Abstract
To detect the region and bleeding frame in the wireless capsule endoscopy video, an automatic computer-aided technique is highly demanded to reduce the burden of physicians. The wireless capsule endoscopy (WCE), is an imaging technology which is recently established and doesn’t require any wired device. This device detects abnormalities in GI tract, i.e. (colon, esophagus, small intestine, and stomach). A WCE video consists of 57,000 images. It is very hard to examine by clinicians. To determine bleeding photos out of fifty-seven thousand WCE images makes the task very hard and expensive. The main goal is to develop an automatic obscure bleeding detection method by using superpixel segmentation and naive Bayes classifier. Naive Bayes and superpixel segmentation are used for this problem.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
One of the primary goals of Wireless Capsule Endoscopy (WCE) is to detect the mucosal abnormalities such as blood, ulcer, polyp, and so on in the gastrointestinal tract. Only less than 5% of total 55,000 frames of a WCE video typically have abnormalities, so it is critical to developing a technique to discriminate abnormal findings from normal ones [1] automatically. We introduce "Bag-of-Visual-Words" method which has been successfully used in particular for image classification in non-medical domains. Initially, the training image patches are represented by color and texture features, and then the bag of words model is constructed by K-means clustering algorithm. Subsequently, the document is described as the histogram of the visual words which is the feature vector of the image. Finally, an SVM classifier is trained using these feature vectors to distinguish pictures with abnormal regions from ones without them [1]. Experimental results on our current dataset show that the proposed method achieves promising performances. Wireless Capsule Endoscopy is a recently established technology to help physicians identify gastrointestinal diseases and abnormalities in the human small intestine [2].
Case endoscopy is a system used to record inward pictures of the gastrointestinal tract for use in therapeutic analysis. The container is comparable fit as a fiddle to a standard pharmaceutical case, despite the fact that somewhat bigger, and contains a minor camera and a variety of LEDs fueled by a battery. After a patient swallows the case, it goes along the gastrointestinal tract taking various pictures every second which are transmitted remotely to a variety of beneficiaries associated with a versatile chronicle gadget conveyed by the patient. The essential utilization of case endoscopy is to look at regions of the small digestive system that can’t be seen by different sorts of endoscopy, for example, colonoscopy or esophagogastroduodenoscopy (EGD).
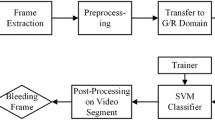
The detection of bleeding regions in the small intestine still an open and crucial issue. Moreover, a good detection classification approach of the blood-based abnormalities in the intestine is missing by the techniques used in this research area according to the gastroenterologists. Thus, this paper offers a novel approach to automatically detect and categorize blood-based abnormalities in the intestine by starting with their definitions. The measure of therapeutic imaging information produced has encountered exponential development, making a tremendous interest in information stockpiling. Cloud services symbolize a massive opportunity. Storing, archiving, partaking and retrieving images in the cloud may allow the industry to handle data more efficiently and cost-effectively while overwhelming many of the legal, regulatory and technological disputes that data needs a pose. Fig. 1 shows the overall storing of medical data.
2 Methods overview
Two common classification methods: SVM and KNN to evaluate their powers in differentiating standard images and blending images. SVM is a supervised machine learning method based on the foundation of statistical learning. The basic idea of the SVM is to find the optimal hyperplane that separates the points of different classes. KNN is a simple and intuitive method widely used in pattern recognition and data mining. This classifier decides on comparing a new testing data with the training data. A two-stage saliency extraction method is proposed to localize the bleeding areas in WCE images. Since these two-stage saliency maps highlight the bleeding regions and separate bleeding mucosa from the uninformative parts that could obtain the bleeding area candidates successfully.
3 Color histogram
A color histogram is a representation of the distribution of colors in an image. The frame shown in Fig. 2 is converted into a grayscale image. Each pixel value present in Fig. 3 characterizes only an amount of light, i.e., it carries only intensity information. The histogram of the image is plotted in Fig. 4. For digital images, a color histogram represents the number of pixels that have colors in each of a fixed list of color ranges that span the image’s color space, the set of all possible colors. Color Histogram is shown in Fig. 5
If the set of possible color values is sufficiently small, each of those colors may be placed on a range by itself; then the histogram is merely the count of pixels that have each possible color. Most often, space is divided into an appropriate number of ranges, often arranged as a regular grid, each containing many similar color values. The color histogram may also be represented and displayed as a smooth function defined over the color space that approximates the pixel counts. Like other kinds of histograms, the color histogram is a statistic that can be viewed as an approximation of an underlying continuous allocation of colors values. It focuses only on the proportion of the number of different types of colors, regardless of the spatial location of the colors. The values of a color histogram are from statistics. They show the statistical distribution of colors and the essential tone of an image.
A large-scale comparison of the naive Bayes classifier with state-of-the-art algorithms for decision tree induction, instance-based learning, and rule induction on standard benchmark datasets, and found it to be sometimes superior to the other learning schemes, even on datasets with substantial feature dependencies. The basic independent Bayes model has been modified in various ways in attempts to improve its performance [6]. Attempts to overcome the independence assumption are mainly based on adding extra edges to include some of the dependencies between the features. In this case, the network has the limitation that each feature can be related to only one other feature. The semi-naive Bayesian classifier is another important attempt to avoid the independence assumption.
The resulting classifier is then used to assign class labels to the testing instances where the values of the predictor features are known, but the value of the class label is unknown. This paper describes various supervised machine learning classification techniques [7]. Of course, a single article cannot be a complete review of all supervised machine learning classification algorithms (also known induction classification algorithms), yet we hope that the references cited will cover the major theoretical issues, guiding the researcher in interesting research directions [3] and suggesting possible bias combinations that have yet to be explored.
The resulting classifier is then used to assign class labels to the testing instances where the values of the predictor features [8] are known, but the value of the class label is unknown. This paper describes various supervised machine learning classification techniques. A single article cannot be a complete review of all supervised machine learning classification algorithms [9] (also known induction classification algorithms), yet that the references cited will cover the major theoretical issues, guiding the researcher in interesting research directions [10] and suggesting possible bias combinations that have yet to be explored.
4 Salient region identification
Detection of salient image regions is useful for applications like image segmentation, adaptive compression [13], and region-based image retrieval. In this paper, we present a novel method to determine salient regions in images using low-level features of luminance and color. Figure 6 shows the segmented output and the bleeding area localized is shown in Fig. 7. The method is fast, easy to implement and generates high-quality saliency maps of the same size and resolution as the input image. We demonstrate the use of the algorithm [15] in the segmentation of semantically meaningful whole objects from digital images. In this case, it is a Naive Bayesian classifier. Although some of the features users have correlation thus ruining the assumption of feature independence, the performance of this classifier [11] is satisfactory. Classification is done per segment. Thus the output of the classifier holds saliency estimation [14] for each segment that is further used to reconstruct the corresponding saliency map using segmentation data. Learning was done using expectation-maximization (EM) learning method [5]. Various features like centroid, area, and eccentricity, etc., are extracted and fed to the classifier.
5 Naïve Bayes classifier
Naïve Bayes classification is a kind of simple probabilistic classification methods based on Bayes’ theorem [17] with the assumption of independence between features [3]. The model is trained on training dataset to make predictions by predict function. This article introduces two functions Naïve Bayes and train for the performance of Naïve Bayes classification. Bayes’ theorem can be used to make a prediction based on prior knowledge and current evidence. With accumulating evidence, the prediction is changed. In technical terms, the prediction is the posterior probability [18] that investigators are interested in. The prior knowledge is termed prior probability that reflects the most probable guess on the outcome without additional evidence.
A complex image registration issue arising while the dependencies between intensities of images to be registered are not spatially homogeneous. Such a situation is frequently encountered in medical imaging when pathology present in one of the images modifies locally intensity dependencies observed on normal tissues. Fig. 8 shows the clustered output from which features are extracted and then fed to the classifier. Usual image registration models, which are based on a single global intensity similarity criterion, fail to register such images, as they are blind to local deviations of intensity dependencies. Such a limitation is also encountered in contrast-enhanced images where there exist multiple pixel classes having different properties of contrast agent absorption. In this paper, we propose a new model in which the similarity criterion is adapted locally to images by classification of image intensity [16] dependencies. Defined in a Bayesian framework, the similarity criterion is a mixture of probability distributions describing dependencies on two classes (Table 1).
The model also includes a class map which locates pixels of the two classes and weighs the two mixture components. The registration problem is formulated both as an energy minimization problem and as a maximum a posteriori estimation problem. It is solved using a gradient descent algorithm [18]. In the problem formulation and resolution, the image deformation and the class map are estimated simultaneously, leading to an original combination of registration and classification that we call image classifying registration. Whenever sufficient information about the class location is available in applications, the registration can also be performed on its own by fixing a given class map. Finally, we illustrate the interest of our model on two real applications from medical imaging: template-based segmentation of contrast-enhanced images and lesion detection in mammograms. We also conduct an evaluation of our model on simulated medical data and show its ability to take into account spatial variations of intensity dependencies while keeping good registration accuracy [4].
6 Simulation results
Input Frame

Gray Image

Histogram

7 Conclusion
An effective method to find the bleeding frame in an Endoscopic video is obtained. Our result shows that the proposed method has obtained more accurately. The CMYK color format helps in identification of the bleeding in a clear manner. The method is applied for numerous frames of few endoscopic videos and found effective.
References
Fu, Y., Zhang, W., Mandal, M., Meng, M.Q.: Computer-aided bleeding detection in WCE video. IEEE J. Biomed. Health Inform. 18(2), 636–42 (2014)
Pan, G., Yan, G., Qiu, X., Cui, J.: Bleeding detection in wireless capsule endoscopy based on probabilistic neural network. J. Med. Syst. 35(6), 1477–1484 (2011)
Al-Rahayfeh, A.A., Abuzneid, A.A.: Detection of bleeding in wireless capsule endoscopy images using range ratio color. Int. J. Multimed. Appl. 2(2), 1–10 (2010)
Hachama, M., Desolneux, A., Richard, F.J.: Bayesian technique for image classifying registration. IEEE Trans. Image Process. 21(9), 4080–4091 (2012)
Amin, M.M., Kermani, S., Talebi, A., Oghli, M.G.: Recognition of acute lymphoblastic leukemia cells in microscopic images using K-means clustering and support vector machine classifier. J. Med. Signals Sens. 5(1), 49–58 (2015)
Dilna, C., Gopi, V.P.: A novel method for bleeding detection in Wireless Capsule Endoscopic images. In: IEEE International Conference on Computing and Network Communications (CoCoNet), (2015). https://doi.org/10.1109/CoCoNet.2015.7411289
Maghsoudi, O.H., Alizadeh, M., Mirmomen, M.A.: A computer-aided method to detect bleeding, tumor, and disease regions in Wireless Capsule Endoscopy. In: IEEE Conference on Signal Processing in Medicine and Biology Symposium (SPMB), (2016). https://doi.org/10.1109/SPMB.2016.7846852
Yuan, Y., Li, B., Meng, M.Q.: Bleeding frame and region detection in the wireless capsule endoscopy video. IEEE J. Biomed. Health Inform. 20(2), 624–630 (2016)
Suman, S., Malik, A.S., Riegler, M., Ho, S.H., Hilmi, I., Goh, K.L.: Detection and classification of bleeding region in WCE images using color feature. In: Proceedings of the 15th International Workshop on Content-Based Multimedia Indexing, Article No. 17 (2017)
Adam, N., Tachecí, I., Sulík, L., Bureš, J., Krejcar, O.: Automatic blood detection in capsule endoscopy video. J. Biomed. Opt. 21(12), 126007 (2016)
Pan, G.B., Yan, G.Z., Song, X.S., Qiu, X.L.: Bleeding detection from wireless capsule endoscopy images using improved Euler distance in CIELab. J. Shanghai Jiaotong Univ. (Science) 15(2), 218–223 (2010)
Unnimadhavan, R.: Automated bleeding detection in wireless capsule endoscopy videos. J. Biomed. 5(8), 218–224 (2017)
Maghsoudi, O.H., Talebpour, A., Soltanian-Zadeh, H., Alizadeh, M., Soleimani, H.A.: Informative and uninformative regions detection in WCE frames. J. Adv. Comput. 3(1), 12–34 (2014)
Priya, K., Archana, K.S., Neduncheliyanm, S.: Bleeding detection through wireless capsule endoscopy (WCE). Int. J. Adv. Comput. Technol. (IJACT) 4(1), 5–13 (2015)
Hwang, S., Celebi, M.E.: Polyp detection in wireless capsule endoscopy videos based on image segmentation and geometric feature. In: IEEE International Conference on Acoustics Speech and Signal Processing (ICASSP), (2010)
Murthi, A., Suganya, D.: Automatic bleeding frame and region detection for GLCM using artificial neural network. J. Adv. Chem. 12(24), 5613–5620 (2016)
Ashok, V., Murugesan, G.: Detection of retinal area from scanning laser ophthalmoscope images (SLO) using deep neural network. Int. J. Biomed. Eng. Technol. 23(2–4), 303–314 (2017)
Coimbra, M., Mackiewicz, M., Fisher, M., Jamieson, C., Soares, J., Silva Cunha, J.P.: Computer vision tools for capsule endoscopy exam analysis. Eurasip NewsLetter 18(1), 1–19 (2007)
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Sivakumar, P., Kumar, B.M. A novel method to detect bleeding frame and region in wireless capsule endoscopy video. Cluster Comput 22 (Suppl 5), 12219–12225 (2019). https://doi.org/10.1007/s10586-017-1584-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10586-017-1584-y