Abstract
Centromere sequences exist as gaps in many genome assemblies due to their repetitive nature. Here we take an unbiased approach utilizing centromere protein A (CENP-A) chomatin immunoprecipitation followed by high-throughput sequencing to identify the centromeric repeat sequence in the threespine stickleback fish (Gasterosteus aculeatus). A 186-bp, AT-rich repeat was validated as centromeric using both fluorescence in situ hybridization (FISH) and immunofluorescence combined with FISH (IF-FISH) on interphase nuclei and metaphase spreads. This repeat hybridizes strongly to the centromere on all chromosomes, with the exception of weak hybridization to the Y chromosome. Together, our work provides the first validated sequence information for the threespine stickleback centromere.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Proper segregation of chromosomes is essential for the faithful and equal separation of each chromosome during both mitosis and meiosis. Mis-segregation of chromosomes can lead to aneuploidy, which is often associated with cancer, miscarriage, and birth defects such as viable trisomies (Kops et al. 2005; Morales et al. 2007; Hunt and Hassold 2010; Lister et al. 2010; Revenkova et al. 2010; Gordon et al. 2012; Ricke and van Deursen 2013). Physical separation of chromosomes occurs by the attachment of microtubule spindle fibers via the kinetochore to a primary constriction on each chromosome, called the centromere. Because of the vital role centromeres play in cell division, each chromosome must contain only a single functional centromere. Functional centromeres are defined by the presence of a centromeric histone variant known as either centromeric histone H3 (CenH3) or centromere protein A (CENP-A) (Palmer et al. 1991; Sullivan et al. 1994). CENP-A replaces histone H3 at functional centromeres and localizes to the primary constriction on metaphase chromosome spreads (Earnshaw and Rothfield 1985; Palmer et al. 1989; Sullivan and Schwartz 1995; Warburton et al. 1997; Amor et al. 2004).
While the presence of CENP-A at functional centromeres is almost entirely conserved among eukaryotes (Henikoff et al. 2001; Malik and Henikoff 2009; Drinnenberg et al. 2014), the genetic sequence at centromeres varies dramatically between species (Henikoff et al. 2001; Alkan et al. 2011; Melters et al. 2013) and even among chromosomes of the same species or closely related species (Shang et al. 2010; Tek et al. 2010; Piras et al. 2010; Gong et al. 2012). Despite variation at the sequence level, most centromere sequences have conserved characteristics, such as being highly repetitive and AT-rich (Melters et al. 2013). In addition, many mammalian centromere repeats contain a conserved centromere protein B (CENP-B) box, a 17-bp DNA motif that recruits the CENP-B protein, thought to aid in de novo centromere formation (Masumoto et al. 1989; Ohzeki et al. 2002; Alkan et al. 2011). Previous methods used to identify centromere sequences include isolation of satellite DNA, restriction digest approaches, and bacterial artificial chromosome (BAC) cloning (Maio 1971; Manuelidis 1978; Haaf et al. 1993; Garrido-Ramos et al. 1994; Crollius et al. 2000; Edwards and Murray 2005; Tek et al. 2010; Shang et al. 2010). However, these approaches can be limited by the restriction enzymes used, by biases against cloning repetitive DNA, and assumptions that the most repetitive sequences in the genome will be the centromere. Chromatin immunoprecipitation (ChIP) with a CENP-A antibody followed by cloning of the ChIP DNA has recently been used to both confirm putative centromere sequences (Zhong 2002; Nagaki et al. 2003, 2004; Nagaki and Murata 2005; Houben et al. 2007), and to identify novel repeats (Lee et al. 2005; Edwards and Murray 2005; Nagaki et al. 2008; Tek et al. 2010; Shang et al. 2010; Alkan et al. 2011). However, CENP-A ChIP cloning is limited both by the number of clones that can be sequenced and biases against cloning repetitive DNA. Chromatin immunoprecipitation with a CENP-A antibody followed by high-throughput sequencing (ChIP-seq) is the most unbiased approach to identify functional centromeric associated DNA (Henikoff et al. 2015), but it has not yet been extensively used (Gong et al. 2012; Henikoff et al. 2015).
Here we sought to identify the centromere sequence in threespine stickleback fish (Gasterosteus aculeatus), an emerging model organism used to study the genetic, genomic, and molecular basis for evolution (Kingsley and Peichel 2007). The threespine stickleback has an assembled genome (Jones et al. 2012), yet the centromere sequence is still unknown. Gaps in the genome assembly correspond with the cytological constriction on most chromosomes (Urton et al. 2011), suggesting that the stickleback centromere is comprised of repetitive sequences (Henikoff 2002; Rudd and Willard 2004). To identify the threespine stickleback centromere sequence, we first identified the complete threespine stickleback CENP-A coding sequence and then performed ChIP-seq using a threespine stickleback specific CENP-A antibody. We confirmed that this 186-bp, AT-rich sequence is centromeric using both fluorescence in situ hybridization (FISH) and CENP-A immunofluorescence coupled with FISH (IF-FISH).
Materials and methods
Fish use and care
Three populations of G. aculeatus were used in this study: wild caught fish from Lake Union (Seattle, WA, USA), laboratory-reared Pacific Ocean fish, and laboratory-reared Japan Sea fish. Both of these laboratory populations were derived from wild-caught fish collected in Akkeshi on Hokkaido Island, Japan (Kitano et al. 2007, 2009). Fish were housed in 29-gal aquarium tanks in summer lighting conditions (16-h light, 8-h dark) at approximately 16 °C in 0.35 % saltwater (3.5 g/l Instant Ocean salt (Spectrum Brands, USA); 0.4 ml/l sodium bicarbonate). Fish were fed with live brine shrimp nauplii and frozen Mysis shrimp two times daily. All institutional and national guidelines for the care and use of laboratory animals were followed, and all procedures were approved by the Fred Hutchinson Cancer Research Center Institutional Animal Care and Use Committee (protocol 1575). Fish were caught from Lake Union with permission of the Washington Department of Fish and Wildlife (Scientific Collection permits 12–057, 13–039, 14–065, and 15–033).
Determining the Cenpa coding sequence
To find the putative G. aculeatus Cenpa gene, the Danio rerio CENP-A protein sequence was used to search for a homologous sequence in the threespine stickleback genome assembly (Ensembl BROAD S1; Feb 2006) using BLASTP. The putative threespine stickleback CENP-A protein (ENSGACP00000024534) is a product of the gene (ENSGACG00000018561.1). However, this predicted gene was missing a stop codon. To determine the full Cenpa coding sequence, RNA was extracted using Trizol (Life Technologies, USA) from a Japan Sea male stickleback kidney. An Invitrogen rapid amplification of cDNA ends (3′ RACE) kit (Life Technologies, USA) was used to make cDNA. Internal primer JC06 (5′-GAGAGGTGTGCCAGAGCTTCTC-3′) was used in conjunction with the 3′ RACE kit primer AUAP to amplify and sequence the entire 3′ end of the Cenpa gene. This coding sequence was used to design primers (5′-ATGCGTCACAATTCATCTACC-3′ and 5′-TTACAGGTTGTCCACCCC-3′) to amplify the entire cDNA. To determine whether the Japan Sea and Pacific Ocean Cenpa cDNA sequences are the same, Pacific Ocean male RNA was extracted from liver tissue using Trizol, cDNA was synthesized using the SuperScript III First-Strand Synthesis System Kit (Life Technologies, USA), and the Cenpa cDNA was Sanger-sequenced following polymerase chain reaction (PCR) amplification with the primers above.
Antibody design
A G. aculeatus CENP-A specific antibody was designed to amino acids 1–22 (MRHNSSTSRRKGKTPQHRPPLA) of the N-terminal end of the threespine stickleback CENP-A protein (Supplementary Fig. S1b). The rabbit IgG affinity purified polyclonal antibody was produced by Covance Research Products (USA).
Western blot
Fifty milligrams of Pacific Ocean female kidney and liver tissue was dissected in 400 μl radioimmunoprecipitation assay buffer (RIPA) (150 mM sodium chloride, 1 % NP-40, 0.5 % sodium deoxycholate, 0.1 % sodium dodecyl sulfate (SDS), 50 μM Tris pH 8.0, 0.3 mM phenylmethanesulfonylfluoride (PMSF)). Using a pellet pestle, tissue was homogenized in 400 μl RIPA and 0.3 mM PMSF on ice. The pestle was rinsed with an additional 600 μl RIPA with PMSF, and then the sample was rotated 2 h at 4 °C. Cells were spun for 20 min at 12,000 rpm and 4 °C. The pellet was resuspended in 30 μl of 2 % SDS, and 6 μl of 6× Laemmli buffer was added. The samples were boiled at 95 °C for 5 min and then run on a 15 % sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gel in running buffer (25 mM Tris, 0.192 M glycine, 0.2 % SDS). Proteins were transferred from the gel to a polyvinylidene difluoride membrane for 1 h at 14 V in transfer buffer (25 mM Tris, 192 mM glycine, 10 % methanol). The membrane was blocked in I-BLOCK (Applied Biosystems, USA) for 1 h. The membrane was incubated with the primary antibody (rabbit, anti-stickleback CENP-A) at 1:1000 in I-BLOCK plus 5 % bovine serum albumin (BSA) on a shaking nutator overnight. The membrane was washed 3 × 5 min with phosphate-buffered saline Tween-20 (PBST) and then incubated with alkaline phosphatase conjugated goat, anti-rabbit secondary antibody (Applied Biosystems, USA) at 1:10,000 in I-BLOCK for 1 h. The membrane was washed 2 × 5 min with PBST and then 2 × 5 min in assay buffer (20 mM Tris pH 9.8, 1 mM magnesium chloride). A few drops of CDP-Star Chemiluminescent Substrate (Sigma-Aldrich, USA) was added to membrane. Film was exposed for 5 s and then developed.
Immunoflourescence on metaphase spreads and interphase nuclei
Immunofluorescence was conducted using a protocol adapted from Blower et al. (2002). For metaphase spreads, Lake Union male spleen tissue was homogenized in 1 ml of 0.56 % potassium chloride (KCl) with a glass dounce. For interphase nuclei, 15 Pacific Ocean embryos at 48 days post-fertilization (dpf) were homogenized in 10 ml of 0.56 % KCl with a glass dounce. Cells were incubated for 45 min on ice to swell, and 350 μl (metaphase spreads) or 250 μl (interphase nuclei) of the cell suspension was spun for 10 min at 1950 rpm onto Fisherfinest Premium Superfrost microscope slides through a Fisher single cytology funnel (Fisher Scientific, USA) using a Cytospin 3 (Shandon, USA). Slides were immediately placed in ice-cold methanol for 20 min followed by a 10-min room temperature incubation in 4 % formaldehyde in phosphate-buffered saline (PBS). Slides were washed 10 min in PBST (PBS, 0.05 % Tween-20) and then incubated with block solution (PBST, 1 % BSA, 0.02 % sodium azide) for 20 min at room temperature. Primary antibody (rabbit, anti-stickleback CENP-A) was added at 1:300 in block solution for 4 h at room temperature. Slides were washed 3 × 5 min in PBST at room temperature. A secondary Alexa Fluor 488 chicken, anti-rabbit IgG antibody (Life Technologies, USA) was added at 1:500 in block solution and incubated overnight at 4 °C. Slides were washed 3 × 5 min in PBST and then counterstained with vectashield 4’,6’-diamidino-2-phenlyindole (DAPI) stain (Vector Laboratories, USA) for 10 min before imaging as described in the “Microscopy” section.
CENP-A chromatin immunoprecipitation
Chromatin preparation from tissue
The ChIP protocol was performed using the SimpleChIP Plus Enzymatic Chromatin IP Kit (Magnetic Beads; Cell Signaling Technology, USA). Approximately 0.1 g of kidney and liver tissue was dissected from two Pacific Ocean females for two independent samples. The tissue was placed into 2 ml of 1× protease inhibitor cocktail (PIC) buffer. The tissue was dounced three times with a glass dounce, and the dounce was rinsed with an additional 2 ml of 1× PIC buffer. Four milliliters of the tissue suspension was incubated with 180 μl of 37 % formaldehyde for 20 min at room temperature on a nutator. From this point, the Simple ChIP protocol was followed with the following modifications: cells were incubated for 30 min at 37 °C with 1.6 μl of micrococcal nuclease (MNase) to create mononucleosomes and dinucleosomes. MNase treatment was stopped with addition of 40 μl 0.5 M ethylenediaminetetraacetic acid (EDTA). Cells were spun at 13,000 rpm for 1 min at 4 °C and resuspended in 400 μl 1× ChIP buffer. Nuclear membranes were disrupted with 3 × 30 s sonication using a probe sonicator on high, with a 30-s incubation on wet ice between pulses. Lysates were clarified by spinning at 10,000 rpm for 10 min at 4 °C. Thirty microliters of the supernatant was taken for chromatin analysis and quantification, and the rest was stored at −80 °C until immunopreciptation (IP).
Immunoprecipitation
For each sample, 10 μg chromatin was diluted in 500 μl 1× ChIP buffer. For each sample, 10 μl was taken and stored at −20 °C for use as the input samples. For each 10 μg IP, chromatin was incubated with 4.64 μg of rabbit, anti-stickleback CENP-A antibody in 1× ChIP buffer overnight at 4 °C with rotation. Thirty microliters of magnetic beads was added to each 10 μg IP. Following a 2-h incubation at 4 °C with rotation, experimental methods were carried out following the Simple ChiP Kit instructions. Input and IP DNA was purified using a MinElute PCR Purification Kit (Qiagen, USA). Two IP and two input samples were each eluted in 11 μl EB buffer. Paired end 150-bp sequencing was performed on the Illumina HiSeq 2500 (Illumina, USA) at the Fred Hutchinson Cancer Research Center Genomics Shared Resource. Fastq files containing the raw sequencing data have been deposited to the NCBI Sequence Read Archive (study accession SRP063504).
ChIP analysis
CENP-A ChIP data was analyzed using the pipeline described by (Henikoff et al. 2015). Paired reads were merged using SeqPrep (https://github.com/jstjohn/SeqPrep). We then discarded any CENP-A ChIP reads that aligned to the BROADS1 assembly of the G. aculeatus genome (Jones et al. 2012), using bwa v0.7.12 (Li and Durbin 2009). The remaining merged pairs (9,182,498 from Pacific Ocean female 1 (POF1) and 10,208,973 from Pacific Ocean female 2 (POF2)) were run through CD-HIT-EST (Li and Godzik 2006; Fu et al. 2012) to create a set of the most abundant IP clusters, with each cluster sharing at least 90 % sequence identity. The longest read for each cluster was used a reference sequence for that cluster. The clusters were ranked by the number of reads, and the top 500 cluster reference sequences were identified. For each cluster reference sequence, we first counted the number of reads in both the total IP and input samples (all mapped and unmapped reads) and then normalized these counts by calculating the reads per million (RPM) for both the IP and input samples. We calculated the fold enrichment of each cluster reference sequence in the IP relative to input by dividing the IP RPM by the input RPM. The top 500 most abundant cluster references were then ranked by order of fold enrichment. Sequences with a fold enrichment of less than 1 were discarded. The total average fold enrichment for all of the top enriched clusters combined was calculated by dividing the total IP RPM by the total input RPM.
The cluster reference sequences determined from above were aligned to the most abundant cluster sequence (Supplementary Fig. S2). The Geneious (Biomatters, New Zealand) sequence global alignment program (parameter: cost matrix 65 % similarity, gap open penalty = 12, gap extension penalty = 3) was used for alignments. From this alignment, a consensus sequence repeating unit was determined using the >50 % identity “strict” parameter in Geneious. A consensus repeating unit was determined for each independent IP sample. The two consensus repeats were then aligned to create a single consensus centromeric repeat sequence (CEN) (Supplementary Fig. S3).
To find the percentage of reads from each ChIP experiment that contained sequences with homology to the CEN, we used BWA-MEM (Li 2013) to align all reads from either the input or the IP to the consensus repeat sequence. Only reads with a score >30 were counted where the score is dependent on the following: B INT ([4]), O INT [,INT] ([6,6]), E INT [,INT] (‘{O} + {E}*k’[1,1], and L INT [,INT] ([5,5]).
To find assembled scaffolds that contain sequence homologous to the centromeric repeat sequence, we conducted a BLAST search for the CEN sequence in the threespine stickleback genome assembly (Ensembl BROAD S1; Feb 2006). We identified sequences with homology to the CEN sequence in scaffolds from regions of the genome that were not assigned to chromosome assemblies and in scaffolds at the edges of gaps in chromosome assemblies that correspond to the putative position of centromeres as determined using the p/q chromosome arm length ratios (Urton et al. 2011). Up to 5 kb of sequence data from each region was extracted, and the consensus centromeric repeat was aligned to these sequences in Geneious (Biomatters, New Zealand) to identify tandem repeats and to determine percent identity between the consensus centromere repeat and the repeats present in the genome assembly.
Fluorescence in situ hybridization on metaphase spreads and interphase nuclei
CEN FISH probe
PCR primers JC103 (5′-GGTGCTAGATTTAGGAAAACA-3′) and JC106 (5′-GTGCATTCATGACTTTTAAGG-3′) were used to amplify the threespine stickleback centromeric repeat from genomic DNA. The PCR product appeared as a ladder. The entire ladder was extracted using the QIAquick gel Extraction Kit (Qiagen, USA). The purified PCR product was cloned into the PCR2.1 vector (Life Technologies, USA). The clone used as a template to make the FISH probe contained 1.5 copies of the centromeric repeat. This clone was amplified using primers JC103 and JC106 with PCR fluorescein labeling mix (Roche Applied Science, Switzerland) to make a fluorescein-12-dUTP-labeled probe of 288 bp. The probe was lyophilized at 55 °C for 3 h and then resuspended in 10 μl hybridization buffer (50 % formamide, 2× saline-sodium citrate (SSC), 10 % dextran sulfate, 0.2 mM ethylenediaminetetraacetic acid (EDTA), 2 mM Tris, pH 8.0). The probe was denatured at 72 °C for 5 min and then stored at 37 °C until use.
Sex chromosome FISH probe
The G. aculeatus BAC clone CHORI-213 101E08 was used as a FISH probe to distinguish the X and Y chromosomes. This BAC clone hybridizes to the end of the p arm of the Y chromosome and to the middle of the p arm of the X chromosome (Ross and Peichel 2008). The BAC FISH probe was labeled with Alexa 568 following the protocol in Urton et al. (2011).
Metaphase spreads and interphase nuclei
Wild-caught fish from Lake Union were injected with 10 μl of 1 % colchicine in PBS for 16 h to arrest mitotic cells. Spleen and liver tissue was dissected and homogenized in 2 ml of 0.56 % KCl using a glass dounce. Metaphase spreads and interphase nuclei were prepared following the protocol described in Ross and Peichel (2008).
Hybridization
Slides were washed in 2× SSC for 5 min, and then in denature solution (2× SSC, 70 % formamide) for 2 min in a 72 °C water bath. Slides were dehydrated in 70 % cold ethanol for 2 min, followed by a room temperature dehydration series for 2 min each in 85, 90, and 100 % ethanol. After the slides were air-dried for 5 min, 10 μl of either the CEN probe, the sex chromosome probe, or both was added. Then, the slides were covered with a coverslip, sealed with rubber cement, and hybridized at 37 °C overnight in a humid chamber. Slides were washed in a 42 °C water bath 3 × 5 min in wash solution 1 (2× SSC, 50 % formamide), followed by 3 × 5 min in 2× SSC. Slides were air-dried for 5 min and then counterstained and mounted with vectashield DAPI stain. Images were taken as described in the “Microscopy” section.
Immunofluorescence-FISH
For IF-FISH, 15 Lake Union embryos at 48 dpf were dounced in 10 ml 0.56 % KCl and 0.1 % Tween-20 using a glass dounce. Metaphase and interphase nuclei slides were prepared following the protocol in the “Immunofluorescence” section, with the following modifications: primary rabbit, anti-stickleback CENP-A antibody was diluted 1:100 in block, and 100 μl was added to the slide, which was covered with a glass coverslip and incubated in a humid chamber overnight at 4 °C. Slides were washed 3 × 5 min with PBST. The secondary goat, anti-rabbit 568 antibody (Invitrogen, USA) was diluted 1:600 in block, and 100 μl was added to the slide and incubated for 4 h at room temperature in the dark. Slides were washed 3 × 5 min in PBST and then post-fixed with 4 % formaldehyde in PBS for 10 min at room temperature. Ten microliters of PCR-labeled CEN probe was lyophilized at 55 °C for 4 h and resuspended in 10 μl of hybridization buffer (50 % formamide, 2× SSC, 10 % dextran sulfate, 20 % TE, pH 8). The probe was denatured at 72 °C for 5 min in a water bath and then put at 37 °C until use. Hybridization with the CEN probe was performed following the protocol in the “Hybridization” section.
Microscopy
Slides were imaged using a Nikon Eclipse 80i microscope (Nikon, Japan) with an automated filter turret (Chroma filters 31000v2 (DAPI), 41001 (FITC), and 41004 (Texas Red); Chroma, USA). Images were taken using the 100× objective, using the Photometrics CoolSNAP ES2 camera (Photometrics, USA). NIS Elements imaging software (BR 3.00, SP7, Hotfix8, Build 548, Nikon, Japan) was used to pseudo-color the images. For CENP-A immunofluorescence images, the Alexa Fluor-488-labeled antibody, and DAPI were pseudo-colored purple and grey, respectively. For FISH images, the Alexa Fluor 568–labeled BAC probe and DAPI were pseudo-colored purple and grey, respectively; the fluorescein-labeled CEN probe is green. For IF-FISH images, the Alexa Flour 568–labeled CENP-A antibody, and DAPI were pseudo-colored purple and grey, respectively.
Results
Identification of the threespine stickleback CENP-A coding sequence
Using 3′ RACE, PCR, and cDNA sequencing, we identified the entire coding sequence of the threespine stickleback CENP-A gene, which is 447 bp and encodes a protein of 148 amino acids (Supplementary Fig. S1a). Interestingly, the amino acid sequences between Pacific Ocean and Japan Sea populations differ by a single amino acid at position 36 (GenBank accession numbers KT321854 (Pacific Ocean), KT321855 (Japan Sea)). The Pacific Ocean sequence is used as the reference here. Like other known CenH3-like proteins, the threespine stickleback CENP-A protein has an N-terminal tail that is highly diverged from the threespine stickleback H3 protein (Supplementary Fig. S1b). The N-terminal tail is also highly diverged among CENP-A proteins from different fish species, while the C-terminal histone fold domain is more conserved (Fountain and Kral 2011).
Threespine stickleback CENP-A is centromere specific
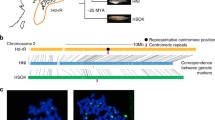
To determine whether the threespine stickleback CENP-A protein is a centromeric histone variant, we designed an antibody that would recognize the N-terminal end of the CENP-A, but not the H3, protein (Supplementary Fig. S1b). A Western blot on protein lysate from threespine stickleback tissue shows that the CENP-A antibody is specific to a prominent band of around 17 kD, consistent with its predicted atomic weight of 16.7 kD (Supplementary Fig. S1c). The antibody hybridizes to punctate spots on interphase nuclei (Fig. 1a) and to the primary constriction of all chromosomes on metaphase spreads (Fig. 1b, c). Taken together, these data suggest that this is a bona fide centromeric histone.
The threespine stickleback CENP-A antibody localizes to the centromere. Immunofluorescence reveals that the CENP-A antibody (purple) hybridizes to a distinct puncta in interphase nuclei from Pacific Ocean embryos, b a single region on each chromosome in a metaphase spread from a Lake Union male, and c the primary constriction (arrowheads) on each chromosome in a higher magnification view of the boxed region in panel (b). Scale bar, 5 μm
CENP-A-associated sequences in threespine stickleback
To identify the centromere sequence, we performed CENP-A ChIP-seq in two independent female fish from the Pacific Ocean population. Using the cluster analysis pipeline described in Henikoff et al. (2015), we identified the 500 most abundant sequences in each IP sample and then determined that the majority of these sequences (320 in POF1 and 299 in POF2) were enriched in the IP sequences relative to input sequences (Table 1). After aligning these enriched sequences (Supplementary Fig. S2), we found a single consensus repeating unit for each individual IP sample. These two consensus repeats are 99.5 % identical and were aligned to create a single consensus repeat (Supplementary Fig. S3). The consensus repeat is 186 bp with an AT content of 62.9 % (Fig. 2a) and was 17.04- or 17.68-fold enriched in the IP relative to input in two independent ChIP-seq experiments (Table 1). This repeat made up 4.43 and 4.29 % of total reads from each input sample, highlighting its overall abundance in the genome. Like many other centromeric repeats, this consensus repeat is AT-rich (Melters et al. 2013), contains a putative centromere protein B (CENP-B) box (Fig. 2b) (Masumoto et al. 2004; Edwards and Murray 2005; Henikoff et al. 2015), and has a sequence length consistent with wrapping around a single nucleosome (Willard 1991; Shelby et al. 1997; Henikoff et al. 2001).
Threespine stickleback centromere repeat sequence. a The consensus GacCEN repeat is 186 bp and has an AT content of 62.9 %. The putative CENP-B box is highlighted in bold. b The putative CENP-B box in the GacCEN shows sequence similarity to the CENP-B box in human (Homo sapiens), dog (Canis familiaris), platypus (Ornithorhynchus anatinus), and the African clawed frog (Xenopus laevis). Identical nucleotides to the human CENP-B box are in red, and asterisks denote the evolutionary conserved domains in humans (Ohzeki et al. 2002; Alkan et al. 2011). This sequence has been deposited in GenBank (accession number KT321856)
Nearly all of the sequences (316/320 from POF1; 289/299 from POF2) that were enriched in the CENP-A IP relative to the input aligned to the consensus repeat (Table 1). The few enriched sequences that did not align to the consensus repeat (4 from POF1; 10 from POF2) did not align to each other and were low in abundance, with less than 150 total reads in the IP and 84 total reads in the input. It is possible that these low-abundance sequences are bound by CENP-A and are interspersed among long stretches of the consensus tandem repeat. However, our data suggests that the majority of sequences bound to CENP-A in the threespine stickleback are variants of the consensus repeat that we identified (Table 1).
The CENP-A-associated sequence is centromere specific
To determine whether the sequence is the bona fide centromere, we generated a fluorescently labeled probe of the consensus repeat sequence identified by ChIP-seq and found that it hybridizes to distinct loci in interphase nuclei (Fig. 3a), as well as to the constriction on 41 of 42 metaphase chromosomes in male threespine sticklebacks (Fig. 3b, c). IF-FISH also shows that the centromere repeat co-localizes with CENP-A in both interphase nuclei (Fig. 4a), as well as on metaphase spreads (Fig. 4b, c). All of these results suggest that the 186-bp repeat, now referred to as GacCEN (threespine stickleback (G. aculeatus) centromeric repeat sequence; GenBank accession number KT321856), is the centromeric repeat in threespine stickleback fish.
The threespine stickleback CEN repeat hybridizes to the centromere. FISH reveals that the CEN probe (green) hybridizes to a distinct puncta in interphase nuclei from Lake Union embryos, b a single region on each chromosome except one (arrow) in a metaphase spread from a Lake Union male, and c the primary constriction (arrowheads) on each chromosome in a higher magnification view of the boxed region in panel (b). Scale bar, 5 μm
The threespine stickleback centromere repeat colocalizes with CENP-A. IF-FISH with CENP-A antibody (purple) shows co-localization with the CEN probe (green) in a interphase nuclei and b, c metaphase chromosomes from Lake Union embryos. Panel (c) shows a higher magnification view of the boxed region in panel (b). Scale bar, 5 μm
The GacCEN probe shows weak hybridization to the Y chromosome
In mammals, the Y chromosome centromere can differ from the centromere on the other chromosomes. For example, the house mouse Y centromere is comprised of a novel satellite repeat (Pertile et al. 2009), and the human Y centromere is a divergent alpha satellite repeat (Wolfe et al. 1985; Miga et al. 2014). Thus, we hypothesized that the single chromosome that did not hybridize to the GacCEN probe in males was the Y (Fig. 3b). Indeed, when we performed FISH with the GacCEN probe and a BAC probe that distinguishes the X and Y chromosomes (Ross and Peichel 2008), we found that the Y chromosome centromere shows very weak hybridization to this centromeric repeat, while the X chromosome shows strong hybridization (Fig. 5).
The threespine stickleback centromere repeat shows weak hybridization to the Y chromosome centromere. a FISH reveals that the CEN probe (green) shows weak hybridization to the Y chromosome but strong hybridization to the X chromosome on a metaphase spread from a Pacific Ocean male. The X and Y chromosomes can be identified and distinguished by hybridization to a FISH probe from BAC 101E08 (purple) (Ross and Peichel 2008). Higher magnification views of hybridization to the X (b) and Y (c) chromosomes are shown. Scale bar, 5 μm
GacCEN repeat shows a decrease in percent identity outside of the core centromere
We searched for sequences with homology to the consensus centromeric repeat in the current stickleback genome assembly (Jones et al. 2012; Ensembl BROADS1) to see if we could uncover any large scaffolds containing the GacCEN repeat. We found multiple copies of a similar repeat on four scaffolds that did not map to chromosome assemblies and at the edges of ten different scaffolds from nine assembled chromosomes, corresponding to the locations of centromeric constrictions in metaphase spreads (Urton et al. 2011). As predicted (Smith 1976; Schueler et al. 2001; Henikoff 2002; Shepelev et al. 2009; Henikoff et al. 2015), the repeats found of the edge of the assembled contigs show a decrease in percent identity to the GacCEN as they move away from the core centromere (Supplementary Fig. S4). None of the GacCEN containing contigs appear to have any higher-order repeat (HOR) structure. However, until longer centromere-containing reads are obtained, it will be difficult to determine whether stickleback centromeres in fact have a HOR structure.
Discussion
By identifying the complete coding sequence of the threespine stickleback CENP-A protein and designing a species-specific antibody, we were able to perform an unbiased CENP-A ChIP-seq experiment to identify the threespine stickleback centromere repeat. We validated this repeat as centromeric by performing FISH, as well as IF-FISH with the CENP-A antibody on metaphase spreads. This repeat shares similar characteristics to other centromere repeats in its size, AT content, and putative CENP-B box (Alkan et al. 2011; Melters et al. 2013). It should be noted that the putative G. aculeatus CENP-B box shows weak conservation to the evolutionarily conserved human CENP-B domains (Alkan et al. 2011) yet shows the same number of conserved bases as the putative dog, platypus, and Xenopus CENP-B boxes (Fig. 2b). The lack of a fully conserved CENP-B box was consistent across all individual cluster repeats, suggesting there are no small subsets of active CENP-B box containing repeats. Interestingly, we also find no evidence for a CENP-B gene in the G. aculeatus genome assembly. Although it is not clear whether the CENP-B box in the GacCEN is functional, it is interesting that the repeat shares similar sequence characteristics with other known centromeric repeats.
A previous study (Melters et al. 2013) used a bioinformatic pipeline to identify candidate centromere sequences in 282 species, including two putative centromere sequences of 313 and 186 bp in threespine stickleback (Table S4 in Melters et al. 2013). While they could not identify the source of the discordance between the two potential repeats, they assumed that the 313-bp sequence was the centromere because of its higher abundance in the genome. However, we tested this repeat using FISH and found that it does not hybridize to the centromere on metaphase chromosomes (data not shown). In fact, the 186-bp repeat that they identified is 93.0 % similar to the GacCEN repeat identified in our study (D. Melters personal communication, July 7, 2015). These results underscore the importance of FISH and/or CENP-A ChIP validation to discern between multiple candidate sequences identified by other methods.
We also previously attempted to identify the G. aculeatus centromere sequence using methods that do not rely on a species-specific CENP-A antibody. We used the RepeatNet program (Alkan et al. 2011), which identifies the most abundant sequences in a sequenced genome. In addition, we used a restriction digest approach to identify abundant repeats in the genome. While both computational and restriction digest methods are easy to use and cheaper than ChIP-seq, they generated a large list of potential repeats. To narrow down our list of putative sequences, we assumed, as is common, that the centromeric repeat would not map to the assembled regions of the genome and that it would be one of the most abundant repeats in the unassembled genome sequences (Alkan et al. 2011; Melters et al. 2013). Based on these criteria, we made a list of candidates to test by FISH, but none hybridized to the centromere. However, after performing the ChIP-seq experiment, we realized that the GacCEN repeat was actually identified using both the computational and the restriction digest methods. Because the edges of many genome assembly scaffold edges contained a few, more diverged, copies of this repeat, our analysis showed that the putative sequences mapped to distinct chromosomes. Furthermore, the GacCEN repeat was not as abundant as other repeats we identified. For these two reasons, the GacCEN repeat sequence was not among the first to be tested. Our experience highlights that other approaches can be used to identify centromeric repeats in organisms without a known CENP-A antibody or an available epitope-tagged CENP-A protein. However, CENP-A ChIP-seq is still the most unbiased way to identify or validate a centromeric repeat.
Our work also demonstrates that it is possible in some cases to identify remnants of centromere repeats on the edges of genome assembly gaps. We found some repeats on the edges of assembled scaffolds that were slightly more divergent than the core centromere sequences identified by ChIP-seq (Supplementary Fig. S4). This is consistent with what has been seen in human CENP-A ChIP-seq experiments in which the sequence homology among α-satellite repeats decreases with distance from the core functional centromere (Henikoff et al. 2015). Unequal crossing over at nearly identical repeats is thought to lead to the homogenization of the core centromere, with mutation leading to divergence and unique differences in repeats outside of the core centromere (Smith 1976; Schueler et al. 2001; Henikoff 2002; Shepelev et al. 2009; Henikoff et al. 2015). These differences allow for the assembly of the centromeric repeats. However, Henikoff et al. 2015 showed that more diverged repeats may lose their centromere function, implying that the core, unassembled centromere repeats make up functional centromeres. Our initial filtering step to remove all CENP-A ChIP reads that mapped to the assembled genome actually discarded the repeats we later found on the edges of genome assembly gaps. While these similar repeats may still have centromeric function, by discarding any repeats that mapped to the genome, we ensured that our methods enriched for the core, functional centromere sequences.
Finally, while we have determined that 41 out of the 42 threespine stickleback chromosomes show hybridization with the GacCEN probe in males, the Y centromere appears to show weak hybridization. This could be due to either a different or divergent Y specific centromere repeat (Pertile et al. 2009; Miga et al. 2014), or to a decrease in the number of repeats on the Y centromere array. Future work will use a similar unbiased ChIP-seq approach in both males and females to identify the Y chromosome centromere repeat.
Abbreviations
- BAC:
-
Bacterial artificial chromosome
- BSA:
-
Bovine serum albumin
- CEN:
-
Centromeric repeat sequence
- CenH3:
-
Centromeric histone H3
- CENP-A:
-
Centromere protein A
- CENP-B:
-
Centromere protein B
- ChIP:
-
Chromatin immunopreciptiation
- ChIP-seq:
-
Chromatin immunopreciptiation sequencing
- DAPI:
-
4’,6’-Diamidino-2-phenlyindole
- dpf:
-
Days post-fertilization
- EDTA:
-
Ethylenediaminetetraacetic acid
- FISH:
-
Fluorescence in situ hybridization
- GacCEN:
-
Threespine stickleback (Gasterosteus aculeatus) centromeric repeat sequence
- H3:
-
Histone H3
- HOR:
-
Higher-order repeat
- IF-FISH:
-
Immunofluorescence combined with FISH
- IP:
-
Immunoprecipitation
- KCl:
-
Potassium chloride
- MNase:
-
Micrococcal nuclease
- PBS:
-
Phosphate-buffered saline
- PBST:
-
Phosphate-buffered saline Tween-20
- PCR:
-
Polymerase chain reaction
- PMSF:
-
Phenylmethanesulfonylfluoride
- POF1:
-
Pacific Ocean female 1
- POF2:
-
Pacific Ocean female 2
- RPM:
-
Reads per million
- SDS:
-
Sodium dodecyl sulfate
- SDS-PAGE:
-
Sodium dodecyl sulfate polyacrylamide gel electrophoresis
- SSC:
-
Saline-sodium citrate
- 3′ RACE:
-
Rapid amplification of cDNA ends
References
Alkan C, Cardone MF, Catacchio CR et al (2011) Genome-wide characterization of centromeric satellites from multiple mammalian genomes. Genome Res 21:137–145
Amor DJ, Bentley K, Ryan J et al (2004) Human centromere repositioning “in progress”. Proc Natl Acad Sci U S A 101:6542–6547
Blower MD, Sullivan BA, Karpen GH (2002) Conserved organization of centromeric chromatin in flies and humans. Dev Cell 2:319–330
Crollius HR, Jaillon O, Dasilva C et al (2000) Characterization and repeat analysis of the compact genome of the freshwater pufferfish Tetraodon nigroviridis. Genome Res 10:939–949
Drinnenberg IA, deYoung D, Henikoff S, Malik HS (2014) Recurrent loss of CenH3 is associated with independent transitions to holocentricity in insects. eLife 3:e03676
Earnshaw WC, Rothfield N (1985) Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma 91:313–321
Edwards NS, Murray AW (2005) Identification of Xenopus CENP-A and an associated centromeric DNA repeat. Mol Biol Cell 16:1800–1810
Fountain DM, Kral LG (2011) Isolation and characterization of the Etheostoma tallapoosae (Teleostei: Percidae) CENP-A gene. Genes 2:829–840
Fu L, Niu B, Zhu Z et al (2012) CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28:3150–3152
Garrido-Ramos MA, Jamilena M, Lozano R et al (1994) Cloning and characterization of a fish centromeric satellite DNA. Cytogenet Cell Genet 65:233–237
Gong Z, Wu Y, Koblizkova A et al (2012) Repeatless and repeat-based centromeres in potato: implications for centromere evolution. Plant Cell 24:3559–3574
Gordon DJ, Resio B, Pellman D (2012) Causes and consequences of aneuploidy in cancer. Nat Rev Genet 13:189–203
Haaf T, Schmid M, Steinlein C et al (1993) Organization and molecular cytogenetics of a satellite DNA family from Hoplias malabaricus (Pisces, Erythrinidae). Chromosom Res 1:77–86
Henikoff S (2002) Near the edge of a chromosome's “black hole”. Trends Genet 18:165–167
Henikoff S, Ahmad K, Malik HS (2001) The centromere paradox: stable inheritance with rapidly evolving DNA. Science 293:1098–1102
Henikoff JG, Thakur J, Kasinathan S, Henikoff S (2015) A unique chromatin complex occupies young α -satellite arrays of human centromeres. Sci Adv 1:e1400234
Houben A, Schroeder-Reiter E, Nagaki K et al (2007) CENH3 interacts with the centromeric retrotransposon cereba and GC-rich satellites and locates to centromeric substructures in barley. Chromosoma 116:275–283
Hunt P, Hassold T (2010) Female meiosis: coming unglued with age. Curr Biol 20:R699–R702
Jones FC, Grabherr MG, Chan YF et al (2012) The genomic basis of adaptive evolution in threespine sticklebacks. Nature 484:55–61
Kingsley DM, Peichel CL (2007) The molecular genetics of evolutionary change in sticklebacks. In: Östlund-Nilsson S, Mayer I, Huntingford FA (eds) Biology of the Three-Spined Stickleback. CRC press, Boca Raton, pp 41–81
Kitano J, Mori S, Peichel CL (2007) Phenotypic divergence and reproductive isolation between sympatric forms of Japanese threespine sticklebacks. Biol J Linn Soc 91:671–685
Kitano J, Ross JA, Mori S et al (2009) A role for a neo-sex chromosome in stickleback speciation. Nature 461:1079–1083
Kops GJPL, Weaver BAA, Cleveland DW (2005) On the road to cancer: aneuploidy and the mitotic checkpoint. Nat Rev Cancer 5:773–785
Lee H-R, Zhang W, Langdon T et al (2005) Chromatin immunoprecipitation cloning reveals rapid evolutionary patterns of centromeric DNA in Oryza species. Proc Natl Acad Sci U S A 102:11793–11798
Li H (2013) Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760
Li W, Godzik A (2006) Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22:1658–1659
Lister LM, Kouznetsova A, Hyslop LA et al (2010) Age-related meiotic segregation errors in mammalian oocytes are preceded by depletion of cohesin and Sgo2. Curr Biol 20:1511–1521
Maio JJ (1971) DNA strand reassociation and polyribonucleotide binding in the African green monkey, Cercopithecus aethiops. J Mol Biol 56:579–595
Malik HS, Henikoff S (2009) Major evolutionary transitions in centromere complexity. Cell 138:1067–1082
Manuelidis L (1978) Chromosomal localization of complex and simple repeated human DNAs. Chromosoma 66:23–32
Masumoto H, Masukata H, Muro Y et al (1989) A human centromere antigen (CENP-B) interacts with a short specific sequence in alphoid DNA, a human centromeric satellite. J Cell Biol 109:1963–1973
Masumoto H, Nakano M, Ohzeki J-I (2004) The role of CENP-B and alpha-satellite DNA: de novo assembly and epigenetic maintenance of human centromeres. Chromosom Res 12:543–556
Melters DP, Bradnam KR, Young HA et al (2013) Comparative analysis of tandem repeats from hundreds of species reveals unique insights into centromere evolution. Genome Biol 14:R10
Miga KH, Newton Y, Jain M et al (2014) Centromere reference models for human chromosomes X and Y satellite arrays. Genome Res 24:697–707
Morales C, Sánchez A, Bruguera J et al (2007) Cytogenetic study of spontaneous abortions using semi-direct analysis of chorionic villi samples detects the broadest spectrum of chromosome abnormalities. Am J Med Genet 146A:66–70
Nagaki K, Murata M (2005) Characterization of CENH3 and centromere-associated DNA sequences in sugarcane. Chromosom Res 13:195–203
Nagaki K, Talbert PB, Zhong CX et al (2003) Chromatin immunoprecipitation reveals that the 180-bp satellite repeat is the key functional DNA element of Arabidopsis thaliana centromeres. Genetics 163:1221–1225
Nagaki K, Cheng Z, Ouyang S et al (2004) Sequencing of a rice centromere uncovers active genes. Nat Genet 36:138–145
Nagaki K, Kashihara K, Murata M (2008) A centromeric DNA sequence colocalized with a centromere-specific histone H3 in tobacco. Chromosoma 118:249–257
Ohzeki J-I, Nakano M, Okada T, Masumoto H (2002) CENP-B box is required for de novo centromere chromatin assembly on human alphoid DNA. J Cell Biol 159:765–775
Palmer DKD, O'Day KK, Margolis RLR (1989) Biochemical analysis of CENP-A, a centromeric protein with histone-like properties. Prog Clin Biol Res 318:61–72
Palmer DK, O'Day K, Trong HL et al (1991) Purification of the centromere-specific protein CENP-A and demonstration that it is a distinctive histone. Proc Natl Acad Sci U S A 88:3734–3738
Pertile MD, Graham AN, Choo KHA, Kalitsis P (2009) Rapid evolution of mouse Y centromere repeat DNA belies recent sequence stability. Genome Res 19:2202–2213
Piras FM, Nergadze SG, Magnani E et al (2010) Uncoupling of satellite DNA and centromeric function in the genus Equus. PLoS Genet 6:e1000845
Revenkova E, Herrmann K, Adelfalk C, Jessberger R (2010) Oocyte cohesin expression restricted to predictyate stages provides full fertility and prevents aneuploidy. Curr Biol 20:1529–1533
Ricke RM, van Deursen JM (2013) Aneuploidy in health, disease, and aging. J Cell Biol 201:11–21
Ross JA, Peichel CL (2008) Molecular cytogenetic evidence of rearrangements on the Y chromosome of the threespine stickleback fish. Genetics 179:2173–2182
Rudd MK, Willard HF (2004) Analysis of the centromeric regions of the human genome assembly. Trends Genet 20:529–533
Schueler MG, Higgins AW, Rudd MK et al (2001) Genomic and genetic definition of a functional human centromere. Science 294:109–115
Shang WH, Hori T, Toyoda A et al (2010) Chickens possess centromeres with both extended tandem repeats and short non-tandem-repetitive sequences. Genome Res 20:1219–1228
Shelby RD, Vafa O, Sullivan KF (1997) Assembly of CENP-A into centromeric chromatin requires a cooperative array of nucleosomal DNA contact sites. J Cell Biol 136:501–513
Shepelev VA, Alexandrov A, Yurov YB et al (2009) The evolutionary origin of man can be traced in the layers of defunct ancestral alpha satellites flanking the active centromeres of human chromosomes. PLoS Genet 5:e1000641
Smith GP (1976) Evolution of repeated DNA sequences by unequal crossover. Science 191:528–535
Sullivan BA, Schwartz S (1995) Identification of centromeric antigens in dicentric Robertsonian translocations: CENP-C and CENP-E are necessary components of functional centromeres. Hum Mol Genet 4:2189–2197
Sullivan KF, Hechenberger M, Masri K (1994) Human CENP-A contains a histone H3 related histone fold domain that is required for targeting to the centromere. J Cell Biol 127:581–592
Tek AL, Kashihara K, Murata M, Nagaki K (2010) Functional centromeres in soybean include two distinct tandem repeats and a retrotransposon. Chromosom Res 18:337–347
Urton JR, McCann SR, Peichel CL (2011) Karyotype differentiation between two stickleback species (Gasterosteidae). Cytogenet Genome Res 135:150–159
Warburton PE, Cooke CA, Bourassa S et al (1997) Immunolocalization of CENP-A suggests a distinct nucleosome structure at the inner kinetochore plate of active centromeres. Curr Biol 7:901–904
Willard HF (1991) Evolution of alpha satellite. Curr Opin Genet Dev 1:509–514
Wolfe J, Darling SM, Erickson RP et al (1985) Isolation and characterization of an alphoid centromeric repeat family from the human Y chromosome. J Mol Biol 182:477–485
Zhong CX (2002) Centromeric retroelements and satellites interact with maize kinetochore protein CENH3. Plant Cell 14:2825–2836
Acknowledgments
We thank Sue Biggins and Steve Henikoff for reading this manuscript. We thank Ryan Basom of the Fred Hutchinson Cancer Research Center Genomics Shared Resource for help with ChIP-seq data analysis, Jaki Braggin for help with the Western blots, and the Henikoff, Malik, and Peichel labs for helpful discussions. This research was supported by a National Science Foundation Graduate Research Fellowship (DGE-1256082), the National Institutes of Health Chromosome Metabolism and Cancer Training Grant (T32 CA009657), and the Fred Hutchinson Cancer Research Center.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Rachel O'Neill, Ph.D.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. S1
Threespine stickleback Pacific Ocean CENP-A cDNA, protein, and antibody. a Full-length cDNA sequence for the threespine stickleback CENP-A protein. This sequence has been deposited in GenBank (accession number KT321854). b Alignment between the Danio rerio CENP-A protein (D.re CENP-A), the threespine stickleback CENP-A protein (G.ac CENP-A), and the threespine stickleback H3 protein (G.ac H3) reveals extensive divergence in the N-terminal tail (red). The threespine stickleback H3 protein sequence is a product of the gene ENSGACG00000005779 (Ensembl BROAD S1; Feb 2006). The antibody was raised against the peptide sequence highlighted with a box. c Western blot of the CENP-A antibody on protein lysates from threespine stickleback liver and kidney. (PDF 183 kb)
Fig. S2
Alignment of the enriched repeats found in the most abundant clusters from both ChIP-seq samples. All repeats that were enriched in the IP relative to input from the 500 most abundant clusters in each sample were aligned in Geneious (Biomatters, New Zealand), but only the 44 repeats (Pacific Ocean female 1 (POF1)) and 41 repeats (Pacific Ocean female 2 (POF2)) that were enriched in the IP relative to input from the 50 most abundant clusters are shown here for simplicity. The blue box indicates the 186 bp repeating unit found in all enriched clusters. (PDF 1016 kb)
Fig. S3
Alignment of the consensus repeating unit from each independent ChIP-seq sample. The consensus repeat sequences from the Pacific Ocean female 1 (POF1) and Pacific Ocean female 2 (POF2) were aligned to create the final consensus centromere repeat sequence shown in Fig. 2. Nucleotide ambiguities: R = A or G; W = A or T. (PDF 92 kb)
Fig. S4
Assembled scaffolds containing centromere repeats were found on both edges of the gap in the chromosome 9 assembly corresponding to the position of the centromere, and on a single edge of the gaps corresponding to the centromeres of chromosomes 1, 3, 5, 7, 17, 18, 20, and the X chromosome (Urton et al. 2011). Additional GacCEN like repeats were found on four scaffolds from unassembled regions of the genome. Each arrow shows the size of the repeat variant, and the percent identity to GacCEN is represented by color: blue <90 %; green 90-95 %; red >95 %. The direction of the arrow points towards the center of the genome assembly gap. The dark shaded regions in each chromosome drawing depicts the region where the GacCEN containing scaffold is found; relative position of the centromeres is based on Urton et al. 2011. (PDF 170 kb)
Rights and permissions
About this article
Cite this article
Cech, J.N., Peichel, C.L. Identification of the centromeric repeat in the threespine stickleback fish (Gasterosteus aculeatus). Chromosome Res 23, 767–779 (2015). https://doi.org/10.1007/s10577-015-9495-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10577-015-9495-3