Abstract
Transmissible spongiform encephalopathies (TSEs) are caused by the accumulation of the abnormal prion protein scrapie (PrPSc). Prion protein aggregation, misfolding, and cytotoxicity in the brain are the major causes of neuronal dysfunction and ultimate neurodegeneration in all TSEs. Parkin, an E3 ubiquitin ligase, has been studied extensively in all major protein misfolding aggregating diseases, especially Parkinson’s disease and Alzheimer’s disease, but the role of parkin in TSEs remains unknown. Here we investigated the role of parkin in a prion disease cell model in which neuroblastoma2a (N2a) cells were treated with prion peptide PrP106–126. We observed a gradual decrease in the soluble parkin level upon treatment with PrP106–126 in a time-dependent manner. Furthermore, endogenous parkin colocalized with FITC-tagged prion fragment106–126. Overexpression of parkin in N2a cells via transfection repressed apoptosis by enhancing autophagy. Parkin-overexpressing cells also showed reductions in apoptotic BAX translocation to the mitochondria and cytochrome c release to the cytosol, which ultimately inhibited activation of proapoptotic caspases. Taken together, our findings reveal a parkin-mediated cytoprotective mechanism against PrP106–126 toxicity, which is a novel potential therapeutic target for treating prion diseases.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Prion diseases, also known as transmissible spongiform encephalopathies (TSEs), are a class of neurodegenerative diseases that affect both humans and animals worldwide. A central event in these diseases is the conformational modification of the normal cellular prion protein (PrPC) from a soluble, predominant alpha-helical conformation to a disease-causing, infectious form (PrPSc) that is β sheet rich, insoluble, and protease resistant (Caughey et al. 2009). The misfolding, aggregation, and accumulation of PrPSc on the neuronal cell surface or within endosomes lead to amyloid deposition, eventually resulting in cytotoxicity and cell death (Olzscha et al. 2011; Stefani and Dobson 2003; Winklhofer et al. 2008).
Parkin is a 53-kDa cytosolic protein that contains 465 amino acid residues. Parkin comprises a RING-IBR-RING motif at its C-terminus, a ubiquitin-like (Ubl) domain at its N-terminus, and a middle segment that links the two domains (Kitada et al. 1998; Morett and Bork 1999). Parkin is a unique RING/HECT hybrid that includes both major classes of E3 ligases in one protein, which for E2 binding may function with a catalytic cysteine and a classical RING motif (Riley et al. 2013). Parkin plays a vital role in the degradation of misfolded proteins (Shimura et al. 2000) and has been reported to be capable of mediating K48-linked polyubiquitination and proteasomal degradation (Hattori and Mizuno 2004; Moore 2006) as well as catalyzing monoubiquitination and K63-linked polyubiquitination to accomplish degradation of several putative substrates (Chen and Sun 2009; Olzmann and Chin 2008). After the discovery of parkin, a number of studies have been carried out to elucidate its function in different neurodegenerative diseases. Autosomal recessive juvenile Parkinson’s disease (ARJPD) is linked to mutations in the PARK2 gene that is responsible for parkin expression (Kitada et al. 1998; Lücking et al. 2000). Parkin deletion in a tau model of AD aggravates accumulation of amyloidogenic proteins (Rodríguez-Navarro et al. 2008), whereas overexpression of parkin decreases the β-amyloid load, downregulates amyloidogenic protein expression, and reduces inflammation (Hong et al. 2013). Another study showed that overexpression of parkin reduces oxidative stress (Hyun et al. 2002), whereas parkin knockdown resulted in increased oxidative damage in other studies (Greene et al. 2005; Palacino et al. 2004). Parkin has broad neuroprotective properties against a wide range of toxic insults (Darios et al. 2003; Hyun et al. 2002, 2005; Manfredsson et al. 2007; Staropoli et al. 2003). Parkin mediates the selective autophagy of dysfunctional mitochondria by initiating mitophagy through the autophagy–lysosomal pathway (Chu 2010; Geisler et al. 2010; Vives-Bauza et al. 2010; Youle and Narendra 2011). Parkin expression prevents the production of reactive oxygen species, ultimately reducing mitochondria-dependent apoptosis, whereas parkin knockdown exacerbates the situation (Darios et al. 2003; Palacino et al. 2004).
Autophagic defects are well documented in neurodegenerative diseases (Boland et al. 2008; Kegel et al. 2000; Lonskaya et al. 2013c; Nixon et al. 2005; Ravikumar et al. 2002; Stefanis et al. 2001; Webb et al. 2003), and such defects include accumulation of autophagosomes and undegraded autophagic vacuoles in neuronal cells (Kegel et al. 2000; Lonskaya et al. 2013c; Nixon et al. 2005). Parkin ameliorates the autophagic defects and enhances autophagic clearance of autophagosomes and autophagic vacuoles (Lonskaya et al. 2013c). The expression of exogenous parkin clears Aβ through ubiquitination and autophagy in Alzheimer’s disease (AD) models (Burns et al. 2009; Khandelwal et al. 2011).
Several research groups have investigated the role of parkin in neurodegenerative diseases such as Parkinson’s disease (PD), AD, and Huntington’s disease (HD), and they have reported that the multifunctional neuroprotective role of this peptide can be exploited as a therapeutic strategy in neurodegenerative pathologies (Chung and Dawson 2004; Darios et al. 2003; Moore 2006; Petrucelli et al. 2002; Um et al. 2010). To date, most parkin-related research has been conducted in either PD or AD models, but no work has been performed to determine the role of parkin in prion diseases. The current study was designed to evaluate the possible neuroprotective role of parkin in prion disease and to elucidate the parkin-mediated cytoprotective mechanism(s). We observed that parkin expression ameliorated the toxic effects of PrP106–126 and enhanced cell survival by promoting autophagy.
Materials and Methods
Cell Culture
All the experimental procedures were carried out on mouse N2a cells obtained from a national platform of experimental cell resources for Sci-Tech, Beijing, China. N2a cells were selected due to their close similarity to primary hippocampal neurons in terms of the mechanisms mediating neuronal cell activation (Kemmerling et al. 2007; Stamer et al. 2002). The N2a cells were cultured as described previously (Pan et al. 2014; Yuan et al. 2013).
Prion Peptide and Other Reagents
Prion peptide PrP106–126 (sequence KTNMKHMAGAAAAGAVVGGLG) and fluorescein isothiocyanate (FITC)-labeled PrP106–126 were synthesized by Sangon Bio-Tech (Shanghai, China) with a purity >98 % according to data from the synthesizer. The peptides were dissolved in phosphate-buffered saline (PBS) to a concentration of 1 mM and then allowed to aggregate at 37 °C for 24 h. The presence of amyloid aggregates was confirmed by ThT fluorometric assay. Experiments were conducted with a final peptide concentration of 150 µM as described previously (Pan et al. 2014; Song et al. 2014; Zhu et al. 2016).
Bafilomycin A1 was purchased from InvivoGen (San Diego, CA, USA); 3-methyladenine (3-MA) was from Selleckchem (USA); and rapamycin was obtained from Beyotime Biotechnology (Wuhan, Hubei, China). Parkin (PRK8) antibody was obtained from Santa Cruz Biotechnology (Santa Cruz, CA, USA), and ubiquitin antibody was obtained from Sangon Bio-Tech (Shanghai, China). All other primary antibodies used were purchased from Proteintech (Chicago, IL, USA), and anti-rabbit and anti-mouse secondary antibodies were from ZSGB-Bio (Beijing, China).
Cell culture and Western Blot Analysis
Mouse neuroblastoma N2a cells (seeding density 1 × 106) were grown in 6-well plates to 70–80 % confluency in Dulbecco’s Modified Eagle Medium (DMEM, Thermo Scientific) containing 10 % fetal bovine Serum (FBS, Life Technologies, Grand Island, NY, USA) and 1 % penicillin/streptomycin at 37 °C and 5 % CO2. The cells were treated with 150 µM PrP106–126 and incubated for 6, 12, 24, 36, and 48 h before being harvested and lysed.
For western blot analysis, harvested cells were lysed in 1× STEN buffer (50 mM Tris [pH 7.6], 150 mM NaCl, 2 mM EDTA, 0.2 % NP-40, 0.2 % bovine serum albumin (BSA), 20 mM phenylmethanesulfonyl fluoride (PMSF), and protease cocktail inhibitor (Lonskaya et al. 2013c), before centrifugation at 10,000×g for 20 min at 4 °C, and the supernatants containing the soluble fraction were collected. The collected supernatants were analyzed by western blotting on sodium dodecyl sulfate (SDS) NuPAGE 12 or 6 % (in case of Ub-proteins) Bis–Tris gel (Solarbio Life Sciences). Parkin was immunoprobed with (1:1000) mouse PRK8 antibody purchased from Santa Cruz Biotechnology as previously described (Burns et al. 2009). Ubiquitinated proteins were probed by rabbit anti-ubiquitin antibody (1:500; Sangon Bio-Tech. Shanghai, China), whereas autophagic flux was measured by probing with LC3-B (1:500) and P62/SQSTM1 (1:1000) antibodies (Proteintech, Chicago, IL, USA). GAPDH (1:2000) was used as a loading control. The western blot results were quantified by densitometric analysis using Quantity One 4.6.9 software (Bio-Rad).
Immunocytochemistry and Confocal Microscopy
To determine the subcellular localization of parkin with FITC-PrP106–126, 1 × 105 N2a cells were seeded on presterilized, polylysine-coated cover slips in a 24-well plate 1 day before treatment. After attachment, the cells were treated with FITC-tagged PrP106–126 in DMEM + fetal bovine serum (FBS) + 1 % penicillin/streptomycin. After 24 h, the cells were washed with PBS and fixed in 4 % paraformaldehyde (W/V), pH 7.4, for 20 min at room temperature. The fixed cells were then washed three times with PBS and blocked for 1 h at 37 °C in 1 % BSA in PBS with Tween 20 (PBST). The cells were then incubated with primary anti-parkin antibody (PRK8) (1:50) overnight at 4 °C. Next, the cells were washed three times with PBS and incubated with secondary anti-mouse IgG antibody (Alexa Fluro 594) for 1 h at 37 °C. Cells were then washed with PBS three times and counterstained with DAPI for 2 min at room temperature. After washing with PBS, cover slips were mounted on glass slides, and images were obtained with an Olympus Fluoview FV1000 confocal microscope. All reagents such as DAPI and fixation and blocking reagents were purchased from Beyotime Biotechnology (Beyotime Institute of Biotechnology, China).
Cell Culture and Transfection
N2a cells were plated at 2 × 105 cells/well in 24-well dishes 24 h prior to transfection. When the cells reached 80–90 % confluency, they were washed with PBS, and the medium was replaced with Opti-MEM (Invitrogen). Cells were then transfected with pCMV–HA–Parkin or pCMV–HA empty vector control using 2 µl Lipofectamine 2000 reagent (Invitrogen) in Opti-MEM (Invitrogen) without serum according to the manufacturer’s instructions. The culture medium was replaced with the same volume of DMEM containing 10 % FBS 6 h after transfection. At 24 h post transfection, the cells were treated with 150 µM PrP106–126 with or without 100 nM Bafilomycin (Baf) A1, 10 mM 3-MA, and 200 nM rapamycin. After 12 h, the cells were harvested and lysed, and the lysates were subjected to western blot analyses.
Annexin V Assay
Apoptosis was assessed in detached N2a cells using an Annexin V-FITC apoptosis detection kit (Beyotime Institute of Biotechnology, China) according to the manufacturer’s instructions. As a positive control, apoptosis was induced using 200 nM staurosporine 12 h before assay. Annexin V-positive cells were determined by measuring the fluorescence at 488 nm excitation and with the 530/30 nm emission wavelengths using a BD FACSCalibur flow cytometer (BD Biosciences, San Jose, CA, USA).
Preparation of Cytosolic and Mitochondrial Fractions
Cytosolic and mitochondrial proteins were collected using a cell mitochondrial isolation kit (Beyotime Institute of Biotechnology, China) in accordance with the manufacturer’s instructions. Cells were transfected as previously described and treated with 150 µM PrP106–126 for 12 h, after which the cells were detached and briefly washed twice with ice-cold PBS. After counting, the cells were resuspended in cell lysis buffer (250 mM sucrose, 1 mM DTT, 10 mM KCl, 1 mM EDTA, 1 mM ethylene glycol bis (aminoethylether)-tetraacetic acid [EGTA], 1.5 mM of MgCl2, 1mM phenylmethylsulfonyl fluoride; 20 mM HEPES, pH 7.4), at 4 °C for 10–15 min, and then the cells were homogenized with a glass homogenizer. The lysates were centrifuged at 600g and 4 °C for 10 min, and the supernatant was collected and centrifuged at 11,000g for 10 min at 4 °C, before being stored as the cytosolic fraction. At the same time, the pellet was resuspended in mitochondrial lysis buffer at 4 °C and centrifuged at 12,000g for 10 min to remove the nuclei.
Statistical Analysis
All data are expressed as mean ± standard deviation (SD) and compared through one-way analysis of variance (ANOVA) with Dunnett’s post test for multiple comparisons or unpaired two-tailed t test for comparison between two groups. P < 0.05 was considered significant. GraphPad Prism version 5.00 (GraphPad Software, San Diego, CA, USA) was used for statistical analysis.
Results
PrP106–126 Treatment Reduces Soluble Parkin Level
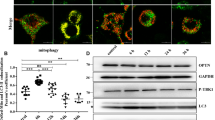
A decrease in the level of soluble parkin compromises its protective function (Wang et al. 2005a) and is associated with autophagic defects in AD (Lonskaya et al. 2013c). Previous research has shown that α-synuclein aggregates interfere with the parkin level in PD (Kawahara et al. 2008), and the stress-induced variation in the available amount of soluble parkin also compromises its cytoprotective functioning (Wang et al. 2005a). To determine the soluble parkin level in response to PrP fragment treatment, N2a cells were incubated with 150 µM PrP106–126, and after 6, 12, 24, 36, or 48 h, the cells were harvested and homogenized. Western blot analysis with anti-parkin antibody (PRK8) revealed the gradual and significant decrease in the soluble parkin level (P < 0.05; Fig. 1a, b), suggesting that the level of soluble parkin is reduced in prion disease. To elucidate the role of ubiquitination and self-degradation in the decreased level of soluble parkin, total ubiquitinated proteins levels at 6, 12, 24, 36, and 48 h were measured, and a significant (P < 0.05) initial increase was observed followed by a decrease in the amount of ubiquitinated proteins (Fig. 1d, e), suggesting that the decreased soluble parkin level is due to ubiquitination and self-degradation. Furthermore, to measure autophagic flux, western blot analysis with anti-microtubule-associated light chain protein 3-B (LC3-B) and ubiquitin-binding protein P62/SQSTM1 revealed a significant increase in the level of LC3-B at 6 and 12 h post treatment (Fig. 1c, f), indicating the induction of autophagy in response to PrP106–126 treatment. The P62/SQSTM1 levels were also decreased from 6 to 24 h (Fig. 1c 2nd blot, g), and collectively these results suggest the destruction of P62-ubiquitinated proteins.
Decrease in level of soluble parkin in time-dependent manner after treatment with PrP106–126. a N2a cells were treated with PrP106–126 for the indicated time periods, and parkin levels were analyzed by western blotting. b Histogram representing the densitometric quantification of soluble parkin protein levels. c Autophagic marker LC3-B and P62/SQSTM1 levels as analyzed with western blot. d Western blot analysis showing ubiquitinated proteins as parkin’s possible target for degradation. e Histogram representing the densitometric quantification of ubiquitinated proteins. f Bar graph showing the fold changes in the level of LC3-B at different time periods. g Bar graph showing the fold changes in the level of P62/SQSTM1 at different time periods. ANOVA with Dunnett’s multiple comparison test. P values are presented as mean ± SD of experiments, and asterisks (*) indicate significant difference between the control and treatment groups; *P < 0.05; **P < 0.01;***P < 0.001. All bands were quantified relative to GAPDH levels
Parkin colocalizes with Intracellular FITC-PrP106–126 in N2a Cells
Previous studies in AD and PD reported that parkin colocalizes with β-amyloid and α-synuclein aggregates (Kawahara et al. 2008; Lonskaya et al. 2013c; Witte et al. 2009). To investigate whether endogenous parkin also colocalizes with PrP106–126, we used N2a cells, grown on polylysine-coated cover slips in 24-well plates, and when the cells reached 60–70 % confluency, they were treated with FITC-PrP106–126. The cells were washed, fixed, blocked, and then incubated with primary anti-parkin antibody (PRK8), secondary anti-mouse IgG antibody (Alexa Fluro 594), and the nuclear stain DAPI. Staining showed colocalization of parkin with intracellular aggregated FITC-PrP106–126 (Fig. 2), which suggests that this interaction between parkin and aggregated FITC-PrP106–126 may probably lead to ubiquitination and PrP fragment clearance.
Parkin colocalization with FITC-PrP106–126 in N2a cells. Confocal microscopy of N2a cells after treatment with or without FITC-PrP106–126, and untreated control cells showing a untreated cells tagged with anti-parkin antibody (PRK8) nuclear DAPI staining and showing the absence of FITC-PrP106–126. b Cells treated with FITC-PrP106–126 showing anti-parkin antibody (PRK8), FITC-PrP106–126, DAPI staining, and merged image showing the colocalization of parkin with FITC-PrP106–126. c Higher magnification treated cells showing anti-parkin antibody (PRK8), FITC-PrP106–126, nuclear DAPI staining, and merged image showing costaining of parkin with FITC-PrP106–126
Parkin Overexpression Alleviates Neuronal Apoptosis Induced by PrP106–126
To determine the cytoprotective role of parkin against PrP106–126-induced toxicity, N2a cells were transfected with either pCMV–HA–Parkin or the pCMV–HA empty vector control as described in the Materials and Methods section. Measurement of the percentage of apoptotic cells using the Annexin V-FITC assay revealed that treatment with PrP106–126 resulted in cellular neurotoxicity in pCMV–HA empty vector-transfected cells, as identified by a significant increase in Annexin V-positive cells compared to nontreated cells. However, parkin overexpression alleviated PrP106–126-induced apoptosis (Fig. 3a, b). These data suggest a cytoprotective role for parkin against PrP fragment-induced toxicity.
Over expression of parkin ameliorates PrP106–126-induced toxicity. a N2a cells were transfected with pCMV–HA–Parkin or pCMV–HA empty vector and then treated with PrP106–126. Cell viability was measured by the Annexin V-FITC assay. b Bar graph indicating Annexin V-positive cells. One-way ANOVA, Newman–Keuls multiple comparison test. P values are presented as mean ± SD, **P < 0.01 (compared with untreated control group and parkin-expressing cells)
Parkin Overexpression Promotes Autophagy in PrP106–126-Treated N2a Cells
Previous studies have shown that exogenous parkin expression promotes the clearance of Aβ via ubiquitination and autophagy (Burns et al. 2009; Khandelwal et al. 2011; Lonskaya et al. 2013b). To investigate the mechanism of cytoprotection in parkin-overexpressing cells, N2a cells were transfected with pCMV–HA–Parkin or pCMV–HA empty vector and treated with 150 µM PrP106–126. At 12 h post treatment, the cells were harvested and lysed as shown in the “Materials and Methods” section. Western blot analysis showed a reduced level of LC3-B in parkin-overexpressing cells compared to those treated with empty vector (Fig. 4a, 2nd blot, b). To ascertain that the reduction in LC3-B was due to enhanced autophagy, we used 100 nM Baf A1, a proton pump inhibitor that blocks lysosome acidification and autophagosome–lysosome fusion (Bae et al. 2014; Yamamoto et al. 1998), and 10 mM 3-MA, a phosphatidylinositol 3-kinase (PI3K) inhibitor. In the presence of Baf A1 and 3-MA, the levels of LC3-B were significantly increased. The cells treated with 200 nM rapamycin, an inhibitor of mTOR that promotes autophagy, showed a decreased level of LC3-B, suggesting that the decreased level of LC3-B is due to enhanced autophagic degradation of autophagosomes in parkin-expressing cells. To further elucidate our hypothesis, we measured the level of undegraded ubiquitin protein by probing with anti-ubiquitin-binding protein P62/SQSTM1 antibody and lysosomes with an antilysosomal marker LAMP2 antibody. As shown in Fig. 4a, c, d, P62 and LAMP2 levels were decreased in parkin-expressing cells compared to those treated with empty vector. Moreover, treatment with Baf A1 and 3-MA reversed the decreased levels, but 3-MA showed no effect on the Lamp2 level. Upon treatment with rapamycin, the levels of both P62 and Lamp 2 were decreased. These results suggest that parkin enhanced the autophagic degradation of ubiquitinated proteins in parkin-expressing cells. When we compare these results at the same time point (12 h) with those in PrP106–126-treated cells (Fig. 1c, f) without parkin overexpression, we clearly observe that parkin overexpression promotes autophagy and PrP106–126 clearance via lysomes.
Exogenous parkin expression enhances autophagy. a N2a cells were transfected with pCMV–HA–Parkin or pCMV–HA empty vector control and then incubated with PrP106–126 with or without Baf A1, 3-MA, and rapamycin for 12 h. Autophagic markers LC3-B (2nd blot), P62/SQSTM1(3rd blot), and LAMP2 (4th blot) were analyzed by WB. b Bar graph showing LC3-B blot quantification by densitometry. c Graph showing P62/SQSTM1 blot quantification by densitometry. d Graph showing LAMP2 blot quantification by densitometry. Values are presented as mean ± SD of experiments; *P < 0.05; **P < 0.01; ***P < 0.001. All bands were quantified relative to GAPDH levels
Parkin Overexpression Disrupts the Mitochondrial Apoptotic Pathway in PrP106–126-Treated N2a Cells
PrP106–126-induced mitochondrial dysfunction leads to neurotoxicity (Ferreiro et al. 2008; O’Donovan et al. 2001). To explore the possible role played by parkin in the PrP106–126-induced mitochondrial apoptotic pathway, we examined the BAX and cytochrome c levels in mitochondrial and cytosolic fractions of PrP106–126-treated cells expressing either pCMV–HA–Parkin or pCMV–HA empty vector. Empty vector-transfected cells treated with PrP peptide showed BAX translocation from the cytosol to the mitochondria and cytochrome c release from mitochondria to cytosol, whereas pCMV–HA–parkin expression reduced BAX translocation and cytochrome c release (Fig. 5a 2nd blot, b 1st blot). To further elucidate the mitochondrial apoptotic pathway via release of apoptotic activating factors from mitochondria to cytosol and subsequent activation of proapototic caspases, we measured the expression levels of caspases 12, 9, and 3 in the cytosolic fraction of pCMV–HA–parkin or pCMV–HA empty vector-expressing cells. A significant difference was observed in the level of active caspase 12 (Fig. 5, 3rd blot, e) between parkin-overexpressing and empty vector-expressing cells, while caspase 9 and caspase 3 showed nonsignificant reduction in parkin-expressing cells as compared to empty vector control cells (Fig. 5a 4th, 5th blot, f, g). These results are in agreement with the idea that parkin prevents PrP106–126-induced toxicity, leading to apoptosis and regulation of mitochondrial homeostasis.
Parkin expression inhibits the mitochondrial apoptotic pathway. N2a cells were transfected with pCMV–HA–Parkin or pCMV–HA empty vector control and then incubated with PrP106–126. a Cytosolic fraction was analyzed by WB probing for parkin, cytochrome c (2nd blot), and apoptotic caspases. b Mitochondrial fraction was analyzed for BAX translocation and parkin recruitment. c Bar graph showing BAX quantification by densitometry. d Bar graph of densitometric quantification of cytochrome c. e Densitometric quantification of caspase 12. f Graph showing caspase 9 quantification by densitometry. g Caspase 3 densitometric quantification. Values are presented as mean ± SD of multiple experiments; *P < 0.05; **P < 0.01
Discussion
Parkin is one of the major E3 ubiquitin ligases present in mammalian cells. Although little is known about the role of parkin in prion diseases, its function in autophagy and clearance of misfolded proteins is well documented in other neurodegenerative diseases. To investigate the role of parkin in prion diseases, the level of endogenous soluble parkin in cells treated with PrP106–126 was measured. The data showed that the soluble parkin level was decreased in a time-dependent manner. Previous studies demonstrated that a decrease in parkin solubility due to stress, α-synuclein, or Aβ aggregates compromises its protective function and is associated with autophagic defects in AD and PD (Kawahara et al. 2008; Lonskaya et al. 2013a, c; Wang et al. 2005a, b). To confirm that the decrease in parkin level is due to autophagic defects or self-degradation of parkin via autophagy, we examined ubiquitinated proteins in cells treated with PrP106–126 and observed an initial increase followed by a decrease in the level of Ub-proteins, suggesting that the decrease in parkin solubility is not due to autophagic defects but may be due to ubiquitination and self-degradation. These results were also confirmed by LC3-B and ubiquitin-binding protein P62/SQSTM1 analysis, which showed the induction of autophagic flux at 6 and 12 h after treatment, as evidenced by increased conversion of LC3 A to LC3 B and a decrease in P62 levels. P62 can bind to ubiquitylated substrates and LC3 on autophagosomes and is itself degraded by autophagy; this specific degradation by autophagy makes p62 a valuable marker of autophagy (Bjørkøy et al. 2005; Ichimura et al. 2008; Komatsu et al. 2007; Pankiv et al. 2007). Furthermore, our confocal microscopic studies also indicated that parkin colocalized with aggregated FITC-PrP106–126, which further confirms its attachment to prion fragments, suggesting its role in ubiquitination, clearance, and cell survival mechanism against the toxic effects of the PrP fragment. In our confocal microscopic studies, the fluorescent intensity of endogenous parkin seems to be increased in contrast with Fig. 1a, which might be due to the fact that here we tagged total parkin while in Fig. 1a we only checked the available soluble parkin.
Parkin promotes neuronal survival in response to numerous toxic insults (Berger et al. 2009; Darios et al. 2003; Ekholm-Reed et al. 2013; MacCormac et al. 2004; Sun et al. 2013; Wang et al. 2013; Yang et al. 2005). In our study, FITC-Annexin V assay revealed that PrP106–126 resulted in neurotoxicity leading to apoptosis in empty vector-treated cells, whereas parkin overexpression prevented the toxic effects and promoted cell survival, as evidenced by the significantly reduced number of Annexin V-positive cells in parkin-overexpressing cells. These findings indicate the cytoprotective function of parkin after a toxic insult is induced by PrP106–126.
Parkin exerts its neuroprotective function by mediating autophagy (Khandelwal et al. 2011; McKeon et al. 2015; Ye et al. 2015). We investigated the role of parkin in autophagic clearance by transfecting N2a cells with pCMV–HA–Parkin or pCMV–HA empty vector and treating them with PrP106–126 for 12 h. Parkin expression led to LC3-B degradation by autophagy, as evidenced by the decrease in the level of LC3-B in parkin-expressing cells compared to empty vector. To further confirm that this decrease in LC3-B level was due to increased autophagosome clearance via lysosomes and to investigate whether parkin expression leads to degradation of ubiquitinated proteins by autophagy as mentioned by Mizushima and Yoshimori (Mizushima and Yoshimori 2007), we checked the levels of ubiquitin-binding protein P62 and lysosomal membrane marker Lamp 2. P62 is a multifunctional protein that binds with LC3-B and can be degraded by autolysosome. Therefore, the degradation of P62 is regarded as a marker of increased autophagic flux. Our results showed that P62 and LAMP2 levels were decreased in parkin-expressing cells compared to those in cells treated with empty vector, suggesting that parkin actively regulates autophagosome clearance in PrP106–126-treated cells. We further confirmed the above findings by treating the cells with Baf A1, 3-MA, and rapamycin. Cotreatment of PrP fragment with Baf A1 increased the levels of LC3-B, P62, and LAMP2 proteins in both parkin- and empty vector-expressing cells, indicating cessation of autophagy, while 3-MA treatment showed a lesser effect on LAMP2 levels; however, P62 and LC3-B levels were increased, which may be due to the lack of an effect of 3-MA on lysosomal function, or it could promote the autophagic flux as described by Wu et al. (2010). Rapamycin treatment was used as a positive control, because induction of autophagy also showed decreased levels of LC3-B, P62, and LAMP2. Our findings are in line with those of previous studies (Hong et al. 2013; Khandelwal et al. 2011; Lonskaya et al. 2013b, c), showing that parkin overexpression promotes autophagy leading to the clearance of misfolded proteins.
The relationship of cell survival, autophagy, and mitochondrial homeostasis is an important area of research in neurodegenerative diseases. Parkin inhibits apoptosis by inducing selective autophagy/mitophagy of dysfunctioning mitochondria (Darios et al. 2003; Johnson et al. 2012; Narendra et al. 2008). Our results show apoptotic BAX translocation from the cytosol to the mitochondria and cytochrome c release from the mitochondria in PrP106–126-treated cells expressing empty vector. However, in parkin-expressing cells these effects were significantly minimized. Our results are in agreement with those of Charan et al. (Charan et al. 2014) who demonstrated the inhibitory function of parkin in BAX translocation to mitochondria.
PrP106–126 induces endoplasmic reticulum (ER) stress, increasing cytosolic Ca2+ levels, and subsequently leading to ER-resident caspase 12 activation (Ferreiro et al. 2008). Caspase 12 has been reported in many previous studies to be a response caspase activated after PrPSc treatment or by any stress stimuli in the ER (Hetz et al. 2003; Nakagawa et al. 2000; Welihinda et al. 1998). In the current study, we measured the level of caspase 12 as a direct measure of ER stress in response to PrP106–126 treatment. Our results showed a significant difference between caspase 12 levels in parkin-overexpressing and empty vector-expressing cells treated with PrP106–126, suggesting the neuroprotective role for parkin in prion-infected cells. It has been previously shown that caspase 12 and caspase 3 are interrelated, and when the stress goes beyond the maximum tolerable threshold of the ER, an apoptotic surge and activation of caspase 3 occur (Lee et al. 2005). Our results also showed differences in activated caspase 3 levels in empty vector-treated cells as compared to parkin-overexpressing cells, proving the interrelationship of caspase 3 and caspase 12. The increased cytosolic Ca2+ level due to PrP peptide-induced stress also disturbs mitochondrial homeostasis and leads to cytochrome c release, which in turn activates procaspase 9, leading to activation of caspase 9 and 3 and ultimately apoptotic cell death (Ferreiro et al. 2008; Hetz et al. 2003; Shah et al. 2015). Our findings that caspase 3 and caspase 9 levels were not significantly different may be due to the previously reported fact that caspase 12 is a substrate of caspase 3 and caspase 3 levels may not be significantly different due to aggresome formation. Alternatively, another explanation may be the time factor as mentioned in a recent study, suggesting that maximum activation of caspases 9 and 3 occurs at 24 h (Mogi and Kondo 2015). Thus, our results suggest the neuroprotective role of parkin in inhibiting both mitochondrial and ER stress-initiated cytotoxic pathways.
In conclusion, our results indicate that parkin protects against PrP106–126-induced neurotoxicity and cell death by enhancing autophagy, reducing ER stress, suppressing BAX translocation to mitochondria, and inhibiting cytochrome c release to lead to abridged activation of proapoptotic caspases. Although further research is necessary to explore in depth the factors that enable autophagy to regulate distinct cell survival and death response mechanisms, our results suggest that parkin may have therapeutic benefits in the treatment of prion diseases.
References
Bae E, Lee H, Jang Y, Michael S, Masliah E, Min D, Lee S (2014) Phospholipase D1 regulates autophagic flux and clearance of α-synuclein aggregates. Cell Death Differ 21:1132–1141
Berger AK, Cortese GP, Amodeo KD, Weihofen A, Letai A, LaVoie MJ (2009) Parkin selectively alters the intrinsic threshold for mitochondrial cytochrome c release. Hum Mol Genet 18:4317–4328
Bjørkøy G, Lamark T, Brech A, Outzen H, Perander M, Øvervatn A, Stenmark H, Johansen T (2005) p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol 171:603–614
Boland B, Kumar A, Lee S, Platt FM, Wegiel J, Yu WH, Nixon RA (2008) Autophagy induction and autophagosome clearance in neurons: relationship to autophagic pathology in Alzheimer’s disease. J Neurosci 28:6926–6937
Burns MP, Zhang L, Rebeck GW, Querfurth HW, Moussa CE-H (2009) Parkin promotes intracellular Aβ1–42 clearance. Hum Mol Genet 18:3206–3216
Caughey B, Baron GS, Chesebro B, Jeffrey M (2009) Getting a grip on prions: oligomers, amyloids and pathological membrane interactions. Annu Rev Biochem 78:177
Charan R, Johnson B, Zaganelli S, Nardozzi J, LaVoie M (2014) Inhibition of apoptotic Bax translocation to the mitochondria is a central function of parkin. Cell Death Dis 5:e1313
Chen ZJ, Sun LJ (2009) Nonproteolytic functions of ubiquitin in cell signaling. Mol Cell 33:275–286
Chu CT (2010) A pivotal role for PINK1 and autophagy in mitochondrial quality control: implications for Parkinson disease. Hum Mol Genet 19:R28–R37
Chung KK, Dawson TM (2004) Parkin and Hsp70 sacked by BAG5. Neuron 44:899–901
Darios F, Corti O, Lücking CB, Hampe C, Muriel M-P, Abbas N, Gu W-J, Hirsch EC, Rooney T, Ruberg M (2003) Parkin prevents mitochondrial swelling and cytochrome c release in mitochondria-dependent cell death. Hum Mol Genet 12:517–526
Ekholm-Reed S, Goldberg MS, Schlossmacher MG, Reed SI (2013) Parkin-dependent degradation of the F-box protein Fbw7β promotes neuronal survival in response to oxidative stress by stabilizing Mcl-1. Mol Cell Biol 33:3627–3643
Ferreiro E, Costa R, Marques S, Cardoso SM, Oliveira CR, Pereira CM (2008) Involvement of mitochondria in endoplasmic reticulum stress-induced apoptotic cell death pathway triggered by the prion peptide PrP106–126. J Neurochem 104:766–776
Geisler S, Holmström KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ, Springer W (2010) PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol 12:119–131
Greene JC, Whitworth AJ, Andrews LA, Parker TJ, Pallanck LJ (2005) Genetic and genomic studies of Drosophila parkin mutants implicate oxidative stress and innate immune responses in pathogenesis. Hum Mol Genet 14:799–811
Hattori N, Mizuno Y (2004) Pathogenetic mechanisms of parkin in Parkinson’s disease. Lancet 364:722–724
Hetz C, Russelakis-Carneiro M, Maundrell K, Castilla J, Soto C (2003) Caspase-12 and endoplasmic reticulum stress mediate neurotoxicity of pathological prion protein. EMBO J 22:5435–5445
Hong X, Liu J, Zhu G, Zhuang Y, Suo H, Wang P, Huang D, Xu J, Huang Y, Yu M (2013) Parkin overexpression ameliorates hippocampal long-term potentiation and β-amyloid load in an Alzheimer’s disease mouse model. Hum Mol Genet 23:1056–1072
Hyun D-H, Lee M, Hattori N, Kubo S-I, Mizuno Y, Halliwell B, Jenner P (2002) Effect of wild-type or mutant Parkin on oxidative damage, nitric oxide, antioxidant defenses, and the proteasome. J Biol Chem 277:28572–28577
Hyun DH, Lee M, Halliwell B, Jenner P (2005) Effect of overexpression of wild-type or mutant parkin on the cellular response induced by toxic insults. J Neurosci Res 82:232–244
Ichimura Y, Kominami E, Tanaka K, Komatsu M (2008) Selective turnover of p62/A170/SQSTM1 by autophagy. Autophagy 4:1063–1066
Johnson BN, Berger AK, Cortese GP, LaVoie MJ (2012) The ubiquitin E3 ligase parkin regulates the proapoptotic function of Bax. Proc Natl Acad Sci 109:6283–6288
Kawahara K, Hashimoto M, Bar-On P, Ho GJ, Crews L, Mizuno H, Rockenstein E, Imam SZ, Masliah E (2008) α-Synuclein Aggregates Interfere with Parkin solubility and distribution role in the pathogenesis of Parkinson disease. J Biol Chem 283:6979–6987
Kegel KB, Kim M, Sapp E, McIntyre C, Castaño JG, Aronin N, DiFiglia M (2000) Huntingtin expression stimulates endosomal–lysosomal activity, endosome tubulation, and autophagy. J Neurosci 20:7268–7278
Kemmerling U, Munoz P, Müller M, Sánchez G, Aylwin ML, Klann E, Carrasco MA, Hidalgo C (2007) Calcium release by ryanodine receptors mediates hydrogen peroxide-induced activation of ERK and CREB phosphorylation in N2a cells and hippocampal neurons. Cell Calcium 41:491–502
Khandelwal PJ, Herman AM, Hoe H-S, Rebeck GW, Moussa CE-H (2011) Parkin mediates beclin-dependent autophagic clearance of defective mitochondria and ubiquitinated Aβ in AD models. Hum Mol Genet 20:2091–2102
Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, Yokochi M, Mizuno Y, Shimizu N (1998) Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature 392:605–608
Komatsu M, Wang QJ, Holstein GR, Friedrich VL, Iwata J-I, Kominami E, Chait BT, Tanaka K, Yue Z (2007) Essential role for autophagy protein Atg7 in the maintenance of axonal homeostasis and the prevention of axonal degeneration. Proc Natl Acad Sci 104:14489–14494
Lee W, Kim D, Boo J, Kim Y, Park I-S, Mook-Jung I (2005) ER stress-induced caspase-12 activation is inhibited by PKC in neuronal cells. Apoptosis 10:407–415
Lonskaya I, Hebron ML, Algarzae NK, Desforges N, Moussa C-H (2013a) Decreased parkin solubility is associated with impairment of autophagy in the nigrostriatum of sporadic Parkinson’s disease. Neuroscience 232:90–105
Lonskaya I, Hebron ML, Desforges NM, Franjie A, Moussa CEH (2013b) Tyrosine kinase inhibition increases functional parkin-Beclin-1 interaction and enhances amyloid clearance and cognitive performance. EMBO Mol Med 5:1247–1262
Lonskaya I, Shekoyan AR, Hebron ML, Desforges N, Algarzae NK, Moussa CE-H (2013c) Diminished parkin solubility and co-localization with intraneuronal amyloid-β are associated with autophagic defects in Alzheimer’s disease. J Alzheimers Dis 33:231–247
Lücking CB, Dürr A, Bonifati V, Vaughan J, De Michele G, Gasser T, Harhangi BS, Meco G, Denèfle P, Wood NW (2000) Association between early-onset Parkinson’s disease and mutations in the parkin gene. N Engl J Med 342:1560–1567
MacCormac LP, Muqit MM, Faulkes DJ, Wood NW, Latchman DS (2004) Reduction in endogenous parkin levels renders glial cells sensitive to both caspase-dependent and caspase-independent cell death. Eur J Neurosci 20:2038–2048
Manfredsson FP, Burger C, Sullivan LF, Muzyczka N, Lewin AS, Mandel RJ (2007) rAAV-mediated nigral human parkin over-expression partially ameliorates motor deficits via enhanced dopamine neurotransmission in a rat model of Parkinson’s disease. Exp Neurol 207:289–301
McKeon JE, Sha D, Li L, Chin L-S (2015) Parkin-mediated K63-polyubiquitination targets ubiquitin C-terminal hydrolase L1 for degradation by the autophagy-lysosome system. Cell Mol Life Sci 72:1811–1824
Mizushima N, Yoshimori T (2007) How to interpret LC3 immunoblotting. Autophagy 3:542–545
Mogi M, Kondo A (2015) Activation of caspase-8 and caspase-9 are required for PC12 cells differentiation. J Immunoass Immunochem 36:547–558
Moore D (2006) Parkin: a multifaceted ubiquitin ligase. Biochem Soc Trans 34:749–753
Morett E, Bork P (1999) A novel transactivation domain in parkin. Trends Biochem Sci 24:229–231
Nakagawa T, Zhu H, Morishima N, Li E, Xu J, Yankner BA, Yuan J (2000) Caspase-12 mediates endoplasmic-reticulum-specific apoptosis and cytotoxicity by amyloid-β. Nature 403:98–103
Narendra D, Tanaka A, Suen D-F, Youle RJ (2008) Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol 183:795–803
Nixon RA, Wegiel J, Kumar A, Yu WH, Peterhoff C, Cataldo A, Cuervo AM (2005) Extensive involvement of autophagy in Alzheimer disease: an immuno-electron microscopy study. J Neuropathol Exp Neurol 64:113–122
O’Donovan CN, Tobin D, Cotter TG (2001) Prion protein fragment PrP-(106–126) induces apoptosis via mitochondrial disruption in human neuronal SH-SY5Y cells. J Biol Chem 276:43516–43523
Olzmann JA, Chin L-S (2008) Parkin-mediated K63-linked polyubiquitination: a signal for targeting misfolded proteins to the aggresome-autophagy pathway. Autophagy 4:85–87
Olzscha H, Schermann SM, Woerner AC, Pinkert S, Hecht MH, Tartaglia GG, Vendruscolo M, Hayer-Hartl M, Hartl FU, Vabulas RM (2011) Amyloid-like aggregates sequester numerous metastable proteins with essential cellular functions. Cell 144:67–78
Palacino JJ, Sagi D, Goldberg MS, Krauss S, Motz C, Wacker M, Klose J, Shen J (2004) Mitochondrial dysfunction and oxidative damage in parkin-deficient mice. J Biol Chem 279:18614–18622
Pan B, Yang L, Wang J, Wang Y, Wang J, Zhou X, Yin X, Zhang Z, Zhao D (2014) C-Abl tyrosine kinase mediates neurotoxic prion peptide-induced neuronal apoptosis via regulating mitochondrial homeostasis. Mol Neurobiol 49:1102–1116
Pankiv S, Clausen TH, Lamark T, Brech A, Bruun J-A, Outzen H, Øvervatn A, Bjørkøy G, Johansen T (2007) p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 282:24131–24145
Petrucelli L, O’Farrell C, Lockhart PJ, Baptista M, Kehoe K, Vink L, Choi P, Wolozin B, Farrer M, Hardy J (2002) Parkin protects against the toxicity associated with mutant α-synuclein: proteasome dysfunction selectively affects catecholaminergic neurons. Neuron 36:1007–1019
Ravikumar B, Duden R, Rubinsztein DC (2002) Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum Mol Genet 11:1107–1117
Riley BE, Lougheed JC, Callaway K, Velasquez M, Brecht E, Nguyen L, Shaler T, Walker D, Yang Y, Regnstrom K et al (2013) Structure and function of Parkin E3 ubiquitin ligase reveals aspects of RING and HECT ligases. Nat Commu 4:1982
Rodríguez-Navarro JA, Gómez A, Rodal I, Perucho J, Martinez A, Furió V, Ampuero I, Casarejos MJ, Solano RM, de Yébenes JG (2008) Parkin deletion causes cerebral and systemic amyloidosis in human mutated tau over-expressing mice. Hum Mol Genet 17:3128–3143
Shah SZA, Zhao D, Khan SH, Yang L (2015) Unfolded protein response pathways in neurodegenerative diseases. J Mol Neurosci 57:529–537
Shimura H, Hattori N, Kubo S-I, Mizuno Y, Asakawa S, Minoshima S, Shimizu N, Iwai K, Chiba T, Tanaka K (2000) Familial Parkinson disease gene product, parkin, is a ubiquitin-protein ligase. Nat Genet 25:302–305
Song ZQ, Yang LF, Wang YS, Zhu T, Zhou XM, Yin XM, Yao HQ, Zhao DM (2014) Overexpression of BAT3 alleviates prion protein fragment PrP106-126-induced neuronal apoptosis. CNS Neurosci Ther 20:737–747
Stamer K, Vogel R, Thies E, Mandelkow E, Mandelkow E-M (2002) Tau blocks traffic of organelles, neurofilaments, and APP vesicles in neurons and enhances oxidative stress. J Cell Biol 156:1051–1063
Staropoli JF, McDermott C, Martinat C, Schulman B, Demireva E, Abeliovich A (2003) Parkin is a component of an SCF-like ubiquitin ligase complex and protects postmitotic neurons from kainate excitotoxicity. Neuron 37:735–749
Stefani M, Dobson CM (2003) Protein aggregation and aggregate toxicity: new insights into protein folding, misfolding diseases and biological evolution. J Mol Med 81:678–699
Stefanis L, Larsen KE, Rideout HJ, Sulzer D, Greene LA (2001) Expression of A53T mutant but not wild-type α-synuclein in PC12 cells induces alterations of the ubiquitin-dependent degradation system, loss of dopamine release, and autophagic cell death. J Neurosci 21:9549–9560
Sun X, Liu J, Crary JF, Malagelada C, Sulzer D, Greene LA, Levy OA (2013) ATF4 protects against neuronal death in cellular Parkinson’s disease models by maintaining levels of parkin. J Neurosci 33:2398–2407
Um JW, Im E, Lee HJ, Min B, Yoo L, Yoo J, Lübbert H, Stichel-Gunkel C, Cho H-S, Yoon JB (2010) Parkin directly modulates 26S proteasome activity. J Neurosci 30:11805–11814
Vives-Bauza C, Zhou C, Huang Y, Cui M, de Vries RL, Kim J, May J, Tocilescu MA, Liu W, Ko HS (2010) PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc Natl Acad Sci 107:378–383
Wang C, Ko HS, Thomas B, Tsang F, Chew KC, Tay S-P, Ho MW, Lim T-M, Soong T-W, Pletnikova O (2005a) Stress-induced alterations in parkin solubility promote parkin aggregation and compromise parkin’s protective function. Hum Mol Genet 14:3885–3897
Wang C, Tan JM, Ho MW, Zaiden N, Wong SH, Chew CL, Eng PW, Lim TM, Dawson TM, Lim KL (2005b) Alterations in the solubility and intracellular localization of parkin by several familial Parkinson’s disease-linked point mutations. J Neurochem 93:422–431
Wang DB, Garden GA, Kinoshita C, Wyles C, Babazadeh N, Sopher B, Kinoshita Y, Morrison RS (2013) Declines in Drp1 and parkin expression underlie DNA damage-induced changes in mitochondrial length and neuronal death. J Neurosci 33:1357–1365
Webb JL, Ravikumar B, Atkins J, Skepper JN, Rubinsztein DC (2003) α-Synuclein is degraded by both autophagy and the proteasome. J Biol Chem 278:25009–25013
Welihinda A, Tirasophon W, Kaufman R (1998) The cellular response to protein misfolding in the endoplasmic reticulum. Gene Expr 7:293–300
Winklhofer KF, Tatzelt J, Haass C (2008) The two faces of protein misfolding: gain-and loss-of-function in neurodegenerative diseases. EMBO J 27:336–349
Witte ME, Bol JG, Gerritsen WH, van der Valk P, Drukarch B, van Horssen J, Wilhelmus MM (2009) Parkinson’s disease-associated parkin colocalizes with Alzheimer’s disease and multiple sclerosis brain lesions. Neurobiol Dis 36:445–452
Wu Y-T, Tan H-L, Shui G, Bauvy C, Huang Q, Wenk MR, Ong C-N, Codogno P, Shen H-M (2010) Dual role of 3-methyladenine in modulation of autophagy via different temporal patterns of inhibition on class I and III phosphoinositide 3-kinase. J Biol Chem 285:10850–10861
Yamamoto A, Tagawa Y, Yoshimori T, Moriyama Y, Masaki R, Tashiro Y (1998) Bafilomycin A1 prevents maturation of autophagic vacuoles by inhibiting fusion between autophagosomes and lysosomes in rat hepatoma cell line, H-4-II-E cells. Cell Struct Funct 23:33–42
Yang H, Zhou H-Y, Li B, Chen S-D (2005) Neuroprotection of Parkin against apoptosis is independent of inclusion body formation. Neuroreport 16:1117–1121
Ye X, Sun X, Starovoytov V, Cai Q (2015) Parkin-mediated mitophagy in mutant hAPP neurons and Alzheimer’s disease patient brains. Hum Mol Genet 24:2938–2951
Youle RJ, Narendra DP (2011) Mechanisms of mitophagy. Nat Rev Mol Cell Biol 12:9–14
Yuan F, Yang L, Zhang Z, Wu W, Zhou X, Yin X, Zhao D (2013) Cellular prion protein (PrPC) of the neuron cell transformed to a PK-resistant protein under oxidative stress, comprising main mitochondrial damage in prion diseases. J Mol Neurosci 51:219–224
Zhu T, Zhao D, Song Z, Yuan Z, Li C, Wang Y, Zhou X, Yin X, Hassan MF, Yang L (2016) HDAC6 alleviates prion peptide-mediated neuronal death via modulating PI3K-Akt-mTOR pathway. Neurobiol Aging 37:91–102
Acknowledgments
This work was supported by the Natural Science Foundation of China (No. 31272532) and the Foundation of Chinese Ministry of Science and Technology (Project No. 2015BAI07B02).
Author information
Authors and Affiliations
Corresponding author
Additional information
S. H. Khan and D. Zhao equally contributed to this work.
Rights and permissions
About this article
Cite this article
Khan, S.H., Zhao, D., Shah, S.Z.A. et al. Parkin Overexpression Ameliorates PrP106–126-Induced Neurotoxicity via Enhanced Autophagy in N2a Cells. Cell Mol Neurobiol 37, 717–728 (2017). https://doi.org/10.1007/s10571-016-0407-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10571-016-0407-7