Abstract
Oxidative stress is a major component of harmful cascades activated in neurodegenerative disorders. Coenzyme Q10 (CoQ10), an essential component in the mitochondrial respiratory chain, has recently gained attention for its potential role in the treatment of neurodegenerative disease. Here, we investigated the possible protective effects of CoQ10 on H2O2-induced neurotoxicity in PC12 cells and the underlying mechanism. CoQ10 showed high free radical-scavenging activity as measured by a DPPH and TEAC. Pre-treatment of cells with CoQ10 diminished intracellular generation of ROS in response to H2O2. H2O2 decreased viability of PC12 cells which was reversed by pretreatment with CoQ10 according to MTT assay. H2O2-induced lipid peroxidation was attenuated by CoQ10 as shown by inhibition of MDA formation. Furthermore, pre-incubation of the cells with CoQ10 also restored the activity of cellular antioxidant enzymes which had been altered by H2O2. Moreover, CoQ10 induced Nrf2 nuclear translocation, the upstream of antioxidant enzymes. These findings suggest CoQ10 augments cellular antioxidant defense capacity through both intrinsic free radical-scavenging activity and activation of Nrf2 and subsequently antioxidant enzymes induction, thereby protecting the PC12 cells from H2O2-induced oxidative cytotoxicity.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Neurodegenerative diseases affect millions of people worldwide, especially in the developed countries (Khandhar and Marks 2007). Neurodegenerative processes have a principle role in several neuronal disorders, such as Alzheimer’s, Parkinson’s, and Huntington’s disease (Glass et al. 2010, Reeve et al. 2008). Oxidative stress is a well-known event in the pathogenesis of neurodegenerative diseases that results from accumulation of free radicals and increase in the formation of reactive oxygen species (ROS) (Saeidnia and Abdollahi 2013). Oxidative stress can cause cell death by damaging cardinal cellular components, such as lipids, proteins, DNA, and RNA. The brain is especially sensitive to oxidative stress, owing to a high oxygen consumption and enrichment in polyunsaturated fatty acids, making it particularly vulnerable to lipid peroxidation. Given the important role of oxidative stress in neurodegeneration, therapeutic strategies which are directed at early interventions targeted at oxidative stress may be one of the promising approaches to preventive interventions for neurodegeneration. Antioxidants play a critical role in neurodegeneration protection by scavenging active oxygen and free radicals and neutralizing lipid peroxides. Thus, an approach which simultaneously enhances various intracellular oxidative defense capacities may be more effective in combating neurodegeneration.
Transcription factor NF-E2-related factor (Nrf2), which plays a crucial role in cellular defense, is a basic leucine zipper transcription factor that resides in the cytoplasm bound to its inhibitor protein, Keap 1, and translocated to the nucleus after stimulation. It is a potent activator of antioxidant response element (ARE)-mediated gene expression, moderating the transcriptional induction of cluster of genes encoding for antioxidative enzymes and cytoprotective proteins (Sen and Packer 1996, Nguyen et al. 2009). Activation of Nrf2 pathway has been demonstrated to be involved in the protection of the nerve cells against oxidative damage in vivo and in vitro (Kensler et al. 2007, Lee et al. 2003, Li et al. 2013). More importantly, the upregulation of Nrf2-driven antioxidant enzymes is beneficial in in vitro and in vivo models of neurodegenerative diseases (Kensler et al. 2007, Pi et al. 2008).
Coenzyme Q10 (CoQ10) is an endogenous antioxidant and powerful free radical scavenger (Gazdík et al. 2003). CoQ10 has been found to have potentially neuroprotective effects on neurodegenerative diseases such as Parkinson’s disease (Shults et al. 2002, Bhat and Weiner 2005), amyotrophic lateral sclerosis (Ferrante et al. 2005), and Huntington disease (Beal 2003). Orally administered CoQ10 reduced neuronal degeneration and increased survival in toxin-induced and transgenic animal models of Parkinson’s and Huntington’s diseases (Ferrante et al. 2002, Matthews et al. 1998). It has also been used in human trials of Parkinson’s and Huntington’s diseases, was well-tolerated, and produced statistically significant or near-significant improvements in clinical rating scales (Feigin et al. 1996, Shults and Haas 2005). Recent study reported that CoQ10 could reduce intracellular depositionof Aβ, which is believed to be an early event in the pathogenesis of AD (LaFerla et al. 2007). However, the role of Nrf2 in the neuroprotective effect of CoQ10 against H2O2-induced oxidative remains an interesting speculation that awaits further investigation.
As part of our study to elucidate the neuroprotective activity of CoQ10, we sought to investigate whether it could protect against H2O2-induced oxidative stress using the rat PC12 pheochromocytoma cell line, which is a useful and widespread model for studying neuronal differentiation into sympathetic neurons and other neurobiochemial and neurobiological events (Guroff 1985), as an in vitro model. Specially, we were interested in the possible correlation among its neuroprotection effect, the redox regulation of Nrf2, and the function of the antioxidant enzyme system.
Materials and Methods
Materials
Coenzyme Q10 (CoQ10) was supplied by Guangdong Runhe Co. Ltd. RPMI Medium1640, penicillin–streptomycin, fetal bovine serum (FBS), and horse serum (HS) were purchased from Gibco (Invitrogen, CA). The assay kits for superoxide dismutase (SOD), catalase (CAT), glutathione peroxidase (GPx), and protein concentration were supplied by Beyotime (China Biotechnology). The assay kit for malondialdehyde (MDA) was purchased from NanJingJianCheng Bioengineering Institute (Nanjing, China). Antibody to Nrf2 (C-20) and Lamin B were from Santa Cruz Biotechnology, Inc (Santa Cruz, CA), and glyceraldehydes-3-phosphate dehydrogenase (GAPDH) rabbit mAb and anti-rabbit IgG AP-linked antibodies were obtained from Cell Signaling Technology (Danvers, MA). All the other reagents were of the highest grade and were obtained from Sigma, unless otherwise indicated.
Cell Culture
The rat pheochromocytoma (PC12) cell line was purchased from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). All cells were plated in culture dishes in RPMI Medium1640 containing 5 % FBS, 10 % HS, and 1 % penicillin–streptomycin. The cells were grown at 37 °C in a humid 5 % CO2 environment, and the medium was changed every other day. Cells were utilized for experiments during exponential growth. PC12 cells were plated at an appropriate density according to each experimental scale. After the cells were attached, the cells exposed to various concentrations of CoQ10 which were diluted in medium for 12 h, followed by incubated with and without H2O2 for 24 h.
Assay of Cell Viability
Briefly, PC12 cells were plated at a density of 5 × 104 cells/ml in 96-well plates, and the cell viability was determined using the conventional MTT assay. After incubation, the cells were treated with the 0.5 mg/ml MTT solution for 4 h at 37 °C. After removal of the MTT solution, cells were treated with 150 μL DMSO to dissolve the dark blue formazan crystals in intact cells, and the plate was shaken for 10 min. The absorbance was measured at 490 nm with a microplate reader (Synergy2, Biotek, USA). Survival of the control groups was defined as 100 %, and cell viability in the treated groups was expressed as a percentage of the control groups.
Assay of Intracellular Reactive Oxygen Species
ROS production in PC12 cells was measured using the redox-sensitive fluorescent dye H2DCF-DA. H2DCF-DA can be deacetylated in cells, where it can react quantitatively with intracellular radicals, mainly H2O2, and convert into its fluorescent products DCF, which are retained within the cell. Briefly, PC12 cells were plated at a density of 5 × 104 cells/ml in 96-well plates, following treatment, the cells were loaded with 10 μM H2DCF-DA at 37 °C for 30 min in the dark, and then washed twice with PBS; finally, the fluorescence intensity was measured at the excitation wavelength of 485 nm and the emission wave length of 530 nm using a fluorescence microplate reader (Synergy2, Biotek, USA). Data were analyzed and expressed as a percentage of the control.
Assay of Lipid Peroxidation
The content of MDA, a compound produced during lipid peroxidation, was determined by the commercially available colorimetric assay kit. This assay is based on the reaction of MDA with thiobarbituric acid (TBA). Two molecule of chromogenic reagent (2-Thiobarbituric acid) with one molecule of MDA yield a stable chromophore at 95 °C, forms a MDA–TBA adduct that absorbs strongly at 532 nm. PC12 cells were plated at a density of 5 × 104 cells/mL in 6-well plates. Briefly, the 100 μL of cell lysates was added to 200 μL 20 % trichloroacetic acid and 100 μL 1 % 2-thiobarbituric acid. The reaction mixture was placed in a water bath at °C for 45 min. After cooling, n-butanol was added and vortexed. The mixture was centrifuged at 3000×g for 10 min. A thiobarbituric acid reacting species, a product of the MDA lipid peroxidation reaction contained in the supernatant, was measured at 532 nm using spectrophotometer and was expressed as nmol of MDA per milligram of protein.
Assay of the Activities of Antioxidant Enzymes
Superoxide dismutase (SOD), Glutathione peroxidase (GPx), and CAT activities were measured according to the manufacturer’s recommendations (Beyotime Biotechnology). PC12 cells were plated at a density of 5 × 104 cells/ml in 6-well plates. Briefly, total SOD activity was detected by the scavenging of superoxide anion generated by xanthine. The remained superoxide anion oxidizes 2-(4-Iodophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium, monosodium salt (WST-1) to WST-1 formazan. Therefore, the total SOD activity in samples is negatively correlative to the absorption of formed WST-1 formazan. The reaction product was measured at 450 nm. One unit of SOD activity was defined as the amount of protein that inhibited the rate of NBT reduction by 50 %. GSH-Px activity was based on the reaction of GSH transformation GSSG. Reduced glutathione in cell homogenate was determined by reaction with 1, 2-dithio-bis nitro benzoic acid (DTNB). 1 M DTNB with 2 M GSH reacted together to form 1 M 5, 5′-dithiobis (2-nitrobenzoic acid) with an intense yellow color. Each lysate was mixed in the solution containing 50 mmol/L Na2HPO4/NaHP2O4, pH 7.0, 1 mmol/L EDTA, 1 mmol/L NaN3, 0.2 mmol/L NADPH, 1 mmol/L glutathione, and 1 U/mL glutathione reductase in 0.1 mL volumn at 25 °C for 5 min. The reaction was initiated by the addition of 1.5 mmol/L cumene hydroperoxide and the absorbance measured at 340 nm. Finally, the amount of GSH-Px vitality was calculated by the formula U/mg = [A340/min (sample) − A340/min (blank)]/0.00622 × dilution factor/protein concentration. The cell extract was collected for measurements of human CAT activity by a CAT analysis kit. Briefly, samples were treated with excess hydrogen peroxide for decomposition by CAT and incubated for an exact time, and the remaining hydrogen peroxide (not decomposed by CAT) was coupled with a substrate that on treatment with peroxidase produced N-4-antipyryl-3-chloro-5-sulfonate-p-benzoquinonemonoimine, which has an absorption maximum at 520 nm and was quantified spectrophotometrically. Catalase activity was then calculated from the assay results. The concentration of total protein was measured with a bicinchoninic acid (BCA) assay kit (Beyotime Biotechnology).
Cytosolic and Nuclear Protein Isolation
Cells treated with various chemicals as detailed in the respective figure legends are harvested, washed with PBS buffer, and kept on ice for 1 min. The suspension was mixed with buffer A (10 mM HEPES, pH 7.5, 10 mM KCl, 0.1 mM EGTA, 0.1 mM EDTA, 1 mM DTT, 0.5 mM phenylmethane sulfonyl fluoride, 5 mg/mL aprotinin, 5 mg/mL pepstatin, and 10 mg/mL leupeptin) and lysed by three freeze-thaw cycles. Cytosolic fractions were obtained after centrifugation at 12,000×g for 20 min at 4 °C. The pellets were resuspended in buffer C (20 mM HEPES, pH 7.5, 0.4 M NaCl, 1 mM EGTA, 1 mM EDTA, 1 mM DTT, 1 mM phenylmethane sulfonyl fluoride, 5 mg/mL aprotinin, 5 mg/mL pepstatin, and 10 mg/mL leupeptin), kept on ice for 40 min, and then centrifuged at 14,000×g for 20 min at 4 °C. The resulting supernatant was used as soluble nuclear fractions for further experiments. All the protein fractions were stored at −70 °C until use, and the protein concentrations were measured with a BCA assay kit (Beyotime Biotechnology).
Western Blot Analysis
Western blotting was performed using the standard method. Equal amounts of proteins were fractionated by SDS-polyacrylamide gel electrophoresis (10 % standard gel) and electrotransferred to Immun-BlotTM PVDF membrane (Bio-Rad). Membranes were blocked for overnight at 4 °C in Tris-buffered saline (TBS), 0.05 % (v/v) Tween-20, 150 mmol/L NaCl and 5 % (w/v) Bovine Serum Albumin (BSA, Santa Cruz), followed by 2-h incubation with primary antibody diluted in the same buffer (Nrf2 1:250, Lamin B 1:250, GAPDH 1:1000). After washing with 0.1 % (v/v) Tween 20 in TBS, the membrane was incubated with anti-rabbit IgG AP-linked secondary antibody for 1 h at room temperature and then washed and developed with 5-bromo-4-chloro-3-indolyl phosphate (BCIP)/nitroblue tetrazolium (NBT) color developing solution. The blots in the samples were quantified by densitometry analysis using Quantity One software. All data from three independent experiments were expressed as the relative intensity compared to the control group for the statistical analyses.
DPPH-Scavenging Capacities
CoQ10 and α-tocopherol were evaluated for their activities to scavenge the stable DPPH radical according to a previously described method (Dinis et al. 1994). Briefly, an aliquot of extract of different concentrations was mixed with 0.8 % of DPPH solution (0.02 mL) in methanol. Reaction mixtures were mixed well and incubated at room temperature for 30 min. Methanol was used as blank, and DPPH solution without addition of extract was used as control. The affinity of the test material to quench the DPPH free radical was evaluated according to the equation scavenging % = (Ac − As)/Ac × 100 %. As and Ac are the absorbance at 517 nm of the reaction mixture with sample and control, respectively.
Trolox Equivalent Antioxidant Capacity (TEAC) Analysis
The ABTS radical cation was prepared by mixing an ABTS stock solution (7 mM in water) with 2.45 mM potassium persulfate. This mixture has to remain for 12–24 h until the reaction is complete and the absorbance is stable. For measurement, the ABTS•+ solution was diluted to an absorbance of 0.700 ± 0.020 at 734 nm. 1 mL ABTS•+ solution and 100 μL of the antioxidant solution were mixed for 45 s, and the absorbance at 734 nm was recorded after 1 min of incubation. TEAC is defined as the concentration (mM) of Trolox having the antioxidant activity equivalent to a 1.0-mM concentration of CoQ10.
Statistics
Data were represented as the mean ± SD (standard deviations) from three experiments with triplicates in all cases. Statistical analyses were performed using one-way analysis variance (ANOVA) followed by Dunnet’s post-hoc test to express the difference among the groups. Data considered statistically significant at p < 0.05.
Results
Effect of CoQ10 on Cell Viability of PC12 Cells
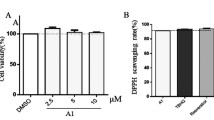
Initially, the cytotoxic potential of CoQ10 on PC12 cells was measured. No cytotoxic effects of CoQ10 were reported up to a concentration of 25 μM using the MTT assay. However, higher amount CoQ10 (50 μM) reduced the viability of the PC12 cells (Fig. 1a). These results indicated that CoQ10 played dual roles in cell death based on its concentrations. Thus, for further experiments, the cells were treated with CoQ10 in the concentration range of 1–25 μM. Furthermore, 25 μM CoQ10 treatment for 24, 48, and 72 h did not show any toxic effect on PC12 cells (Fig. 1b).
Effect of CoQ10 on H2O2-Induced Cytotoxicity and Intracellular ROS Generation in PC12 Cells
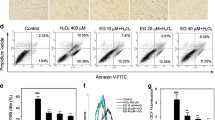
To evaluate the in vitro protective effect of CoQ10, we tested its protective effect on H2O2-induced cytotoxicity in PC12 cells. PC12 cells were treated with CoQ10 for 12 h, followed by further incubation in the presence or the absence of H2O2 for 24 h. Treatment with H2O2 (75 μM) for 24 h induced approximately 50 % cell death, whereas CoQ10, at non-cytotoxic concentrations (1–25 μM), resulted in marked enhancement of survival of the PC12 cells as compared to the H2O2-treated group (Fig. 2a). Maximal rescue occurred at a concentration of 25 μM of CoQ10. These results showed that CoQ10 blocked the injury caused by H2O2 in PC12 cells.
Effect of CoQ10 on H2O2-induced oxidative toxicity and intracellular ROS generation in PC12 cells. Effect of CoQ10 on H2O2-induced oxidative toxicity in PC12 cells. a PC12 cells were pretreated with various concentrations of CoQ10 for 12 h and then incubated with and without 75 μM H2O2 for 24 h. Effect of CoQ10 on H2O2-induced intracellular ROS generation in PC12 cells. b PC12 cells were pretreated with various concentrations of CoQ10 for 12 h and then incubated with and without 75 μM H2O2 for 24 h. c PC12 cells were incubated with CoQ10 (25 μM) for the indicated time and then treated with 75 μM H2O2 for 24 h. Values are mean ± SD (n = 3). ‘*’ and ‘#’ represent significant difference from control and H2O2-treated groups, respectively. *p < 0.05; **p < 0.01; ***p < 0.001; # p < 0.05; ## p < 0.01; ### p < 0.001
The degree of intracellular ROS generation in cells was measured using fluorescence assay with H2DCF-DA probe. As shown in Fig. 2b, treatment with CoQ10 significantly reduced the H2O2-induced ROS generation. Moreover, as shown in Fig. 2c, this effect was dependent on the duration of CoQ10 pretreatment. These results suggest that CoQ10 induced the expression of a gene(s) essential to ROS antagonism.
Effect of CoQ10 on H2O2-Induced Lipid Peroxidation in PC12 Cells
As shown in Fig. 3, when PC12 cells were exposed to H2O2 (75 μM) for 24 h, a significant increase in MDA level was found. Pretreated with CoQ10 for 12 h caused a decrease in MDA production in PC12 cells.
Effect of CoQ10 on H2O2-induced lipid peroxidation in PC12 cells. PC12 cells were pretreated with various concentrations of CoQ10 for 12 h and then incubated with and without 75 μM H2O2 for 24 h. MDA content in cell homogenate was estimated. Values are mean ± SD (n = 3). ‘*’ and ‘#’ represent significant difference from control and H2O2-treated groups, respectively. ***p < 0.001; ## p < 0.01; ### p < 0.001
Effect of CoQ10 on H2O2-Induced Changes in the Activity of Antioxidant Enzymes
As SOD, GPx, and CAT serve as the detoxifying system for preventing damage caused by ROS and play a pivotal role in the scavenging of free radicals, we assessed whether CoQ10 affected the activities of these antioxidant enzymes. H2O2 treatment (75 μM) for 24 h significantly depleted the activities of these antioxidant enzymes in PC12 cells. Whereas exposed to CoQ10 for 12 h caused a dose-dependent increase in the activities of these antioxidant enzymes (Fig. 4) in PC12 cells, which is correlated with its protection against H2O2-induced injury (Fig. 2a). These results suggest that the cytoprotective effect of CoQ10 is mediated through antioxidant enzymes induction.
Effect of CoQ10 on H2O2-induced changes in the activity of antioxidant enzymes. PC12 cells were pretreated with various concentrations of CoQ10 for 12 h and then incubated with and without 75 μM H2O2 for 24 h. Activity of antioxidant enzymes SOD (a), GPx (b), and CAT (c) in cell homogenate was estimated. Values are mean ± SD (n = 3). ‘*’ and ‘#’ represent significant difference from control and H2O2-treated groups, respectively. *p < 0.05; **p < 0.01; ***p < 0.001; # p < 0.05; ## p < 0.01; ### p < 0.001
Effect of CoQ10 on Nrf2 Nuclear Translocation in PC12 Cells
Since activated Nrf2 is an important upstream contributor to the antioxidant enzymes expression, we investigated the possible involvement of CoQ10 on the level of nuclear Nrf2 in PC12 cells using Western blot analysis. As shown in Fig. 5, the H2O2 (75 μM) treatment substantially downregulated the nuclear Nrf2 protein levels. The nuclear fractions of CoQ10-pretreated PC12 cells showed a significant increase in Nrf2 levels, whereas they were decreased in the cytoplasmic fractions.
Effect of CoQ10 on Nrf2 nuclear translocation in PC12 cells. PC12 cells were pretreated with 25 μM CoQ10 for 12 h and then incubated with and without 75 μM H2O2 for 24 h, after which the nuclear and cytosolic Nrf2 protein was determined by Western blot analyses. Values are mean ± SD (n = 3). ‘*’ and ‘#’ represent significant difference from control and H2O2-treated groups, respectively. *p < 0.05; ***p < 0.001; # p < 0.05; ## p < 0.01; ### p < 0.001
Effect of CoQ10 on Free Radical-Scavenging Activities
To evaluate the antioxidant activity of CoQ10, we investigated its DPPH-scavenging actions in an in vitro cell-free system. Figure 6a demonstrates that DPPH-scavenging potentials increased as the concentrations of CoQ10 and α-tocopherol increased. This result indicated that the antioxidant effect of CoQ10 is similar to α-tocopherol for trapping DPPH. The TEAC of CoQ10 was further measured from the decolorization of ABTS•+. Figure 6b shows that CoQ10 has compatible antioxidant potential with positive control Trolox.
Discussion
CoQ10, an essential cofactor involved in mitochondrial oxidative phosphorylation as well as a potent antioxidant, has been studied in multiple in vitro models of neuronal toxicity, with results that overall have supported a neuroprotective effect. Detailed and extensive pre-clinical studies have also strongly supported that CoQ10 provides neuroprotection and slow disease progression in neurodegenerative diseases. In the present study, we established an in vitro damage model of PC12 cells using H2O2 and provided new evidences on the protective effect of CoQ10 against H2O2-induced toxicity. CoQ10 increased the cell viability, attenuated the MDA formation, and restored the activity of cellular antioxidant enzymes in PC12 cells treated with H2O2. In particular, we explored the underlying mechanism and found that CoQ10 induced Nrf2 nuclear translocation. We demonstrated for the first time that the CoQ10-induced Nrf2-associated ARE response readily occurs and accounts for a protective strategy against the oxidative action of H2O2 in PC12 cells.
Oxidative stress refers to the mismatched redox equilibrium between the production of free radicals and the ability of cells to defend against them. One feasible way to prevent free radical-mediated cellular injuries is to augment the oxidative defense capacity through intake of antioxidants. Moreover, the induction of endogenous phase II detoxifying enzymes or antioxidative proteins seems to be a reasonable strategy for delaying disease progression.
DPPH• and ABTS• are stable free radicals, which are frequently used for evaluation of relative antioxidant capacity compared with standard antioxidants. Current research demonstrates that CoQ10 has compatible antioxidant potential with positive control, α-tocopherol, and Trolox (Fig. 6). These results showed that CoQ10 is an excellent candidate antioxidant, which are consistent with the previous reports showing that CoQ10 is an oxygen-free radical scavenger. This was also confirmed with our cell-based antioxidant assay in which CoQ10 significantly reduced the basal intracellular level of ROS (Fig. 2b, c). Treatment with low doses of CoQ10 (1–25 μM) showed a significant protective effect against H2O2-induced neurotoxicity in PC12 cells when measured by MTT assay (Fig. 2a). These results showed a clear and strong correlation among free radical-quenching activities, ROS-scavenging activity, and the enhanced resistance to H2O2-induced oxidative damage. As a consequence, the intrinsic antioxidant capacity may play a role for CoQ10, in contributing towards the partial or total alleviation of cellular oxidative stress.
MDA, a decomposition product of peroxidased polyunsaturated fatty acids (Mukai and Goldstein 1976) that is assessed as an index of lipid peroxidation, and increased MDA levels have been observed in patients with neurodegenerative diseases (Pratico et al. 2002). In the present study, PC12 cells treated with H2O2 underwent peroxidation of its lipid bilayer, leading to increased formation of MDA, which was ameliorated in the presence of CoQ10 pretreatment (Fig. 3). These data indicated that the protective effects of CoQ10 antagonize H2O2-mediated lipid peroxidation in PC12 cells.
Endogenous antioxidant enzymes represent the first line of defense of the brain to counteract the deleterious effects of ROS. The best studied cellular antioxidants are the enzymes SOD, GPx, and CAT, which are three primary endogenous antioxidants involved in direct elimination of ROS. SOD, the first line of defense against free radicals, has ROS-metabolizing activity and can efficiently and specifically catalyze the dismutation of O2 − to O2 and H2O2. GPx catalyzes the decomposition of H2O2 and other peroxides (e.g., lipid peroxides in cell membranes) and then converts H2O2 into O2 and H2O, using reduced glutathione as a substrate. CAT is also a major primary antioxidant defense component sharing similar function to GPx. Induction of antioxidant enzymes is highly recognized as an important therapeutic target for pharmacological intervention of oxidative disorders. In the present study, we showed that the activities of SOD, GPx, and CAT in cell homogenate were decreased, which indicated that a disturbance in the endogenous antioxidant balance occurs in H2O2-induced PC12 cells. And the recovered activities of SOD, GPx, and CAT were observed in PC12 cells pretreated with CoQ10. Furthermore, the increase of antioxidant enzymes by CoQ10 conferred cytoprotection against H2O2-induced oxidative stress (Fig. 2a). These results strongly indicate that in our experimental setting, the cytoprotective effects of CoQ10 may be related to the increase in the activity of antioxidant.
To further elucidate the upstream regulators for the induction of endogenous antioxidant enzymes, we have focused on the Nrf2 signaling pathway. Under normal cellular conditions, cytosolic Nrf2 associates physically with Keap1 which acts as an intracellular redox sensor through its reactive cysteine residues. In response to oxidative stress, Nrf2 is released from Keap1 anchoring and translocates into the nucleus where it binds DNA at the ARE for activation of distinct set of genes encoding detoxifying enzymes including SOD, GPx, and CAT (Itoh et al. 1999, 2003). Recently, Activation of Nrf2 has emerged as key therapeutic targets for treatment of a variety of oxidative stress-related neurodegenerative insults. Nrf2-null mice resulted in a decrease in the basal expression level of detoxifying or antioxidant genes (Johnson et al. 2008). In contrast, Nrf2+/+ mice protects the brain from cerebral ischemia in vivo (Shih et al. 2005), whereas primary neuronal cultures treated with chemical activators of the Nrf2-ARE pathway displayed significantly greater resistance to oxidative stress-induced neurotoxicity (Johnson et al. 2008). Beneficial effects of tBHQ and sulforaphane, both potent inducers of the Nrf2-ARE pathway, have been reported in animal models of neurodegeneration and cerebral ischemia. Using Western blot, we found that Nrf2 was promoted translocation into the nucleus in PC12 cells exposed to CoQ10 (Fig. 5). Therefore, our results suggest that CoQ10 protects PC12 cells against neurotoxicity via transcriptional activation of Nrf2 and upregulation of antioxidant enzyme activity. However, further studies are extensively ongoing to identify the intracellular signaling cascades involved in the protective effect of CoQ10 on the neurotoxicity of H2O2 in PC12 cells.
In summary, the intrinsic free radical-scavenging activity and inducing of the antioxidant enzymes activities through activation of Nrf2 exposed to CoQ10 confer protection against the H2O2-induced oxidative damage in PC12 cells. However, the complete molecular milieu that links all these events needs to be elucidated. Continued attempts to identify novel target molecules responsible for the Nrf2 regulation and to clarify their cross-talk with upstream and downstream signaling molecules will pave the way to exploiting preventive and/or therapeutic strategies for the management of oxidative stress-mediated disorders. These findings indicate that CoQ10 might be a potential candidate for the treatment of oxidative damage-mediated human neurodegenerative diseases and other oxidative stress-related diseases.
References
Beal MF (2003) Bioenergetic approaches for neuroprotection in Parkinson’s disease. Ann Neurol 53(3):39–47
Bhat V, Weiner WJ (2005) Parkinson’s disease. diagnosis and the initiation of therapy. Minerva Med 96:145–154
Dinis TC, Maderia VM, Almeida L (1994) Action of phenolic derivatives (acetaminophen, salicylate, and 5-aminosalicylate) as inhibitors of membrane lipid peroxidation and as peroxyl radical scavengers. Arch Biochem Biophys 315:161–169
Feigin A, Kieburtz K, Como P, Hickey C, Claude K, Abwender D, Zimmerman C, Steinberg K, Shoulson I (1996) Assessment of coenzyme Q10 tolerability in Huntington’s disease. Mov Disord 11:321–323
Ferrante RJ, Andreassen OA, Dedeoglu A, Ferrante KL, Jenkins BG, Hersch SM, Beal MF (2002) Therapeutic effects of coenzyme Q10 and remacemide in transgenic mouse models of Huntington’s disease. J Neurosci 22(5):1592–1599
Ferrante KL, Shefner J, Zhang H, Betensky R, O’Brien M, Yu H, Fantasia M, Taft J, Beal MF, Traynor B, Newhall K, Donofrio P, Caress J, Ashburn C, Freiberg B, O’Neill C, Paladenech C, Walker T, Pestronk A, Abrams B, Florence J, Renna R, Schierbecker J, Malkus B, Cudkowicz M (2005) Tolerance of high-dose (3,000 mg/day) coenzyme Q10 in ALS. Neurology 65(11):1834–1836
Gazdík F, Piják MR, Borová A, Gazdíková K (2003) Biological properties of coenzyme Q10 and its effects on immunity. Cas Lek Cesk 142(7):390–393
Glass CK, Saijo K, Winner B, Marchetto MC, Gage FH (2010) Mechanisms underlying inflammation in neurodegeneration. Cell 140(6):918–934
Guroff G (1985) PC12 cells as a model of neuronal differentiation. Bottenstein JE Cell culture in neurosciences. Plemun Press, New York, pp 245–272
Itoh K, Ishii T, Wakabayashi N, Yamamoto M (1999) Regulatory mechanisms of cellular response to oxidative stress. Free Radical Res 31(4):319–324
Itoh K, Wakabayashi N, Katoh Y, Ishii T, O’Connor T, Yamamoto M (2003) Keap1 regulates both cytoplasmic–nuclearshuttling and degradation of Nrf2 in response to electrophiles. Genes Cells 8(4):379–391
Johnson JA, Johnson DA, Kraft AD, Calkins MJ, Jakel RJ, Vargas MR, Chen PC (2008) The Nrf2–ARE pathway: an indicator and modulator of oxidative stress in neurodegeneration. Ann N Y Acad Sci 1147:61–69
Kensler TW, Wakabayashi N, Biswal S (2007) Cell survival responses to environmental stresses via the Keap1–Nrf2–-ARE pathway. Annu Rev Pharmacol Toxicol 47:89–116
Khandhar SM, Marks WJ (2007) Epidemiology of Parkinson’s disease. Dis Mon 53(4):200–205
LaFerla FM, Green KN, Oddo S (2007) Intracellular amyloid-beta in Alzheimer’s disease. Nat Rev Neurosci 8:499–509
Lee JM, Shih AY, Murphy TH, Johnson JA (2003) NF–E2–related factor–2 mediates neuroprotection against mitochondrial complex I inhibitors and increased concentrations of intracellular calcium in primary cortical neurons. J Biol Chem 278:37948–37956
Li L, Du JK, Zou LY, Wu T, Lee YW, Kim YH (2013) Decursin isolated from Angelica gigas Nakai rescues PC12 Cells from amyloid β–protein–induced neurotoxicity through Nrf2-Mediated upregulation of heme oxygenase-1: potential roles of MAPK. Evid-Based Complement Alternate Med. doi:10.1155/2013/467245
Matthews RT, Yang L, Browne S, Baik M, Beal MF (1998) Coenzyme Q10 administration increases brain mitochondrial concentrations and exerts neuroprotective effects. Proc Natl Acad Sci USA 95(15):8892–8897
Mukai FH, Goldstein BD (1976) Mutagenicity of malondialdehyde, a decomposition product of peroxidised polyunsaturated fatty acids. Science 191:868–869
Nguyen T, Nioi P, Pickett CB (2009) The Nrf2-antioxidant response element signaling pathway and its activation by oxidative stress. J Biol Chem 284:13291–13295
Pi J, Zhang Q, Woods CG, Wong V, Collins S, Andersen ME (2008) Activation of Nrf2-mediated oxidative stress response in macrophages by hypochlorous acid. Toxicol Appl Pharmacol 226:236–243
Pratico D, Clark CM, Liun F, Rokach J, Lee VY, Trojanowski JQ (2002) Increase of brain oxidative stress in mild cognitive impairment: a possible predictor of Alzheimer disease. Arch Neurol 59(6):972–976
Reeve K, Krishnan KJ, Turnbull D (2008) Mitochondrial DNA mutations in disease, aging, and neurodegeneration. Ann N Y Acad Sci 1147:21–29
Saeidnia S, Abdollahi M (2013) Toxicological and pharmacological concerns on oxidative stress and related diseases. Toxicol Appl Pharmacol 273:442–455
Sen CK, Packer L (1996) Antioxidant and redox regulation of gene transcription. FASEB J 10:709–720
Shih AY, Imbeault S, Barakauskas V, Erb H, Jiang L, Li P, Murphy TH (2005) Induction of the Nrf2–driven antioxidant response confers neuroprotection during mitochondrial stress in vivo. J Biol Chem 280(24):22925–22936
Shults CW, Haas R (2005) Clinical trials of coenzyme Q10 in neurological disorders. BioFactors 25(1–4):117–126
Shults CW, Oakes D, Kieburtz K, Beal MF, Haas R, Plumb S, Juncos JL, Nutt J, Shoulson I, Carter J, Kompoliti K, Perlmutter JS, Reich S, Stern M, Watts RL, Kurlan R, Molho E, Harrison M, Lew M (2002) Effects of coenzyme Q10 in early Parkinson disease: evidence of slowing of the functional decline. Arch Neurol 59(10):1541–1550
Acknowledgments
We gratefully acknowledge financial support of this work by Guangdong Medical College foundation (No. B2011011), Zhanjiang Science and Technology Planning Project (No. 2012C3104018), Shenzhen Science and Technology Planning Project (No. 201302173), Innovation Experiment Program for University Students of Guangdong Medical College (LZDM011).
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical Statement
This article does not contain any studies with human participants or animals performed by any of the authors.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Li, L., Du, J., Lian, Y. et al. Protective Effects of Coenzyme Q10 Against Hydrogen Peroxide-Induced Oxidative Stress in PC12 Cell: The Role of Nrf2 and Antioxidant Enzymes. Cell Mol Neurobiol 36, 103–111 (2016). https://doi.org/10.1007/s10571-015-0224-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10571-015-0224-4