Abstract
Understanding population structure is important for guiding ongoing conservation and restoration efforts. The greater sage-grouse (Centrocercus urophasianus) is a species of concern distributed across 1.2 million km2 of western North America. We genotyped 1499 greater sage-grouse from 297 leks across Montana, North Dakota and South Dakota using a 15 locus microsatellite panel, then examined spatial autocorrelation, spatial principal components analysis, and hierarchical Bayesian clustering to identify population structure. Our results show that at distances of up to ~240 km individuals exhibit greater genetic similarity than expected by chance, suggesting that the cumulative effect of short-range dispersal translates to long-range connectivity. We found two levels of hierarchical genetic subpopulation structure. These subpopulations occupy significantly different elevations and are surrounded by divergent vegetative communities with different dominant subspecies of sagebrush, each with its own chemical defense against herbivory. We propose five management groups reflective of genetic subpopulation structure. These genetic groups are largely synonymous with existing priority areas for conservation. On average, 85.8 % of individuals within each conservation priority area assign to a distinct subpopulation. Our results largely support existing management decisions regarding subpopulation boundaries.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The evolution of species and their ecological communities is influenced by the effect of historic physiogeographic features (Carrol et al. 2007; Lomolino et al. 2006; Wiens 2007). Across northwestern North America, the advance and recession of glaciers, formation of mountains, and carving of river valleys have all sculpted modern landscapes into ecologically distinctive areas composed of unique assemblages of soils, vegetation, and wildlife (Carstens et al. 2005; Shafer et al. 2010; Soltis et al. 1997). Many of these landscapes are being rapidly altered by anthropogenic forces (Ricketts 1999), which may affect genetic connectivity within and among wildlife populations (Short Bull et al. 2011).
Big sagebrush (Artemisia tridentata) likely colonized and diversified in North America from Eurasia via Beringia during the late Tertiary or early Quaternary (McArthur and Plummer 1978; Stanton et al. 2002). Big sagebrush is a topographic climax dominant species that provides soil stability and ground cover and functions as critical habitat for at least 350 species of birds, reptiles, and mammals (Allen et al. 1984; Green et al. 2001; Green and Flinders 1960; Monsen and Shaw 2000; Wambolt 1996). However, the geographic extent of sagebrush and the plant and animal communities it supports has been drastically reduced and fragmented by anthropogenic disturbances including cultivation, energy development, invasive species, wildfire, and exurban development (Braun 1998; Braun et al. 2002; Copeland et al. 2009; Knick et al. 2003; Murphy et al. 2013; Naugle et al. 2004, 2006, 2011). The combined effect of sagebrush fragmentation and loss poses a major threat to the greater sage-grouse (Centrocercus urophasianus) a sentinel species for sagebrush ecosystem integrity (Smits and Fernie 2013).
Greater sage-grouse, henceforth sage grouse, is a species of conservation concern, an icon of sage-steppe ecotypes, an umbrella species for shrub-grassland communities (Rowland et al. 2006), and an indicator species for landscape scale connectivity (Aldridge et al. 2008). Sage grouse are sagebrush obligates—relying on sagebrush for every aspect of their life history: food (Thacker et al. 2012; Doherty et al. 2008; Wallestad and Eng 1975), nesting (Holloran and Anderson 2005), brood rearing (Hagen et al. 2007), and spring breeding congregations known as leks (Connelly et al. 2000; Wallestad and Schladweiler 1974). Males battle with one another to claim the center of the lek and energetically display to potential mates. Females appear to select their mate based on phenotypic traits (Gibson et al. 1991); this mate selection process can occur many times with multiple mates during a single breeding season (Semple et al. 2001). Most females nest within 5 km of the lek (Holloran and Anderson 2005). Lek attendance by males is significantly correlated with female lek attendance (Bradbury et al. 1989) and despite long seasonal migratory movements (up to 240 km; Smith 2012) and large home ranges (4–195 km2; Connelly et al. 2011a, b), fidelity to leks and stability in lek location is well documented (Dalke et al. 1963; Dunn and Braun 1985; Emmons and Braun 1984; Patterson 1952; Wallestad and Schladweiler 1974). However, sage grouse may shift or abandon leks because of persistent disturbance or alteration of sagebrush cover (Holloran et al. 2010; Walker et al. 2007).
Sage grouse once occupied over 1.2 million km2 (Edminster 1954; Schroeder et al. 2004). The species now occupies less than 0.67 million km2 across 11 western states and two Canadian provinces (Patterson 1952; Schroeder et al. 2004)—56 % of its range compared to pre-western settlement (Schroeder et al. 2004). An additional 29 % of the remaining species’ range is likely at risk of extirpation (Aldridge et al. 2008). Increased geographic isolation and declines of sage grouse populations range-wide coincides with fragmentation and loss of sagebrush (Copeland et al. 2009; Schrag et al. 2011). Due to loss of habitat and subsequent population declines, the U.S. Fish and Wildlife Service was petitioned to consider listing the species under the Endangered Species Act in 2010. The species was found warranted for listing but precluded by higher priority actions (U.S. Fish and Wildlife Service 2010); however, as a condition of a court approved settlement agreement, a status review was required. In September 2015, the USFWS determined the species is not warranted for listing, due to the species’ relative abundance (which increased since 2010), widespread distribution, and reduced extinction threat (U.S. Fish and Wildlife Service 2015).
In light of the USFWS listing decisions, state and federal agencies have drafted comprehensive conservation planning strategies. Using the current large-scale understanding of population structure, expert opinion, and published research, management agencies in all western states have collaborated to draft management plans that identify and protect the areas deemed most important for sage grouse survival and reproduction (U.S. Fish and Wildlife Service 2013). As part of their planning strategy, the Western Association of Fish and Wildlife Agencies (WAFWA), whose membership is composed of twenty-three states, has delineated seven Management Zones (MZs; Stiver et al. 2006) by grouping sage grouse populations and subpopulations which occur within common floristic provinces identified by Connelly et al. (2004). In addition, Montana Fish, Wildlife & Parks (MTFWP), North Dakota Game and Fish (NDGF) and South Dakota Game, Fish & Parks (SDGFP) have collectively delineated 20 Priority Areas for Conservation (PACs) to protect the highest densities of sage grouse based on male lek attendance.
The goal of these PACs is to protect important lek complexes and to conserve associated habitat (Montana Fish, Wildlife & Parks 2014). On public lands, the BLM and USFS completed their largest planning effort in history to implement regulatory mechanisms that safeguard sage-steppe habitats (U.S. Fish and Wildlife Service 2015). Since 2010 the NRCS-led Sage Grouse Initiative (SGI) has reduced fragmentation of large and intact sage-steppe habitats, increasing the acquisition of conservation easements by 1809 %, totaling 361,984 acres (NRCS 2015b). Through 2018 SGI has committed another US$211 million to conserve an additional 3.5 million acres range-wide (NRCS 2015a). In Montana and the Dakotas, 870,000 acres of priority habitat will be conserved by acquiring additional conservation easements, prioritizing restoration of intervening croplands and by grazing livestock sustainably across the landscape (NRCS 2015a. In Montana, the implementation of conservation actions within and among these PACs is backed by an executive order and US$10 million for conservation projects to benefit sage grouse and sage grouse habitat via the Greater Sage Grouse Stewardship Act (State of Montana 2014, 2015). The agencies involved have recognized the need for continued incorporation of the best available science in executing their conservation strategies such that their focus is not only directed at conservation management within individual PACs, but also at planning for connectivity among PACs to prevent isolation and divergence of existing populations in the future (Finch et al. 2016).
Relatively little is known about sage grouse genetic variation, genetic population structure, and population connectivity at a high-resolution, regional scale relevant to state and regional federal managers. At the broad scale, mitochondrial DNA analysis yielded 80 haplotypes, distributed into two distinct monophyletic clades, both of which showed signals of allopatric fragmentation and gene flow restricted by distance (IBD; Oyler-McCance et al. 2005). All of the populations across the northeastern range belonged to clade II, which is hypothesized to have expanded northward after the last glacial maximum (Zink 2014). The most comprehensive evaluation of population structure and genetic diversity examined range-wide genetic structure using samples from 46 locations. Nuclear marker analysis showed that geographic distance was the most significant factor shaping ten genetic subpopulations across the species’ range (Oyler-McCance et al. 2005). Three of the subpopulations occupy the northeastern portion of the species’ range (hereafter “the northeastern range”)—two in Montana, and one in North Dakota and South Dakota (Oyler-McCance et al. 2005). In a high-resolution, small-scale population structure analyses of northern Montana and southern Canada two subpopulations were discovered that were located north and south of the Milk River (Bush et al. 2011).
Our primary objective was to quantify sage grouse genetic population structure and gene flow at a high resolution. Our secondary objective was to compare existing management group boundaries (MZs and PACs) to genetic population structure and characterize gene flow among management groups. Lastly, we wanted to initiate exploration of the relationship between population structure and major landscape features (vegetation and elevation) across the northeastern range of the species. To address our objectives we used a combination of individual and population-based approaches.
Methods
Study area and sampling
We used 3481 spatially-referenced sage grouse feather and blood samples representative of the northeastern range of the species in the United States of America (154,800 km2 of sagebrush dominated ecosystems in Montana, North Dakota, and South Dakota; Fig. 1). Feather samples were collected non-invasively (Bush et al. 2005, Segelbacher 2002) from leks where they were dropped or plucked by sage grouse during lekking activity, while blood samples were collected from sage grouse on leks as part of radio telemetry research efforts. Samples were collected from 351 leks (mean of 9.91 samples per lek ± 8.58 [SD]) between 2009 and 2012 by field biologists and technicians with the Bureau of Land Management, MTFWP, and the Montana Audubon Society.
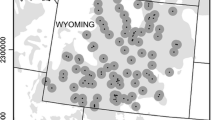
Primary and Secondary genetic population structure for greater sage-grouse sampled in Montana, North Dakota and South Dakota as determined using STRUCTURE. Points show greater than 70 % membership of individuals to each of the primary K = 3 clusters [N (northern): red, SE (southeastern): blue/yellow, and SW (southwestern): orange/purple], and the primary/secondary K = 5 clusters [N (northern): red, SE-E (southeastern-east): blue, SE-W (southeastern-west): yellow, SW-N (southwestern-north): purple, SW-S (southwestern-south): orange, and unassigned: white]. Individuals with <70 % assignment to any subpopulation are unassigned. Individuals and PACs (colored polygons) are colored by suggested management group membership in accordance with genetic subpopulations. PACs, listed listed from west to east by centroid: B3 (Beaverhead 3), B1 (Beaverhead 1), B2 (Beaverhead 2), GV (Golden Valley), C3 (Carbon 3), F (Fergus), M (Musselshell), SP (South Phillips), NR (North Rosebud), NV (North Valley), PRB (Powder River Basin 1, 2 & 3), MG (McCone-Garfield), C (Carter), CC (Cedar Creek), ND (North Dakota), SD (South Dakota). Also shown are, state lines (dashed grey lines). Q-plots are shown for the primary K = 3 clusters b and the secondary K = 2 clusters discovered within both the southeastern c and southwestern d primary subpopulations. Q-plot subpopulation abbreviations are as listed above, and colors correspond to map (a)
Laboratory analysis
DNA extraction
Feather DNA was extracted from the quill (calamus) using QIAGEN’s DNeasy Blood and Tissue Kit and the user developed protocol for purification of total DNA from nails, hair, or feathers. We modified the protocol by incubating samples for a minimum of 8 h after addition of Proteinase K and by eluting DNA with 100 µl of Buffer AE. Feather samples were extracted in a lab used only for non-invasive DNA extraction in order to avoid potential contamination from samples with higher DNA concentrations. Blood samples were extracted using QIAGEN’s DNeasy Blood and Tissue Kit and the protocol for nucleated blood.
Microsatellite DNA amplification and electrophoresis
We amplified 15 variable microsatellite loci and one sex-diagnostic locus in eight multiplex polymerase chain reactions (PCR) (Table 1 in Supplementary Material). We initially used 16 microsatellite loci; however, we removed TUD3 due to a significant heterozygote excess. Results were nearly identical with and without the inclusion of TUD3. The total PCR volume of 10 µl contained 2.0 µl of DNA template and 8 µl reagent mix. Locus specific reaction mixes, annealing temperatures, and thermal cycler profiles are listed in Tables 2 and 3 in the Supplementary Material. PCR mixes included 1 µM dye-labeled forward primer (IDT® Custom DNA Oligos), 1 µM reverse primer (Eurofins MWG Operon Custom DNA Oligos), 1 U Taq polymerase (Invitrogen™), 1 × reaction buffer (Invitrogen™), 200 µM of each dNTP, 2.0 mM MgCl2 (Invitrogen™), and 1.5 mg/ml BSA. Each PCR included two samples of known genotype to allow for calibration of genotypes across gels, for identification of PCR-generated stutter, and for identification of sample contamination.
We loaded PCR product into a 6 % polyacrylamide gel in a Li-Cor Biosciences 4300 DNA Analyzer (Li-Cor Biosciences, Lincoln, Nebraska USA) and electrophoresed for 2 h 30 min at 1500 V, with a current of 40 mA, and a power of 40 W. PCR products were visualized and genotypes were determined using Li-Cor Biosciences’ e-Seq software.
Genotyping
To ensure correct genotypes from low quality and low quantity feather DNA samples, each sample was PCR amplified at least twice across the 15 variable microsatellite loci to screen for allele dropout, stutter artifacts and false alleles. To minimize genotyping error, each genotype was scored by two independent observers. If there was any discrepancy between the first two genotypes or if samples failed to amplify in both replicates, samples were PCR amplified and genotyped an additional two times. If successful in this repeat analysis, genotypes were retained. If the repeat analysis failed, the sample was assigned a missing score for that locus.
To screen samples for quality control, we removed from analysis any individual for which amplification failed at five or more loci. After removal of poor quality samples, genotypes were screened to ensure consistency between allele length and length of the microsatellite repeat motif. We used program DROPOUT v2.3 (McKelvey and Schwartz 2005) and package ALLELEMATCH v2.5 (Galpern et al. 2012) in R (R Core Team 2016) to screen for genotyping error and to identify and remove multiple captures of the same individual from the same lek in the same year. We quantified the power of our microsatellite locus panel to discern individuals using probability identity (PID; Evett and Weir 1998): the probability that two individuals drawn at random from the population could have the same genotype across all loci.
Population genetic descriptive statistics
We calculated the average number of alleles across 15 loci (A), expected heterozygosity (H e ), and observed heterozygosity (H o ) in the R package GSTUDIO (Dyer 2014). We calculated F IS for each locus, and tested for deviation from Hardy–Weinberg proportions (HWP) and gametic disequilibrium among loci correcting for multiple tests for significance using Bonferroni corrected p-values using program GENEPOP v4.5.1 (Rousset 2008). Finally, we calculated allelic richness (AR; El Mousadik and Petit 1996) using program FSTAT v2.9.3.2 (Goudet 1995).
Sampling scheme has been shown to have an effect on Bayesian clustering analyses in the presence of spatial autocorrelation (Schwartz and McKelvey 2009). Therefore, we tested for IBD using Mantel tests for correlation (Mantel 1967) between matrices of genetic distance (i.e., individual-based AMOVA distance calculated in the R package GSTUDIO) and Euclidean geographic distance (calculated in the R package VEGAN; Oksanen et al. 2015) using the Pearson product-moment correlation coefficient (R). We calculated R for 80 even-distance classes and tested for significance of spatial autocorrelation using 999 permutations of the distance matrices with a Bonferoni corrected α of 0.05 in the R package VEGAN. We visualized spatial autocorrelation between samples using Mantel correlograms. Within the correlogram, each individual bin distance was determined using the Sturges equation (Sturges 1926) to optimize the number of data points in each bin.
Individual-based analyses
We tested for spatial structure using an individual-based spatial principal components analysis (sPCA) calculated in the R package ADEGENET (Jombart and Ahmed 2011). We also conducted a group-based principal component analysis (PCA) conducted in the R package GSTUDIO. The group-based PCA was calculated by computing the mean component scores for all individuals located within each PAC (n = 1116; individuals located outside of PACs were excluded from this analysis). To estimate the number of subpopulations within the study area, we used the Bayesian clustering program STRUCTURE v2.3.4 (Pritchard et al. 2000). STRUCTURE groups samples so that gametic disequilibrium and deviation from HWP are both minimized within each cluster (K). Therefore, it can be used to evaluate the most probable K from the pool of individual genotypes, and to provide admixture or assignment scores (Q-values). We used STRUCTURE to analyze all individuals for values of K from 1 to 7. We used settings recommended by Falush et al. (2003) for detecting subtle population subdivision using the admixture model, correlated allele frequencies among populations, and with the allele frequency distribution parameter (λ) set to 1. We allowed STRUCTURE to infer the value of the model’s Dirichlet parameter (α), for the degree of admixture, from the data. We set the length of burn-in period before the start of data collection to 1,000,000, the number of MCMC reps after burn-in to 1,000,000, and did not use user-defined population-of-origin for each individual, nor user-defined sampling location for each individual (non-informative prior). We completed ten replicate runs for each value of K.
To determine the most probable value of K, we used STRUCTURE HARVESTER v0.6.94 (Earl and vonHoldt 2012) to plot the mean and standard deviation of the natural log of the probability of each value of K (Ln p(K)) and to plot ∆K, a second-order statistic formulated by Evanno et al. (2005). We selected the most probable K by examining the plots generated and selecting the value of K at which ΔK is greatest and/or at which the Ln p(K) plot asymptotes. The most probable number of K clusters was determined as one if the ΔK plot indicated a K of two but the Ln p(K) plot clearly exhibited the highest Ln p(K) at K = 1. Once we determined the most probable value of K, we used program CLUMPP v 1.1.2 (Jakobsson and Rosenberg 2007) to average individual assignment to each of the K clusters across the ten STRUCTURE replicates for each value of K. In CLUMPP we used 30,000 repeats of the greedy method with greedy option two and the pairwise matrix similarity statistic, G.
We examined hierarchical subpopulation structure following determination of the primary level of population structure. First, we split the pool of sample genotypes by majority percent population assignment (Q-value). Next, we independently analyzed each of the primary hierarchical clusters for additional substructure using STRUCTURE and the same settings as above. We continued our hierarchical analysis in this way until we inferred that the most likely number of clusters was one (K = 1) for each sample pool analyzed (according to our interpretation of the Ln p(K) plot), as suggested by Coulon et al. (2008). Within each inferred primary and secondary cluster we calculated HWP and gametic disequilibrium among loci, A, AR, H e , H o , and F IS as described above. Among clusters, we calculated F ST (Weir and Cockerham 1984) using program GENEPOP v4.5.1 (Rousset 2008), and constructed F ST based dendrograms by first transforming the pairwise F ST matrix to a maximum distance matrix (using the supremum norm) and then via the single linkage method (closely related to a minimal spanning tree) using the R package STATS (R Core Team 2016).
As an independent verification of subpopulation structure, we compared the results of STRUCTURE to those of program TESS v. 2.3 (Francois et al. 2006; e.g., Chen et al. 2007), a second Bayesian analysis which uses not only each sample’s genotype but also a prior informed by both global trend surfaces and spatial autocorrelation to determine the most probable number of population clusters. In TESS we completed 10 iterations for each value of K from two to ten, allowing admixture, using the conditional auto-regressive (CAR) Gaussian model, the default program values for spatial interaction parameter (0.6) and degree of trend (Linear (1)), with 12,000 MCMC sweeps and a burn-in of 2,000 sweeps for each run. From the output of TESS, we selected K by calculating the change in mean Deviance Information Criterion (ΔDIC)—averaged across all ten replicates for each value of K—between successive K values and selecting the value of K for which we calculated the greatest value of ΔDIC (largest decrease). After selecting the most probable value of K, we again used CLUMPP to average individual assignment to each of the K clusters across the 20 TESS replicates for the most probable value of K. Here we used the same CLUMPP parameter options as stated above.
Comparison of population structure to priority areas for conservation and landscape characteristics
After we determined the most probable hierarchical population structure via the individual-based analyses described above, we calculated the percent of each genetic subpopulation within each of the current sage grouse PACs recognized by the U.S. Fish and Wildlife Service (2013) and within management zones recognized by WAFWA (Stiver et al. 2006). We then compared genetic population structure to landscape characteristics to explore factors that may have influenced population structure, as follows. First, we buffered all sampled leks by 5 km, a management buffer suggested for the protection of nesting habitat (Connelly et al. 2000), then we calculated the percent area within the buffer covered by the three big sagebrush subspecies (basin big sagebrush: Artemisia tridentata tridentata, mountain big sagebrush: A. t. vaseyana, and Wyoming big sagebrush: A. t. wyomingensis) and associated vegetation, basing our calculations on a broad-scale remote-sensed vegetation map of the western USA (Comer et al. 2002). Second, we used a digital elevation map (Gesch et al. 2002) to measure elevation—a proxy for abiotic factors such as temperature and precipitation–-within each 5 km buffer. We calculated the range and variability of elevation across all 5 km buffers in each subpopulation. Finally, we compared mean elevation across all 5 km buffers in each subpopulation using the Kruskal–Wallis rank sum test.
Results
Genotyping
After removing inferior quality samples (those that did not genotype across at least 2/3 of all loci; n = 718) and all duplicate genotypes (the same individual sampled from the same lek in the same year; n = 1260) we retained 1499 of 3481 samples genotyped (43.1 %). The 1499 genotypes from feathers (n = 1445) and bloods (n = 54) were from 324 leks (mean of 4.6 samples per lek ± 3.69 [SD]): 1393 samples from 297 leks in Montana (mean = 4.69 samples per lek ± 3.75 [SD]), 15 samples from eight leks in North Dakota (mean = 1.88 samples per lek ± 0.99 [SD]), 69 samples from 17 leks in South Dakota (mean = 4.06 samples per lek ± 2.98 [SD]), and 22 samples from three leks across the northern border of Wyoming (mean = 7.33 samples per lek ± = 2.08 [SD]). Samples sizes per lek in North Dakota are low; however, our sampling reflects the lek counts, where there was an average of 4.82 males per lek counted on an average of 14 leks during the time we collected samples (Robinson 2014). Using our 15 locus panel, PID was 8.24 × 10−18, providing substantial power to discern individuals. We were able to determine sex for 1487 (99.2 %) of the final individual genotypes (Table 1). Each occurrence of an individual recaptured at more than one lek in the same year (n = 22), or in more than one year at the same (n = 18) or at different leks (n = 7) was retained for all analyses.
Population genetic descriptive statistics
Before taking into account any population structure, 12 of the 15 loci showed a deficit of heterozygotes (Table 2), and 47 out of 120 pairwise locus comparisons showed gametic disequilibrium. Within the sample pool there were an average of 13.60 alleles per locus ranging from seven alleles at TUT3 to 32 alleles at MSP11 (Table 1 in Supplementary Material), with an expected heterozygosity of 0.752, and an F IS of 0.045 (Table 3a).
Individual-based analyses
We found substantial genetic spatial structure using the PAC-based PCA. The sampled PACs are clearly sorted by geographic location from west to east as PC1 values increase and from north to south as PC2 values increase (Fig. 2). Together, the first (35.6 %) and second (17.2 %) principal components (PC) captured 52.8 % of the variance in the data.
PCA of the mean PC scores for all individual sample genotypes within each of the 16 PACs. PAC abbreviations listed alphabetically: Beaverhead 1 (B1), Beaverhead 2 (B2), Beaverhead 3 (B3), Carter (C), Carbon 3 (C3), Cedar Creek (CC), Fergus (F), Golden Valley (GV), Musselshell (M), McCone-Garfield (MG), North Dakota (ND), North Rosebud (NR), North Valley (NV), Powder River Basin (1, 2 & 3) (PRB), South Dakota (SD), South Phillips (SP)
The first eigenvector (Fig. 1a in Supplementary Material) and second eigenvector (Fig. 1b in Supplementary Material) of the sPCA show the major north–south and east–west genetic divergence patterns. We retained both the first and second eigenvectors, as they composed 74.2 and 61.2 % of the variation in the data respectively. Furthermore, both eigenvectors were highly spatially autocorrelated, with a Moran’s I of 0.226 and 0.205, respectively (Fig. 2 in Supplementary Material).
We used the Bayesian clustering results—both the natural log of the probability of K (Ln p(K)) and ∆K statistic plots to infer that the sample genotypes represented three spatially distinct population clusters (Fig. 3 in Supplementary Material): a subpopulation in southwestern Montana (SW), a subpopulation in northern Montana (N), and a subpopulation in southeastern Montana, North Dakota, and South Dakota (SE). Genetic admixture was present among these subpopulations (Fig. 1b).
After assigning each individual to one of the three primary subpopulations by maximum Q-value, we proceeded with STRUCTURE analysis of each subpopulation independently. We discovered secondary hierarchical population structure within both the SE and the SW subpopulations (Fig. 1c and d), but not within the N subpopulation (Fig. 4a and d in Supplementary Material). The SE group divided into secondary eastern (SE-E) and western (SE-W) subpopulations (Figs. 4b, e and 5b all in Supplementary Material) while the SW subpopulation split into secondary northern (SW-N) and southern (SW-S) subpopulations (Figs. 4c, f and 5c all in Supplementary Material).
We discovered no evidence for additional hierarchical levels of population substructure. The cumulative primary and secondary subpopulations are shown in Fig. 1a, where each individual is depicted as a member of a subpopulation if its population admixture—Q-value—was at least 70 % for that cluster. Despite the disparity in sample sex ratio (Table 1), the STRUCTURE results for females and males analyzed independently were consistent with substructure identified with the full dataset (results not shown).
After grouping samples into primary hierarchical subpopulations based on maximum Q-value, no loci were out of HWP in more than one subpopulation (Table 2). However, five loci were out of HWP in the southeastern subpopulation and one locus was out of HWP in the southwestern subpopulation. Only one locus pair was in significant gametic disequilibrium in more than one subpopulation (BG16/SGCA5 in the N and SE subpopulations; α = 4.167 × 10−4; p < 0.001). After grouping samples into the secondary hierarchical subpopulations, no loci were out of HWP in more than one subpopulation (Table 2, α = 3.333 x 10−3), and only one locus pair was significantly in gametic disequilibrium in more than one population (MS06.8/MSP11 in both the N and SW-S subpopulations). When grouping samples by PAC, no loci were found significantly out of HWP in more than one PAC (only BG16 in Carter, and SGCTAT1 in McCone-Garfield), and none of the locus pairs was in gametic disequilibrium in more than one PAC.
When both hierarchical levels of population structure were considered, the average number of alleles per locus, per population ranged from 8.73 in the SW-N subpopulation to 11.73 in the N subpopulation (mean = 10.39 ± 4.12 [SD]). Averages for H e across all subpopulations 0.747 ± 0.16 [SD] (Table 3a). The average number of alleles per locus, per PAC ranged from 5.27 in the North Dakota (ND) PAC to 10.07 in the South Phillips (SP) PAC (mean = 8.31 ± 3.17 [SD]). Within PACs, average H e was 0.723 ± 0.17 [SD] (Table 3b).
We found a significant positive correlation between increasing geographic distance between individuals and increasing genetic divergence (Mantel test: r = 0.271, p = 0.001; Fig. 6 in Supplementary Material). Genotypes were significantly more similar than expected by chance for samples up to 242.6 km apart and significantly more dissimilar than expected by chance for samples over 311.9 km apart.
Divergence among geographically proximal subpopulations and PACs, as measured by F ST, was low compared to those which were distal (Fig. 3a, b; raw values in Table 4a, b in Supplementary Material, respectively). Dendrograms reveal divergence relationships among the subpopulations detected using Bayesian clustering analyses. The greatest divergence was found between the two most distal subpopulations, the SE-E and SW-N subpopulations (F ST = 0.0777), while the least divergence was found between the two most proximal subpopulations, the SE-E and SE-W subpopulations (F ST = 0.0174). Mean F ST among all genetic subpopulations discovered using STRUCTURE was 0.0473 ± 0.0243 [SD]. Mean F ST among all PACs was 0.0380 ± 0.0216 [SD]. The dendrogram of the PACs (Fig. 3b) highlights that while the Beaverhead 1 and Beaverhead 2 PACs are less diverged (F ST = 0.004) than are the Beaverhead 2 and Beaverhead 3 PACs (F ST = 0.005), it is the greater divergence between Beaverhead 1 and Beaverhead 3 (F ST = 0.009) that drives Beaverhead 2 and Beaverhead 3 to cluster more closely.
Genetic divergence among the five greater sage-grouse subpopulations detected using STRUCTURE (subpopulation abbreviations provided in caption to Fig. 1) (a), and among the 16 PACs sampled (PAC abbreviations provided in caption to Fig. 2) (b). Divergence is measured in pairwise comparisons using Wright’s F ST (Wright 1949). Greater divergence—higher F ST—is shown as darker shades of grey. Subpopulation abbreviations listed alphabetically: N (northern), SE-E (southeastern-east), SE-W (southeastern-west), SW-N (southwestern-north), SW-S (southwestern-south). PAC abbreviations listed alphabetically: B1 (Beaverhead 1), B2 (Beaverhead 2), B3 (Beaverhead 3), C (Carter), C3 (Carbon 3), CC (Cedar Creek), F (Fergus), GV (Golden Valley), M (Musselshell), MG (McCone-Garfield), ND (North Dakota), NR (North Rosebud), NV (North Valley), PRB (Powder River Basin 1, 2 & 3), SD (South Dakota), SP (South Phillips)
Finally, we found strong concordance between the resulting individual assignment of TESS and that of the primary level of hierarchical analysis in STRUCTURE (Fig. 7 in Supplementary Material). In TESS, three clusters (K = 3) showed the largest drop in DIC between successive values of K = 2–3 (∆DIC = 2748.2).
Comparison of population structure to priority areas for conservation and landscape characteristics
Management zones and priority areas for conservation (PACs)
The N and SE-E subpopulations are located almost entirely within WAFWA MZI (99.4 and 99.7 %, respectively; Fig. 8 in Supplementary Material). Similarly, the SW-N subpopulation is entirely contained and the SW-S subpopulation is nearly entirely contained within MZIV (96.1 %). The SE-W subpopulation spans MZI (74.7 %), II (21.1 %) and IV (4.2 %).
On average, 85.8 % of the individuals within each PAC belong to the same subpopulation (Fig. 4). Six PACs encompass a majority of the N subpopulation (mean = 80.6 %), five PACs envelop a majority of the SE-E subpopulation (mean = 88.7 %), two PACs envelop a majority of the SE-W subpopulation (mean = 87.7 %), one PAC encompasses a majority of the SW-N subpopulation, and two PACs envelop a majority of the SW-S subpopulation (mean = 92.0 %).
Subpopulation composition within 16 PACs sorted by majority assignment percentage. PACs listed in order of display: NV (North Valley), SP (South Phillips), F (Fergus), MG (McCone-Garfield), M (Musselshell), NR (North Rosebud), SD (South Dakota), CC (Cedar Creek), C (Carter), ND (North Dakota), PRB (Powder River Basin 1, 2 & 3), C3 (Carbon 3), GV (Golden Valley), B3 (Beaverhead 3), B1 (Beaverhead 1), B2 (Beaverhead 2)
Vegetation
Within 5 km of leks in the N subpopulation the landscape was dominated by grasses (53 %) with the dominant sagebrush subspecies being Wyoming big sagebrush (19 %; Table 5; Fig. 9 in Supplementary Material). Within 5 km of leks in the SE-E subpopulation the landscape was dominated by grasses (61 %) with the dominant sagebrush being Wyoming big sagebrush (10 %). The 5 km buffer of the SE-W subpopulation leks contained mixed xeric shrubland (39 %) and grasses (31 %) with Wyoming big sagebrush (15 %) and mountain big sagebrush (2 %) being the dominant structural elements. In the SW-N subpopulation, mountain big sagebrush dominates the 5 km buffer of lekking grounds (63 %) and is interspersed with basin big sagebrush (24 %). Finally, within 5 km of the leks in the SW-S subpopulation, mountain big sagebrush (77 %) and Wyoming big sagebrush (15 %) are the most prevalent subspecies of sagebrush with a minority cover of grasses (4 %).
Elevation
Mean elevation within 5 km of leks in each subpopulation was significantly different among subpopulations (Kruskal–Wallis rank sum test, H = 642.77, 4 d.f., p < 0.001), and there was little to no overlap of interquartile ranges (IQR; Fig. 10 in Supplementary Material). Most notably, the southwestern subpopulations occupy far higher elevations than do the other three subpopulations. Elevation increased from N to SE-E to SE-W to SW-N to SW-S subpopulations (IQR: 781–998, 910–1022, 1007–1258, 1858–2003, 2043–2202 m). The subpopulations spanned an elevation range of 671–2411 meters.
Discussion
We detected discontinuities in sage grouse genetic connectivity across the northeastern range of the species. These discontinuities indicate divergence in allele frequencies between three primary subpopulations, and secondary hierarchical subpopulations within two of the primary subpopulations (Fig. 4). The greater divergence among the primary subpopulations compared to divergence among the secondary subpopulations may be indicative of the chronology in which these subpopulations diverged.
We detected the pattern of population structure using both spatially independent (PCA: Fig. 2, and STRUCTURE: Fig. 1a) and spatially dependent (sPCA: Fig. 1 in Supplementary Material, and TESS: Fig. 7 in Supplementary Material) analyses. Concordance between the aspatial and spatial analyses supports the patterns of genetic subpopulation structure detected, as does the concordance between the two Bayesian clustering methods (STRUCTURE and TESS) and the two ordination methods (PCA and sPCA). Using the hierarchical approach to analyze genetic structure, we detected genetic substructure within the SE and SW subpopulations that otherwise would have remained cryptic. Had we not independently analyzed the primary level subpopulations we would not have found the substructure resulting from low levels of population divergence among secondary subpopulations. This oversight may occur if divergence among primary subpopulations is far greater than divergence among secondary subpopulations (Latch et al. 2006).
The primary population structure we detected (i.e., three subpopulations across the northeastern range: N, SE, SW) supports results of Oyler-McCance et al. (2005). However, our sampling intensity allowed us to define spatial subpopulation structure with greater resolution. Furthermore, we detected additional substructure within two of these primary subpopulations: both in the southwestern Montana subpopulation (SW) and in the southeastern Montana/North Dakota/South Dakota subpopulation (SE).
Within the southeastern portion of our study area, our results align with those of Schulwitz et al. (2014) who discovered hierarchical structure within sage grouse populations in northwest Wyoming and southeastern Montana. They found that their Southeast Montana, South Powder River Basin, and North Powder River Basin sampling locations constituted a single subpopulation (that which we have called the SE-E subpopulation). Furthermore, their results indicate that there is genetic discontinuity between this subpopulation and subpopulations throughout the rest of Wyoming. The magnitude of genetic divergence among the subpopulations identified in both studies were similar (see Table 4b in Supplementary Material and Table 2 in Schulwitz et al. 2014).
We did not find the same subpopulation substructure that Bush et al. (2011) found within the N subpopulation via their analysis of thirteen microsatellite loci. Our results may differ due to our lack of samples from sage grouse in Alberta and Saskatchewan. Alternatively, there may have been a change in substructure between the years during which Bush et al. sampled and the years during which we sampled.
Similar to prior studies, we detected a signal of IBD. In prior research, IBD has been suggested as one of the primary drivers of genetic divergence for sage grouse (Bush et al. 2011; Davis et al. 2015; Oyler-McCance et al. 2005; Schulwitz et al. 2014). We found IBD, but also that up to 242 km there is greater genetic similarity than would be expected by chance. This suggests that despite the relatively short dispersal distances documented (7–9 km; Dunn and Braun 1985), the cumulative effect of these dispersals translates into long-range connectivity.
Physiogeographic correlates with genetic subpopulations
We found evidence for the impact of pre-European physiogeographic landscape processes on contemporary subpopulation structure. First, elevations occupied by the subpopulations are significantly different (Fig. 10 in Supplementary Material). The two southeastern populations—most notably the SE-W subpopulation—occupy elevations more similar to the two southwestern subpopulations, which may facilitate gene flow among these subpopulations if individuals are locally adapted to habitats at this elevation. The smaller divergence among these subpopulations (F ST = 0.0508) compared to the divergence of the N and southwestern subpopulations (F ST = 0.0590) may be due to gene flow among the southwestern and southeastern clusters through subpopulations located in Idaho and Wyoming which are outside the geographic extent of this study. Future research could examine whether gene flow among the southeastern and southwestern subpopulations is fostered by genetic connectivity through subpopulations in Idaho and Wyoming.
Second, there are stark differences in the vegetative community assemblages concomitant with subpopulations. Most notable are the differences in dominant subspecies of sagebrush, a plant vital to the species’ requirements for food, cover, and nest success (Connelly et al. 2011a, b). We found that the two southwestern subpopulations occur on a landscape dominated by mountain big sagebrush, a subspecies that grows on high-elevation mountain slopes (Jaeger et al. 2016). These two subpopulations are also surrounded by basin big sagebrush, a subspecies that grows in deep-soil drainage basins (Jaeger et al. 2016). Basin big sagebrush does not occur within proximity of leks in any other subpopulation and mountain big sagebrush only composes 2 % of the vegetation proximal to leks in the SE-W subpopulations. The SW-N subpopulation has far more basin big sagebrush proximal to leks (24 %) than does the SW-S subpopulation (3 %) which occupies significantly higher elevation.
The three sagebrush subspecies within our study area have been shown to differ in terpene composition and quantity. Terpenes are chemical compounds found in plants that have been shown to function as anti-herbivory agents (Byrd et al. 1999; Thacker et al. 2012; Jaeger et al. 2016). Differences in terpenes among sagebrush subspecies could be an important factor in diet selection (Frye et al. 2013). Prior work has shown that sage grouse prefer the palatability of mountain big, over Wyoming big, over basin big sagebrush (Rosentreter 2004), and that this diet selection may well be linked to both lower monterpene content and higher crude protein content within the preferred subspecies (Remington and Braun 1985).
Given the stark differences in both elevation and in prevalent sagebrush subspecies among subpopulations, it is possible that these physiogeographic factors have impacted both subpopulation divergence and adaptive divergence. For example, it is possible that adaptations to terpene metabolism have arisen within subpopulations, allowing for improved digestion of the most prevalent sagebrush subspecies. Similar to our findings, Schulwitz et al. (2014) found that sage grouse subpopulations across northwest Wyoming occupied distinct ecoregions, composed of distinct assemblages of species and shaped by different environmental processes.
Major river-highway corridors
In the Rocky Mountain west, the landscape is composed of basin and range topography. Major highways are routed along large river valleys across this landscape. The combination of direct and indirect effects of these river-highway corridors and associated landscape alteration may have shaped sage grouse genetic population structure. The vast majority (95.9 %) of the N subpopulation is north of the Interstate 90/Interstate 94/Yellowstone and Bighorn River corridor, while 89 % of the southern subpopulations (SE-E, SE-W, SW-N and SW-S) were found south of the corridor (individually, 69.4 % of the southeastern subpopulation and 100 % of the southwestern subpopulation).
We found the greatest amount of admixture within individuals on leks closest to these major river-highway corridors. That is, there was a significant negative correlation between distance from major highways that co-occur with major rivers and percent admixture (r(1541) = −0.1638556, p < 0.001, 95 %CI −0.212–−0.115). This indicates that gene flow is occurring across these corridors but that these areas appear to be where subpopulations interface. However, it is difficult to discern whether it is the effects of transportation or that of the natural landscape features composing the corridors that has influenced genetic structure.
Major river-highway corridors may have exacerbated genetic divergence among subpopulations. Divergence among the N and southern subpopulations (Figs. 2 and 5a in Supplementary Material) may have been influenced by impeded gene flow across the Yellowstone River and Interstate 94 corridor, an area largely converted to cultivation in recent human history and where declines in lek attendance and lek extirpation have both been observed. The effect of highways on genetic connectivity and genetic diversity are detailed across multiple taxa (Holderegger and Di Giulio 2010; Jackson and Fahrig 2011; Proctor et al. 2005), especially that of wider highways with high traffic volume such as the Interstate 90/Interstate 94 corridor. Highways surrounded by cultivated land are a documented barrier to sage grouse gene flow (Bush et al. 2011). Distance to highways has been shown to lead to nest failure (Webb et al. 2012) and has been shown to decrease peak male sage grouse lek attendance by over 70 % (Blickley et al. 2012). Furthermore, road noise has been linked to the reduction of breeding bird densities in multiple avian species (Parris and Schneider 2009; Slabbekoorn and Ripmeester 2008).
Management and conservation implications
The five subpopulations we detected span the boundaries of the MZs recognized by WAFWA. The main discrepancy we found between genetic population structure and the boundaries of these MZs is that the SE-W population overlaps the boundaries of the three northeastern MZs: MZI, MZII and MZIV (Fig. 8 in Supplementary Material). Depending on the goal of managing populations, it may be useful to use genetic structure in addition to common floristic provinces to set boundaries of MZs.
MTFWP, NDGFD & SDGFP delineated management areas, called PACs, to protect areas with the greatest abundance of birds (U.S. Fish and Wildlife Service 2013). Our results provide insight into how these units can be functionally managed at multiple scales that reflect genetic connectivity. Existing PACs align very well with genetic population structure, allowing relatively simple assembly into management groups (Fig. 1a). These managment groups could serve as the conservation units used when setting conservation goals, modifying policy, targeting conservation resources, assessing connectivity, translocation, and regulation of harvest. For example, translocations may wish to use sage grouse from within a genetically identified subpopulation. We suggest grouping PACs into five management groups based on genetic structure: a Northern management group composed of the Fergus, McCone-Garfield, Musselshell, North Rosebud, North Valley, and South Phillips PACs; a Southeastern-East management group composed of the Carter, Cedar Creek, PRB, North Dakota and South Dakota PACs; a Southeastern-West management group composed of the Carbon 3 and Golden Valley PACs; a Southwestern-North management group composed of solely the Beaverhead 3 PAC; and a Southwestern-South management group composed of the Beaverhead 1 and Beaverhead 2 PACs (Fig. 4).
Each PAC encompasses a group of individuals almost entirely from a single subpopulation, with the exception of the North Rosebud PAC (Fig. 4). North Rosebud is the most diverse PAC by population membership, being composed mostly of individuals in the N subpopulation and nearly equal parts SE-E and SE-W (Fig. 4). When measuring divergence among PACs, values of F ST were smaller than when measuring divergence among genetic subpopulations. This is likely the result of mixed genetic subpopulation membership when pooling individuals by PAC.
In the PAC-based dendrogram (Fig. 3b), the Golden Valley PAC, located in the Southeastern-West management group, aligns more closely with PACs in the Northern management group (Fergus, McCone-Garfield, Musselshell, North Rosebud, North Valley, and South Phillips) than it does with the PACs in the Southeastern-East management group (Carter, Cedar Creek, PRB, North Dakota, and South Dakota). We might expect Golden Valley to align most closely with the PACs within the southeastern primary subpopulation. However, the unexpected dendrogram alignment is likely due to grouping individuals from within each PAC, which for Golden Valley means 30 % of the individuals in the pack assign to the N subpopulation. Despite these few caveats, state-delineated PACs align very well with genetic subpopulation structure.
Future research directions
We sampled genetic data from sage grouse subpopulations located across multiple landscapes with different physiogeographic histories. These subpopulations exist at different elevations, surrounded by different assemblages of vegetation, and persist in spite of novel landscape disturbances. Maintaining connectivity and minimizing habitat fragmentation within and among these subpopulations is critical to the persistence of the species. A thorough analysis of which landscape features and disturbances have the greatest effect on genetic connectivity would lead to a better understanding of which areas on the landscape are essential to maintaining connectivity. These areas could be the targets for management action. Therefore, we suggest that it would prove invaluable for further study to discern whether it is the anthropogenic landscape alteration or natural features that have had the greatest influence on genetic structure.
The coincidence of breaks in genetic continuity and underlying physiogeography suggests that it is possible that adaptive differences have arisen within subpopulations. These adaptive differences could in turn restrict contemporary gene flow among subpopulations. Therefore, quantification of adaptive variation across not only the northeastern portion, but also the entire range could lend a much better understanding of the species’ phylogeography. A comparison of our analysis of subpopulation structure, an understanding of adaptive variation, and an analysis of fine-scale connectivity among leks within and among subpopulations—using network theory or a similar approach—would provide a powerful set of tools to assist managers as they set management objectives to preserve genetic diversity at across multiple scales (Manel et al. 2010).
References
Aldridge CL, Nielsen SE, Beyer HL, Boyce MS, Connelly JW, Knick ST, Schroeder MA (2008) Range-wide patterns of greater sage-grouse persistence. Divers Distrib 14:983–994
Allen AW, Cook JG, Armbruster MJ (1984) Habitat suitability index models: Pronghorn. U.S. Fish and Wildlife Service FWS/OBS-82/10.65:1–22
Blickley JL, Word KR, Krakauer AH, Phillips JL, Sells SN, Taff CC, Wingfield JC, Patricelli GL (2012) Experimental chronic noise is related to elevated fecal corticosteroid metabolites in lekking male greater sage-grouse (Centrocercus urophasianus). PLoS One 7:e50462
Bradbury J, Vehrencamp S, Gibson R (1989) Dispersion of displaying male sage grouse. Behav Ecol Sociobiol 24:1–14
Braun CE (1998) Sage grouse declines in western North America: what are the problems? Proc West Assoc State Fish Wildl Agencies 78:139–156
Braun CE, Oedekoven OO, Aldridge CL (2002) Oil and gas development in western North America: effects on sagebrush steppe avifauna with particular emphasis on sage grouse. Trans N Am Wildl Nat Resour Conf 67:337–349
Bush KL, Vinsky MD, Aldridge CL, Paszkowski CA (2005) A comparison of sample types varying in invasiveness for use in DNA sex determination in an endangered population of greater Sage-Grouse (Centrocercus uropihasianus). Conserv Genet 6:867–870
Bush KL, Dyte CK, Moynahan BJ, Aldridge CL, Sauls HS, Battazzo AM, Walker BL, Doherty KE, Tack J, Carlson J (2011) Population structure and genetic diversity of greater sage-grouse (Centrocercus urophasianus) in fragmented landscapes at the northern edge of their range. Conserv Genet 12:527–542
Byrd DW, McArthur ED, Wang H, Graham JH, Freeman DC (1999) Narrow hybrid zone between two subspecies of big sagebrush, Artemisia tridentata (Asteraceae). VIII. Spatial and temporal pattern of terpenes. Biochem Syst Ecol 27:11–25
Caizergues A, DuBois S, Loiseau A, Mondor G, Rasplus JY (2001) Isolation and characterization of microsatellite loci in black grouse (Tetrao tetrix). Mol Ecol Notes 1:36–38
Caizergues A, Rätti O, Helle P, Rotelli L, Ellison L, Rasplus JY (2003) Population genetic structure of male black grouse (Tetrao tetrix L.) in fragmented vs. continuous landscapes. Mol Ecol 12:2297–2305
Carrol SP, Hendry AP, Reznick DN, Fox CW (2007) Evolution on ecological time-scales. Funct Ecol 21:387–393
Carstens B, Brunsfeld S, Demboski J, Good J, Sullivan J (2005) Investigating the evolutionary history of the Pacific Northwest mesic forest ecosystem: hypothesis testing within a comparative phylogeographic framework. Evolution 59:1639–1652
Chen C, Durand E, Forbes F, François O (2007) Bayesian clustering algorithms ascertaining spatial population structure: a new computer program and a comparison study. Mol Ecol Notes 7:747–756
Comer P, Kagan J, Heiner M, Tobalske C (2002) Current distribution of sagebrush and associated vegetation in the Western United States (excluding NM and AZ). Interagency Sagebrush Working Group. http://SAGEMAP.wr.usgs.gov
Connelly JW, Schroeder MA, Sands AR, Braun CE (2000) Guidelines to manage sage grouse populations and their habitats. Wildl Soc Bull 28:967–985
Connelly JW, Knick ST, Schroeder MA, Stiver SJ (2004) Conservation assessment of greater sage-grouse and sagebrush habitats. Western Association of Fish and Wildlife Agencies
Connelly JW, Hagen CA, Schroeder MA (2011a) Characteristics and dynamics of Greater Sage-Grouse populations. In: Knick ST, Connelly JW (eds) Greater Sage-Grouse: ecology and conservation of a landscape species and its habitats. Studies in Avian Biology, 38th edn. University of California Press, Berkeley, pp 53–67
Connelly J, Rinkes E, Braun C (2011b) Characteristics of greater sage-grouse habitats: a landscape species at micro and macro scales. In: Knick ST, Connelly JW (eds) Greater Sage-Grouse: ecology and conservation of a landscape species and its habitats. Studies in Avian Biology, 38th edn. University of California Press, Berkeley, pp 69–83
Copeland HE, Doherty KE, Naugle DE, Pocewicz A, Kiesecker JM (2009) Mapping oil and gas development potential in the US Intermountain West and estimating impacts to species. PLoS One 4:e7400
Core Team R (2016) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna
Coulon A, Fitzpatrick J, Bowman R, Stith B, Makarewich C, Stenzler L, Lovette I (2008) Congruent population structure inferred from dispersal behaviour and intensive genetic surveys of the threatened Florida scrub-jay (Aphelocoma cœrulescens). Mol Ecol 17:1685–1701
Dalke PD, Pyrah DB, Stanton DC, Crawford JE, Schlatterer EF (1963) Ecology, productivity, and management of sage grouse in Idaho. J Wildl Manag 27:811–841
Davis DM, Reese KP, Gardner SC, Bird KL (2015) Genetic structure of Greater Sage-Grouse (Centrocercus urophasianus) in a declining, peripheral population. Condor 117:530–544
Doherty KE, Naugle DE, Walker BL, Graham JM (2008) Greater sage-grouse winter habitat selection and energy development. J Wildl Manag 72:187–195
Dunn PO, Braun CE (1985) Natal dispersal and lek fidelity of sage grouse. Auk 102:621–627
Dyer RJ (2014) GSTUDIO: Analyses and functions related to the spatial analysis of genetic marker data. R package version 3.2.1
Earl DA, vonHoldt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:1–3
Edminster FC (1954) American game birds of field and forest: their habits, ecology, and management. Scribner, New York
El Mousadik A, Petit R (1996) High level of genetic differentiation for allelic richness among populations of the argan tree [Argania spinosa (L.) Skeels] endemic to Morocco. Theor Appl Genet 92:832–839
Emmons SR, Braun CE (1984) Lek attendance of male sage grouse. J Wildl Manag 48:1023–1028
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Evett IW, Weir BS (1998) Interpreting DNA evidence: statistical genetics for forensic scientists. Sinauer, Sunderland
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587
Finch DM, Boyce DA, Chambers JC, Colt CJ, Dumroese K, Kitchen SG, McCarthy C, Meyer SE, Richardson BA, Rowland MM, Rumble MA, Schwartz MK, Tomosy MS, and Wisdom MJ. 2016 Conservation and restoration of sagebrush ecosystems and sage-grouse: An assessment of USDA Forest Service Science. RMRS-GTR, 348: 1–54
Francois O, Ancelet S, Guillot G (2006) Bayesian clustering using hidden Markov random fields in spatial population genetics. Genetics 174:805–816
Frye GG, Connelly JW, Musil DD, Forbey JS (2013) Phytochemistry predicts habitat selection by an avian herbivore at multiple scales. Ecology 94:308–314
Galpern P, Manseau M, Hettinga P, Smith K, Wilson P (2012) ALLELEMATCH: an R package for identifying unique multilocus genotypes where genotyping error and missing data may be present. Mol Ecol Resour 12:771–778
Gesch D, Oimoen M, Greenlee S, Nelson C, Steuck M, Tyler D (2002) The National Elevation Dataset. Photogramm Eng Remote Sens 68:5–11
Gibson RM, Bradbury JW, Vehrencamp SL (1991) Mate choice in lekking sage grouse revisited: the roles of vocal display, female site fidelity, and copying. Behav Ecol 2:165–180
Goudet J (1995) FSTAT (version 1.2): a computer program to calculate F-statistics. Am J Heredity 86:485–486
Green JS, Flinders JT (1960) Habitat and Dietary Pygmy Rabbit. Soc Range Manag 33:136–142
Green GA, Livezey KB, Morgan RL (2001) Habitat selection by northern sagebrush lizards (Sceloporus graciosus graciosus) in the Columbia Basin, Oregon. N West Nat 82:111–115
Hagen CA, Connelly JW, Schroeder MA (2007) A meta-analysis of greater sage-grouse Centrocercus urophasianus nesting and brood-rearing habitats. Wildl Biol 13:42–50
Holderegger R, Di Giulio M (2010) The genetic effects of roads: a review of empirical evidence. Basic Appl Ecol 11:522–531
Holloran MJ, Anderson SH (2005) Spatial distribution of greater sage-grouse nests in relatively contiguous sagebrush habitats. Condor 107:742–752
Holloran MJ, Kaiser RC, Hubert WA (2010) Yearling greater sage-grouse response to energy development in Wyoming. J Wildl Manag 74:65–72
Jackson ND, Fahrig L (2011) Relative effects of road mortality and decreased connectivity on population genetic diversity. Biol Conserv 144:3143–3148
Jaeger DM, Runyon JB, Richardson BA (2016) Signals of speciation: volatile organic compounds resolve closely related sagebrush taxa, suggesting their importance in evolution. New Phytologist
Jakobsson M, Rosenberg NA (2007) CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23:1801–1806
Jombart T, Ahmed I (2011) adegenet 1.3-1: new tools for the analysis of genome-wide SNP data. Bioinformatics 27:3070–3071
Kahn N, St. John J, Quinn TW (1998) Chromosome-specific intron size differences in the avian CHD gene provide an efficient method for sex identification in birds. Auk 115(4):1074–1078
Knick ST, Dobkin DS, Rotenberry JT, Schroeder MA, Vander Haegen WM, Van Riper C III (2003) Teetering on the edge or too late? Conservation and research issues for avifauna of sagebrush habitats. Condor 105:611–634
Latch EK, Dharmarajan G, Glaubitz JC, Rhodes OE Jr (2006) Relative performance of Bayesian clustering software for inferring population substructure and individual assignment at low levels of population differentiation. Conserv Genet 7:295–302
Lomolino MV, Riddle BR, Brown JH, Whittaker RJ (2006) Biogeography. Sinauer Associates, Sunderland
Manel S, Joost S, Epperson BK, Holderegger R, Storfer A, Rosenberg MS, Scribner KT, Bonin A, Fortin MJ (2010) Perspectives on the use of landscape genetics to detect genetic adaptive variation in the field. Mol Ecol 19:3760–3772
Mantel N (1967) The detection of disease clustering and a generalized regression approach. Cancer Res 27:209–220
McArthur ED, Plummer AP (1978) Biogeography and management of native western shrubs: a case study, section Tridentatae of Artemisia. Gt Basin Nat Mem 2:229–243
McKelvey K, Schwartz M (2005) dropout: a program to identify problem loci and samples for noninvasive genetic samples in a capture-mark-recapture framework. Mol Ecol Notes 5:716–718
Monsen SB, Shaw NL (2000) Big sagebrush (Artemesia tridentata) communities—ecology, importance and restoration potential. In: Billings Land Reclamation Symposium, 2000: Striving for Restoration, Fostering Technology, and Policy for Reestablishing Ecological Function: March 20-24, 2000, Sheraton Billings Hotel, Billings, Montana. Bozeman: Montana State University, Publication No. 00-01
Montana Fish, Wildlife & Parks (2014) Metadata for Greater sage-grouse core areas. http://fwp.mt.gov/gisData/metadata/sgcore.htm
Murphy T, Naugle DE, Eardley R, Maestas JD, Griffiths T, Pellant M, Stiver SJ (2013) Trial by fire. Rangelands 35:2–10
Natural Resources Conservation Service (2015a) Outcomes in conservation: sage grouse initiative. U.S. Department of Agriculture, February 2015
Natural Resources Conservation Service (2015b) Sage grouse initiative 2.0: Investment Strategy, FY 2015–2018. U.S. Department of Agriculture, August 2015
Naugle DE, Aldridge CL, Walker BL, Cornish TE, Moynahan BJ, Holloran MJ, Brown K, Johnson GD, Schmidtmann ET, Mayer RT (2004) West Nile virus: pending crisis for greater sage-grouse. Ecol Lett 7:704–713
Naugle DE, Walker BL, Doherty KE (2006) Sage grouse population response to coal-bed natural gas development in the Powder River Basin: interim progress report on region-wide lek-count analyses. University of Montana, Missoula
Naugle DE, Doherty KE, Walker BL, Copel HE, Holloran MJ, Tack JD (2011) Sage grouse and cumulative impacts of energy development. In: Naugle DE (ed) Energy development and wildlife conservation in western North America. Island, Washington, pp 55–70
Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O’Hara RB, Simpson GL, Solymos P, Stevens MH, Wagner H (2015) VEGAN: Community Ecology Package. https://cran.r-project.org/web/packages/vegan/index.html
Oyler-McCance SJ, St. John J (2010) Characterization of small microsatellite loci for use in non-invasive sampling studies of Gunnison sage-grouse (Centrocercus minimus). Conserv Genet Resour 2:17–20
Oyler-McCance SJ, Taylor SE, Quinn TW (2005) A multilocus population genetic survey of the greater sage-grouse across their range. Mol Ecol 14:1293–1310
Parris KM, Schneider A (2009) Impacts of traffic noise and traffic volume on birds of roadside habitats. Ecol Soc 14:29–50
Patterson RL (1952) The sage grouse in Wyoming. Sage Books, Denver
Piertney SB, Höglund J (2001) Polymorphic microsatellite DNA markers in black grouse (Tetrao tetrix). Mol Ecol Notes 1:303–304
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Proctor MF, McLellan BN, Strobeck C, Barclay RM (2005) Genetic analysis reveals demographic fragmentation of grizzly bears yielding vulnerably small populations. Proc R Soc B 272:2409–2416
Remington TE, Braun CE (1985) Sage grouse food selection in winter, North Park, Colorado. J Wildl Manag 49:1055–1061
Ricketts TH (1999) Terrestrial ecoregions of North America: a conservation assessment. Island Press
Robinson AC (2014) Management plan and conservation strategies for greater sage-grouse in North Dakota. North Dakota Game and Fish Department, Bismark
Rosentreter R (2004) Sagebrush identification, ecology, and palatability relative to sage grouse. In: Shaw NL, Monsen SB, Pellant M (eds) Sage grouse habitat restoration symposium proceedings; 4–7 June 2001; Boise, ID. Proceedings RMRS-P-000. Department of Agriculture, Forest Service, Rocky Mountain Research Station, Ft. Collins
Rousset F (2008) Genepop’007: a complete re-implementation of the genepop software for Windows and Linux. Mol Ecol Resour 8:103–106
Rowland MM, Wisdom MJ, Suring LH, Meinke CW (2006) Greater sage-grouse as an umbrella species for sagebrush-associated vertebrates. Biol Conserv 129:323–335
Schrag A, Konrad S, Miller S, Walker B, Forrest S (2011) Climate-change impacts on sagebrush habitat and West Nile virus transmission risk and conservation implications for greater sage-grouse. GeoJournal 76:561–575
Schroeder MA, Aldridge CL, Apa AD, Bohne JR, Braun CE, Bunnell SD, Connelly JW, Deibert PA, Gardner SC, Hilliard MA (2004) Distribution of sage grouse in North America. Condor 106:363–376
Schulwitz S, Bedrosian B, Johnson JA (2014) Low neutral genetic diversity in isolated greater sage-grouse (Centrocercus urophasianus) populations in Northwest Wyoming. Condor 116:560–573
Schwartz MK, McKelvey KS (2009) Why sampling scheme matters: the effect of sampling scheme on landscape genetic results. Conserv Genet 10:441–452
Segelbacher G (2002) Noninvasive genetic analysis in birds: testing reliability of feather samples. Mol Ecol 2:367–369
Segelbacher G, Paxton RJ, Steinbrück G, Trontelj P, Storch I (2000) Characterization of microsatellites in capercaillie Tetrao urogallus (AVES). Mol Ecol 9:1919–1952
Semple K, Wayne R, Gibson R (2001) Microsatellite analysis of female mating behaviour in lek-breeding sage grouse. Mol Ecol 10:2043–2048
Shafer A, Cullingham CI, Cote SD, Coltman DW (2010) Of glaciers and refugia: a decade of study sheds new light on the phylogeography of northwestern North America. Mol Ecol 19:4589–4621
Short Bull AR, Cushman SA, Mace R, Chilton T, Kendall KC, Landguth EL, Schwartz MK, McKelvey K, Allendorf FW, Luikart G (2011) Why replication is important in landscape genetics: American black bear in the Rocky Mountains. Mol Ecol 20:1092–1107
Slabbekoorn H, Ripmeester EA (2008) Birdsong and anthropogenic noise: implications and applications for conservation. Mol Ecol 17:72–83
Smith R (2012) Conserving Montana’s sagebrush highway: long distance migration in sage grouse. Master’s Thesis, The University of Montana
Smits JEG, Fernie KJ (2013) Avian wildlife as sentinels of ecosystem health. Comp Immunol Microbiol Infect Dis 36:333–342
Soltis DE, Gitzendanner MA, Strenge DD, Soltis PS (1997) Chloroplast DNA intraspecific phylogeography of plants from the Pacific Northwest of North America. Plant Syst Evol 206:353–373
Stanton DJ, McArthur ED, Freeman DC, Golenberg EM (2002) No genetic substructuring in Artemisia subgenus Tridentatae despite strong ecotypic subspecies selection. Biochem Syst Ecol 30:579–593
State of Montana (2014) Executive order creating the Montana sage grouse oversight team and the Montana sage grouse habitat conservation program. Executive Order No. 10-2014, pp1–29
State of Montana (2015) Executive order amending and providing for implementation of the Montana sage grouse conservation strategy. Executive Order No. 12–2015, pp 1–38
Stiver SJ, Apa AD, Bohne JR, Bunnell SD, Deibert PA, Gardner SC, Hilliard MA, McCarthy CW, Schroeder MA (2006) Greater Sage-grouse comprehensive conservation strategy. Western Association of Fish and Wildlife, Cheyenne
Sturges HA (1926) The choice of a class interval. J Am Stat Assoc 21:65–66
Taylor SE, Oyler-McCance SJ, Quinn TW (2003) Isolation and characterization of microsatellite loci in Greater sage-grouse (Centrocercus urophasianus). Mol Ecol Notes 3:262–264
Thacker ET, Gardner DR, Messmer TA, Guttery MR, Dahlgren DK (2012) Using gas chromatography to determine winter diets of greater sage-grouse in Utah. J Wildl Manag 76:588–592
U.S. Fish and Wildlife Service (2010) 12-month findings for petitions to list the greater sage-grouse (Centrocercus urophasianus) as threatened or endangered. Fed Reg 75:13910–14014
U.S. Fish and Wildlife Service (2013) greater sage-grouse (Centrocercus urophasianus) conservation objectives: final report. U.S. Fish and Wildlife Service, Denver, February 2013
U.S. Fish and Wildlife Service (2015) 12-Month finding on a petition to list greater sage-grouse (Centrocercus urophasianus) as an endangered or threatened species. Fed Reg 80:59858–59942
Walker BL, Naugle DE, Doherty KE (2007) Greater sage-grouse population response to energy development and habitat loss. J Wildl Manag 71:2644–2654
Wallestad R, Eng RL (1975) Foods of adult sage grouse in Central Montana. J Wildl Manag 39:628–630
Wallestad R, Schladweiler P (1974) Breeding season movements and habitat selection of male sage grouse. J Wildl Manag 38:634–637
Wambolt CL (1996) Mule deer and elk foraging preference for 4 sagebrush taxa. J Range Manag 49:499–503
Webb SL, Olson CV, Dzialak MR, Harju SM, Winstead JB, Lockman D (2012) Landscape features and weather influence nest survival of a ground-nesting bird of conservation concern, the greater sage-grouse, in human-altered environments. Ecol Process 1:1–15
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370
Wiens JA (2007) Foundation papers in landscape ecology. Columbia University Press, New York, p 582
Wright S (1949) The genetical structure of populations. Ann Eugen 15:323–354
Zink RM (2014) Comparison of patterns of genetic variation and demographic history in the greater sage-grouse (Centrocercus urophasianus): relevance for conservation. Open Ornithol J 7:19–29
Acknowledgments
This research would not be possible without the many state, federal, and NGO biologists and technicians who have spent countless hours working tirelessly in the field and behind desks for the conservation of the greater sage-grouse. We would specifically like to acknowledge Jake Chaffin (BLM, now USFS), Ben Deeble (Montana Audubon), Brad Fedy (University of Waterloo), Rick Northrup (MTFWP), Sara Oyler-McCance (USGS), Aaron Robinson (NDGFD), Travis Runia (SDGFP), Dale Tribby (BLM), Catherine Wightman (MTFWP), and David Wood (BLM). Our molecular genetic analyses and manuscript preparation would not have been possible without the assistance of Kevin McKelvey, Kristy Pilgrim, Cory Engkjer and the tireless laboratory efforts of Kara Bates, Nasreen Broomand, Taylor Dowell, Scott Hampton, Randi Lesagonicz, Inga Ortloff, Sara Schwarz, Kate Welch, and Katie Zarn, all of the USFS Rocky Mountain Research Station National Genomics Center for Wildlife and Fish Conservation. Use of trade names does not imply endorsement by the U.S. Government. The views in this article are those of the authors and do not necessarily reflect those of their employers.
Funding
This study was supported by grants from the Montana and Dakotas Bureau of Land Management (07-IA-11221643-343, 10-IA-11221635-027, 14-IA-11221635-059), the Great Northern Landscape Conservation Cooperative (12-IA-11221635-132), and the Natural Resources Conservation Service—Sage Grouse Initiative (13-IA-11221635-054).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Cross, T.B., Naugle, D.E., Carlson, J.C. et al. Hierarchical population structure in greater sage-grouse provides insight into management boundary delineation. Conserv Genet 17, 1417–1433 (2016). https://doi.org/10.1007/s10592-016-0872-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-016-0872-z