability of assignment (vertical axis) to four classifications: Candy Darter (Etheostoma osburni), Variegate Darter (E. variatum), F1 hybrid (E. osburni × E. variatum), advanced-generation hybrid (F2+, backcross with Candy Darter, backcross with Variegate Darter) for samples collected in a 2004 and b 2014. Site numbers are indicated in brackets. The relative position of Summersville Dam is indicated with a dashed line
Proportions of Variegate Darter (Etheostoma variatum) alleles (open diamonds) per site detected during this study in a 2004 and b 2014. Solid circles indicate the presence of pure Candy Darters (E. osburni) in a sample. Open circles indicate the collection location of first generation (F1) hybrids (E. osburni × E. variatum). Lines (dams) and asterisks (waterfalls) represent important landmarks referred to in the text. The proportion of Variegate Darter alleles within each sample where they were detected is labeled
In 2014, Variegate Darter alleles were detected at all of the sampling locations in the Greenbrier River drainage, including locations upstream of site 15, except for site 19 (Figs. 3b, 4b). Two individuals appeared to be F1 hybrids that were collected from sites 16 and 18 (P = 0.652 and P = 0.660, respectively). Pure Candy Darter individuals were collected at all sites upstream of site 15, but no pure Candy Darter individuals were collected at site 15 or downstream, indicating that genetic swamping has occurred in that portion of the Greenbrier River drainage (Table 1; Fig. 5b). Small percentages of Variegate Darter alleles also were detected at two locations above Summersville Dam at sites 1 and 6 (7.0% and 1.0%, respectively; Table 1). The Gauley River drainage population of Candy Darters upstream of Summersville Dam appeared to be pure, with the exception of this low-frequency occurrence of Variegate Darter alleles. The other sites upstream of Summersville Dam produced samples comprised entirely of pure Candy Darters, including sites 3, 4, and 5. Pure Candy Darter individuals were present at site 7 immediately below Summersville Dam, but hybrids were also present with a low percentage (2.0%) of Variegate Darter alleles. Sites 8 and 9, the other two sites downstream of Summersville Dam, were characterized by high proportions of Variegate Darter alleles and a lack of pure Candy Darters, indicating that genetic swamping has occurred in this lower portion of the Gauley River. Two hybrid individuals in the Greenbrier River drainage, one from site 16 and one from site 18, were genotyped as F1 hybrids (P = 0.652 and P = 0.660; Fig. 4b). Pure Candy and Variegate darters were both present at site 16. Two hybrid individuals from site 8 downstream of Summersville Dam were genotyped as F1 hybrids (P = 0.655 and P = 0.701). Genetic swamping with the effective loss of Candy Darters appears to have occurred in the Gauley River drainage from site 8 downstream and in the Greenbrier River drainage from site 15 downstream.
Morphologic characterization of F1 hybrid
Lateral line scale count and the presence or absence of supratemporal, opercular (collectively referred to as cephalic squamation), and exposed breast squamation allowed for the visual identification of F1 hybrid individuals. Dorsolateral saddle and dark lateral bar counts also were useful for reinforcing hybrid individual identification. One F1 hybrid had Variegate Darter lateral line scale counts of 55 and 54 (left and right, respectively), 4 dorsolateral saddles, and 7 dark lateral bars, but lacked cephalic and exposed breast squamation that would be expected to be present on a Variegate Darter. Three individuals had Candy Darter lateral line scale counts, but possessed cephalic squamation. These individuals also had species character conflicts among dorsolateral saddle and dark lateral bar counts. All F1 hybrids possessed characters of both parental species (Table 2). A photograph of an individual that expresses hybrid characters is shown in Fig. 6. Photographs of Candy and Variegate darters are given in Figs. 7 and 8.
Discussion
We have detected an expansion of the hybridization zone in the span of 10 years, with the replacement of native Candy Darter alleles by introduced Variegate Darter alleles. Only four sites that were surveyed in 2014 appear to contain exclusively pure Candy Darter individuals, three of which were upstream of Summersville Dam in the Gauley River drainage. The other site (Site 19) in the Greenbrier River drainage was adjacent to one site (20) upstream where hybridization was detected. Morphological traits (e.g., lateral line scale count, presence/absence of cephalic squamation, exposed breast squamation) were evaluated that allow for the visual identification of F1 hybrids in the field.
The Variegate Darter has invaded the native range of Candy Darter, resulting in widespread introgression and the loss of native Candy Darter populations. Since at least 1982, the introduced Variegate Darter has expanded its range within the New River drainage below Bluestone Dam. Our results support the inference of an increase in introgressive hybridization and genetic swamping from 2004 to 2014. The collection of F1 hybrids during both time-periods indicates that such hybridization is ongoing. Site 15 (Anthony Creek) was the upstream-most extent of hybridization in the Greenbrier River drainage in 2004, but hybridization was detected at all sites except a small sample at Site 19 upstream of Site 15 in 2014. No hybridization was detected upstream of Summersville Dam in 2004, but a low percentage of Variegate Darter alleles was detected at two locations upstream of Summersville Dam in 2014. The presence of an active hybrid zone was not detected upstream of Summersville Dam, but this drainage should be monitored for the possible development of such a phenomenon (Allendorf et al. 2001). There are also four populations of Candy Darter in Virginia that should be monitored for signs of introgressive hybridization. The hybrid zone could be spreading naturally from a single introduced nucleus or could be facilitated by additional introductions (e.g., bait bucket, aquarium release) of Variegate Darter alleles. Introductions may be occurring repeatedly at the same location or sporadically throughout the drainage. Variegate Darter alleles also may be transported by human movement of hybrid individuals.
It seems more likely that the current state of the hybrid zone is a natural expansion of the zone of contact. Introgressive hybridization is one mechanism that facilitates dispersal of invasive fish species (Hitt et al. 2003; Walters et al. 2008; Ward et al. 2012). Expansion, following introduction and establishment, is characteristic of some hybrid swarms (Kolar and Lodge 2001). Candy Darter alleles were detected in samples from sites 9, 10, and 13, for which no records of Candy Darters exist. The presence of Candy Darter alleles in these samples suggests natural dispersal of hybrid individuals to this area from a former population of Candy Darters. The presence of Variegate Darters and hybrids in the New River drainage, the thoroughness of genetic swamping of Candy Darters by Variegate Darters within the hybrid zone, the presence of hybrids where Candy Darters were previously absent, and the pattern of species’ alleles along a concentration continuum suggest a natural dispersion of hybrids and hybridization in the Greenbrier and lower Gauley river drainages. If dispersion of the hybrids and Variegate Darters was to rely solely on widespread anthropogenic introductions, it may be expected to appear as many concentrated populations of hybrids or Variegate Darters rather than a single, dendritic continuum. This interpretation does not preclude the possibility of past, contemporary, or future Variegate Darter introductions simultaneous to natural dispersion, and does not explain the presence of Variegate Darter alleles in the Gauley River drainage above Summersville Dam, which were likely the product of additional and separate introduction events.
Hybridization has the potential to lead to population extinction, most often due to genetic swamping instead of demographic swamping (Todesco et al. 2016). Small populations can be more susceptible to the threat of introgression due to the limited availability of conspecific mate options (Lawson et al. 2017). Introgression in small populations has been observed to result in increased genetic diversity and effective population size, as observed in the endemic mottled duck (Anas fulvigula maculosa) after introgression with introduced mallard ducks (A. platyrhynchos) (Peters et al. 2014). However, these effects are often observed when introgression rates are low (Roberts et al. 2010). At high rates of introgression, genetic swamping can occur, with small native populations being the most threatened. For example, specialist species often exist at smaller population sizes, making them more vulnerable to genetic swamping. This phenomenon was observed when the obligate estuarine Black Bream (Acanthopagrus butcheri) was replaced by the migratory marine Yellowfin Bream (A. australis) through introgression (Roberts et al. 2010). Introgression is a significant threat for rare endemic species, such as Bartram’s Bass, which is endemic to the Savannah River basin and is threatened by extensive introgression with the introduced nonnative Alabama Bass (Bangs et al. 2018). Population viability can be further compromised when the hybrids have reduced fitness relative to the parental species. Examples of this include hybridization between native Westslope Cutthroat Trout (Oncorhynchus clarki lewisi) and introduced Rainbow Trout (O. mykiss) (Muhlfeld 1999), and the native Roanoke Bass (Ambloplites cavifrons) and introduced Rock Bass (A. rupestris) (Eschenroeder and Roberts 2018).
The fitness consequences of introgression between Candy and Variegate darters are unknown. One possible consequence is outbreeding depression. This would occur when hybrid individuals exhibit lower fitness within the Candy Darter geographic range than the parental species. However, it has been argued that outbreeding depression is uncommon, temporary if it does occur, and predictable (e.g., long separation time, different adaptations) (Frankham et al. 2017). Another potential outcome is heterosis, or hybrid vigor, which is the production of hybrid offspring that exhibit fitness traits that are superior to those of the parental species. Such may be the case only in F1 hybrids, and outbreeding depression may occur in subsequent generations (Muhlfeld et al. 2009; Eschenroeder and Roberts 2018). Given the widespread dispersal of hybrids in the New, Gauley, and Greenbrier river drainages, introgression is likely not resulting in outbreeding depression. Variegate Darters appear to be found in warmer habitats than Candy Darters (Jenkins and Burkhead 1994). Cold stream temperatures may favor Candy Darters and decrease the rate of Variegate Darter introgression; however, it is not a complete barrier to Variegate Darter gene flow, as evidenced by the discovery of Variegate Darter alleles in certain headwater locations sampled in the Greenbrier River drainage. Field data on survival and reproduction would be required to assess the fitness consequences of hybridization between Candy and Variegate darters.
The F1 hybrid phenotype exhibited a range of characters that overlap with both parental species. All of the meristic character ranges of the F1 hybrid overlapped with ranges for the Candy Darter as well as with published ranges for the Variegate Darter (Page 1983; Jenkins and Burkhead 1994). No single character was diagnostic for the identification of a hybrid. Identifying a hybrid relied on identifying contradicting characters that may not be the same in other hybrid individuals. A notable character that aided in the identification of a hybrid was the presence of cephalic squamation. Although Page (1983) lists the opercular squamation as variable in Candy Darters, this study found that it, in conjunction with squamation of the supraopercular region, was useful for distinguishing hybrids. F1 hybrids expressed combinations of meristic and pigmentation characters of both parental species, as well as intermediacy of single characters.
There are limitations to using only microsatellite loci to detect hybridization. It is possible for a specimen to possess nuclear genotypes from one parental species but have mitochondrial haplotypes from another parental species (Wilson and Bernatchez 1998; Vilà et al. 2003). Therefore, the rate of introgression may be underestimated in this study. Another study observed a greater range of introgression with non-neutral single nucleotide polymorphisms (SNPs; Fitzpatrick et al. 2009). Inclusion of these additional genetic markers would likely further refine the detection of introgression between these two species. Additionally, inclusion of mitochondrial DNA can identify sex-biased hybridization and help detect historical introgression.
The detrimental effects of human-induced introductions of an invasive species are apparent in the results of this research. This study found strong evidence that the Candy Darter in the Greenbrier River drainage is threatened with genetic swamping by the Variegate Darter. The influence of the hybridization in this drainage is pervasive. A loss of the Greenbrier population of Candy Darters would result in a substantial gap in the geographic range and genetic diversity of this species. Evidence of Variegate Darter introduction into the Gauley River drainage above Summersville Dam was detected as low concentrations of Variegate Darter alleles; however, the presence of a large-scale hybridization has yet to be observed in the upper drainage. Only a small number of pure Candy Darter populations remain, making these populations a priority for preservation and management.
References
Allendorf FW, Leary RF, Spruell P, Wenburg JK (2001) The problems with hybrids: setting conservation guidelines. Trends Ecol Evol 16:11:613–622
Almodóvar A, Nicola GG, Leal S, Torralva M, Elvira B (2012) Natural hybridization with invasive bleak Alburnus alburnus threatens the survival of Iberian endemic calandino Squalius alburnoides complex and southern Iberian chub Squalius pyrenaicus. Biol Invasions 14.11:2237–2242
Anderson EC, Thompson EA (2002) A model-based method for identifying species hybrids using multilocus genetic data. Genetics 160:1217–1229
Bangs MR, Oswald KJ, Greig TW, Leitner JK, Rankin DM, Quattro JM (2018) Introgressive hybridization and species turnover in reservoirs: a case study involving endemic and invasive basses (Centrarchidae: Micropterus) in southeastern North America. Cons Gen 19:57–69
Burns AD (2007) Comparison of two electrofishing gears (backpack and parallel wires) and abundances of fishes of the upper Greenbrier River drainage. MS Thesis, West Virginia University, Morgantown, WV
Cincotta DA, Chambers DB, Messinger T (1999) Recent changes in the distribution of fish species in the New River basin in West Virginia and Virginia. In: Proceedings of the new river symposium, Apr 15–16, Boone North Carolina, US National Park Service, Glen Jean, WV, pp 98–106
Eschenroeder JC, Roberts JH (2018) What role has hybridization played in the replacement of native Roanoke bass with invasive Rock bass? T Am Fish Soc 147:497–513
Fitzpatrick BM, Johnson JR, Kump DK, Shaffer HB, Smith JJ, Voss SR (2009) Rapid fixation of non-native alleles revealed by genome-wide SNP analysis of hybrid tiger salamanders. BMC Evol Biol 9:1:176
Frankham R, Ballou JD, Ralls K, Eldridge MDB, Dudash MR, Fenster CB, Lacy RC, Sunnucks P (2017) Genetic management of fragmented animal and plant populations. Oxford University Press, New York
Halas D, Simons AM (2014) Cryptic speciation reversal in the Etheostoma zonale (Teleostei: Percidae) species group, with an examination of the effect of recombination and introgression on species tree inference. Mol Phylogenet Evol 70:13–28
Hitt NP, Frissell CA, Muhlfeld CC, Allendorf FW (2003) Spread of hybridization between native westslope cutthroat trout, Oncorhynchus clarki lewisi, and nonnative rainbow trout, Oncorhynchus mykiss. Can J Fish Aquat Sci 60:12:1440–1451
Hocutt CH, Wiley EO (1986) The zoogeography of North American freshwater fishes. Wiley, Hoboken
Hubbs CL, Black JD (1940) Percid fishes related to Poecilichthys variatus, with descriptions of three new forms. Occas Pap Mus of Zool Univ Mich, Number 416
Hubbs CL, Trautman MB (1932) Poecilichthys osburni, a new darter from the upper Kanawha River system in Virginia and West Virginia. Ohio J Sci 32:1:31–38
Jelks HL, Walsh SJ, Burkhead NM, Contreras-Balderas S, Diaz-Pardo E, Hendrickson DA, Lyons J, Mandrak NE, McCormick F, Nelson JS, Platania SP, Porter BA, Renaud CB, Schmitter-Soto JJ, Taylor EB, Warren ML Jr (2008) Conservation status of imperiled North American freshwater and diadromous fishes. Fisheries 33:8:372–407
Jenkins RE, Burkhead NM (1994) Freshwater fishes of Virginia. Am Fish Soc, Bethesda
Kirtland JP (1840) Descriptions of four new species of fishes. Boston J Nat Hist 3:273–277
Kolar CS, Lodge DM (2001) Progress in invasion biology: predicting invaders. Trends Ecol Evol 16:4:199–204
Lawson LP, Fessl B, Vargas FH, Farrington HL, Cunninghame HF, Mueller JC, Nemeth E, Sevilla PC, Petren K (2017) Slow motion extinction: inbreeding, introgression, and loss in the critically endangered mangrove finch (Camarhynchus heliobates). Cons Gen 18:159–170
Manaster CJ (2009) ALLELOGRAM v2.2: a program for normalizing and binning microsatellite genotypes. http://code.google.com/p/allelogram/
Matthews WJ, Turner TF, Osborne MJ (2016) Breakdown of a hybrid swarm between two darters (Percidae), Etheostoma radiosum and Etheostoma spectabile, with loss of one parental species. Copeia 104:873–878
Morizot DC, Calhoun SW, Clepper LL, Schmidt ME (1991) Multispecies hybridization among native and introduced centrarchid basses in central Texas. T Am Fish Soc 120.3:283–289
Muhlfeld CC, Kalinowski ST, McMahon TE, Taper ML, Painter S, Leary RF, Allendorf FW (2009) Hybridization rapidly reduces fitness of a native trout in the wild. Biol Lett 5:328–331
Page LM (1983) Handbook of darters. TFH Publications, Neptune City
Page LM, Burr BM (2011) Peterson field guide to freshwater fishes of North America North of Mexico. Houghton Mifflin Harcourt, Boston
Peters JL, Sonsthagen SA, Lavretsky P, Rezsutek M, Johnson WP, McCracken KG (2014) Interspecific hybridization contributes to high genetic diversity and apparent effective population size in an endemic population of mottled ducks (Anas fulvigula maculosa). Cons Gen 15:509–520
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Raymond M, Rousset F (1995) GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J Hered 86:248–249
Rhymer JM, Simberloff D (1996) Extinction by hybridization and introgression. Annu Rev Ecol Syst 27:83–109
Rice WR (1989) Analyzing tables of statistical tests. Evolution 43:223–225
Roberts DG, Gray CA, West RJ, Ayre DJ (2010) Marine genetic swamping: hybrids replace an obligately estuarine fish. Mol Ecol 19:508–520
Seehausen O, Van Alphen JJ, Witte F (1997) Cichlid fish diversity threatened by eutrophication that curbs sexual selection. Science 277:5333:1808–1811
Stauffer JR, Boltz JM, White LR (1995) Fishes of West Virginia. Proc Acad Natl Sci Phila, Philadelphia
Switzer JF (2004) Molecular systematics and phylogeography of the Etheostoma variatum species group (Actinopterygii: Percidae). PhD dissertation, Saint Louis University, St Louis, MO
Switzer JF, Welsh SA, King TL (2008) Microsatellite DNA primers for the candy darter, Etheostoma osburni and variegate darter, Etheostoma variatum, and cross-species amplification in other darters (Percidae). Mol Ecol Resour 8.2:335–338
Taylor EB, Boughman JW, Groenenboom M, Sniatynski M, Schluter D, Gow JL (2006) Speciation in reverse: morphological and genetic evidence of the collapse of a three-spined stickleback (Gasterosteus aculeatus) species pair. Mol Ecol 15.2:343–355
Todesco M, Pascual MA, Owens GL, Ostevik KL, Moyers BT, Hubner S, Heredia SM, Hahn MA, Caseys C, Bock DG, Rieseberg LH (2016) Hybridization and extinction. Evol App 9:892–908
Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P (2003) Micro-Checker, version 2.2.3. Department of Biological Sciences and Department of Computer Science, University of Hull, Hull
Vilà C, Walker C, Sundqvist AK, Flagstad Ø, Andersone Z, Casulli A, Kojola I, Valdmann H, Halverson J, Ellegren H (2003) Combined use of maternal, paternal and bi-parental genetic markers for the identification of wolf–dog hybrids. Heredity 90 1:17–24
Walters DM, Blum MJ, Rashleigh B, Freeman BJ, Porter BA, Burkhead NM (2008) Red shiner invasion and hybridization with blacktail shiner in the upper Coosa River, USA. Biol Invasions 10.8:1229–1242
Ward JL, Blum MJ, Walters DM, Porter BA, Burkhead N, Freeman B (2012) Discordant introgression in a rapidly expanding hybrid swarm. Evol Appl 5:4:380–392
Wellman DI (2004) Post-flood recovery and distributions of fishes in the New River Gorge National River and the Gauley River National Recreation Area. MS Thesis, West Virginia University, Morgantown, WV
Wilson CC, Bernatchez L (1998) The ghost of hybrids past: fixation of arctic charr (Salvelinus alpinus) mitochondrial DNA in an introgressed population of lake trout (S. namaycush). Mol Ecol 7:1:127–132
Acknowledgements
This manuscript is dedicated to the memory of Isaac Gibson. The legacy of Isaac’s dedication and tireless commitment to the conservation of this special species will endure. The late Tim King deserves special recognition for his participation and guidance in the early stages of this research as well as his contributions to the field of conservation genetics; John Switzer for the collection of preliminary data; Angie Burns, Lara Hedrick, Jim Hedrick, Heather Hildebrand, Tim Hodge, Dustin Kimble, David Okernick, Nathaniel Owens, Ken Sheehan, David Thorne, and David Wellman for assistance with specimen collection; and Barbara Douglas for review of the manuscript. Funding for this research was provided by West Virginia Division of Natural Resources, State Wildlife Grant, the National Institute of Food and Agriculture, U.S. Department of Agriculture, Hatch Project WVA00690, and the West Virginia Agricultural and Forestry Experiment Station. This study was performed under the auspices of West Virginia University IACUC protocol 01-0510. Any use of trade, firm, or product names is for descriptive purposes only and does not imply endorsement by the U.S. Government.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Gibson, I., Welsh, A.B., Welsh, S.A. et al. Genetic swamping and possible species collapse: tracking introgression between the native Candy Darter and introduced Variegate Darter. Conserv Genet 20, 287–298 (2019). https://doi.org/10.1007/s10592-018-1131-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-018-1131-2