Abstract
It is well established that wood cellulose has highly ordered architecture at scales from a few nanometres to several microns in wood cell wall. Its preferred orientation could be in-situ investigated by polarized Raman spectroscopy. However, the technique is currently underused due to the challenges in sample preparation, instrumentation access and availability of prediction models. Here, a novel strategy is proposed to analyse the microfibril orientation without requiring prior knowledge of the fiber alignment. We derive the mathematical dependence between the incident light polarization angle and the intensity of Raman signal. An updated prediction model of microfibril orientation was thus constructed to replace the previous empirical functions. The problems of degradation and burning were addressed by adjusting the laser power and shortening the acquisition time. The orientation direction of cellulose microfibrils can be determined by the polarization-dependent 1096 cm−1 intensity without rotating sample. The strategy was successfully used to study the variations in molecular orientation throughout a cell wall of balsa wood. It was found that the cellulose microfibrils are arranged spirally to form the cell wall.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Cellulose in wood is a linear polymer of β-1,4-glucopyranose units, which synthesized as organized microfibrils with lengths of around 1 to 10 μm and diameters of 3 to 4 nm (Agarwal et al. 2016, 2018). As the backbone of wood cell walls, cellulose with highly ordered architecture at scales from a few nanometres to several microns determines the macroscopic mechanical properties of cell walls. Cellulose microfibril orientation are usually described by microfibril angle (MFA) in wood science, referring to the angle between the directions of the helical windings of cellulose microfibrils in the secondary cell wall of fibers and tracheids and the long axis of the cells (Barnett and Bonham 2004). Although it is perhaps the easiest ultrastructural variable to measure for plant cell walls, time-consuming sample preparation and specific techniques are still required: either measurement of individual fibers or tracheids using microscopy, or measurement of bulk samples using X-ray diffraction or near infrared (NIR) spectroscopy (Donaldson 2008). Even some methods require chemical or physical pretreatment of the sample to expose microfibrils. Obviously, detection of individual fibers or tracheids offers more details on how cellulose microfibrils arranged in plant cell wall. This will enable researchers to predict the mechanical properties of plants and to design biomimetic materials according to specific structures.
Raman spectroscopy is a fast and non-destructive characterization technique. It can be utilized to assign the Raman modes based on crystal symmetry and Raman selection rules and also to detect the crystallographic orientation of anisotropic materials (Liu et al. 2017). Atalla and Agarwal first used this technique in 1985 to study the orientation of components in the native wood cell wall (Atalla and Agarwal 1985). They prospectively pointed out that a mapping of variations in molecular orientation throughout an individual cell would allow us to detect nodes in molecular organization and their relation to cell morphology. Unfortunately, such a mapping was not feasible at that time due to the limitations of the instrument and the theory: Raman scattering is a weak phenomenon in nature which is easily masked by the huge fluorescence background; therefore, the sample preparation and data processing are very complex. This problem has been alleviated in recent years by developing novel Raman instruments. The latest confocal Raman microscopy is equipped a pinhole for superior rejection of fluorescence. On top of this platform, Gierlinger et al. reported that the Raman spectra of wood acquired with linear polarized laser light included information about polymer composition as well as the MFA (Gierlinger et al. 2010). They investigated the dependency between cellulose and laser orientation direction, and utilized the multi-variate methods to describe the orientation-dependent changes of band height ratios and spectra. The predicted results of MFA are in coincidence with X-ray diffraction determination. Similarly, Sun et al. employed polarized Raman spectroscopy to determine the microfibril orientation within rice cell walls by performing ellipse fitting (Sun et al. 2016). Basically, the polarization direction of the incident light is closely related to the microfibril orientation. Gierlinger & Schwanninger and Sun et al. provided the models for prediction of microfibril orientation. However, none of them supplied the derivation process of the model. Therefore, the applicability of the model is questionable, especially for determination of MFA in different species of wood.
In this study, we proposed a strategy to determine the microfibril orientation on the basis of polarized Raman spectroscopy, which includes sample preparation method, instrument configuration, and prediction model. The model was derived in detail to prove the process feasibility. As an example, the strategy was employed to determine the microfibril orientation in the cell wall of balsa wood, and visualize the variations in cellulose orientation throughout cell wall layers. The procedure can be theoretically extended to measure the orientation of other molecules, which contributes to investigating the internal structure of wood cell wall.
Materials and methods
Sample preparation
A 7-year-old balsa tree (Ochroma lagopus Swartz) was provided by the arboretum of Beijing Forestry University, China. Wood discs were collected at a height of 1.5 m above ground level and cut into small tissue blocks (about 3 mm × 3 mm × 5 mm). After immersing the blocks into boiling deionized water for 30 min, they were immediately transferred to deionized water with room temperature for 30 min. This step should be repeated until the blocks sink to the bottom of the container, indicating that the air in blocks was removed and that the blocks were softened. Without any embedding routine, one tissue block was fixed on a sliding microtome (Leica 2010R). As shown in Fig. 1, the axial direction of the fiber (\(z{\prime }\)) is at an inclination angle of ~ 5° from the vertical direction (\(z\)) of the microtome. This step is designed to distinguish the morphological region of the cell wall where the orientation of microfibrils is determined. According to the aggregation-induced emission (AIE) mechanism, the intensity of lignin fluorescence is closely related to its aggregation state (Xue et al. 2020). Therefore, the tissue sample is prepared as thin as possible to reduce the lignin content and weaken the fluorescence effect. The 10-µm-thick cross-sections were prepared and then placed on a glass slide with a drop of deionized water. The sample was covered by a coverslip for subsequent Raman investigation. The coverslip was sealed with nail polish to prevent evaporation of water.
Polarization configuration for Raman spectroscopy
The measurements were performed using a LabRam HR Evolution Raman spectroscopy (Horiba Jobin Yvon) equipped with 532 nm laser and objective of 100× (NA 0.90). For point investigation, the acquisition time was 5 s. For mapping measurement, a marked area of 41 × 41 pixels was scanned at a step of 0.5 μm with an acquisition time of 0.5 s. The estimated laser spot size was ~ 0.7 μm (1.22λ/NA). The lateral resolution is approximately ½ of it (0.61λ/NA). The grating was 300 g/mm and the spectral resolution was about 11 cm− 1. The polarization direction of the incident laser was changed by counter clockwise rotating the half-wave plate automatically. Both point and mapping measurement were performed at every polarization direction ranging from 0° to 360° at an interval of 10°. Therefore, for each point 36 spectra were recorded. In mapping mode, 1681 points were sampled and 60,516 spectra were recorded.
The schematic diagram of the polarization configuration for polarized Raman spectroscopy is displayed in Fig. 2. The laboratory coordinates (\(xyz\)) are represented by black arrows. A polarization configuration of Raman measurement is often determined by the laser polarization direction and the analyzer direction (Liu et al. 2017). Here, the sample coincides with the laboratory coordinates without sample rotation. By rotating the fast axis of the half-wave plate with an angle of α/2, the incident laser polarization is rotated from y axis with α. We indicated the laser polarization direction as αL. The analyzer direction was set to vertical (VR). The polarization configuration is defined as αLVR. In this case, the microfibril orientation respecting to the laboratory coordinates is described as β. The Raman intensity depends on the α and β, which reaches the strongest value at β = α, i.e., the microfibril orientation is lying along the direction of laser polarization.
Schematic diagram of the αLVR polarization configuration for Raman spectroscopy. The polarizer is set in the beam path to make the incident laser to be vertically polarized. The analyzer before the spectrometer entrance selects vertically polarized Raman signal to be detected. The half-wave plate is used to change the polarization direction of laser or signal. Laboratory coordinate (xyz) is represented by black arrows. The green two-way arrows stand for the incident laser polarization reaching at sample, and the purple red two-way arrows represent the original polarization of Raman signal corresponding to vertically or horizontally polarized signal selected by the analyzer before spectrometer entrance. The blue arrow represents the microfibril orientation. The white lines in the samples represent microfibrils
Prediction model for determination of microfibril orientation
The microfibril orientation should be automatically analyzed point-by-point. The intensity of a Raman active mode with Raman tensor \({R}_{j}\) is calculated by:
where \({R}_{j}\) is a 3 × 3 Raman tensor, \({e}_{\text{L}}\) and \({e}_{\text{R}}\) are the unit polarization vectors of the incident laser and scattered Raman signal, respectively (Loudon 2001). For a Raman mode with multiple Raman tensors, the total Raman intensity \(I\) is obtained by the summation of Raman intensity from each Raman tensor \({R}_{j}\). We consider an arbitrary Raman tensor for \({R}_{j}\) given in the corresponding crystal coordinates:
In αLVR polarization configuration, the incident laser propagates along \(z\) axis with polarization vector \({e}_{\text{L}}^{T}=(-\text{sin}\alpha \text{cos}\alpha 0)\). The Raman signal is backscattered along \(z\) axis with polarization vector fixed by the analyzer as \({e}_{\text{R}}=\left(0 1 0\right)\). Based on Eq. (1) and Eq. (2), Raman intensity of the arbitrary Raman tensor in the αLVR configuration is:
The Raman intensity is a function of laser incident angle α, which can be fitted by the following equation. Letting \({x}_{1}= {d}^{2}-{e}^{2}\), \({x}_{2}=-ed\), \({x}_{3}={e}^{2}+\epsilon\):
where \(\epsilon\) is the is the residual between the fitting function and the observed data. Letting \(x={[{x}_{1},{x}_{2},{x}_{3} ]}^{T}\), \(B={[{I}_{1},{I}_{2},{I}_{3},\dots ,{I}_{m} ]}^{T}\), \(A = \left[ {\begin{array}{*{20}c} {\begin{array}{*{20}c} {\sin ^{2} \alpha _{1} } \\ {\sin ^{2} \alpha _{2} } \\ \end{array} } & {\begin{array}{*{20}c} {\sin 2\alpha _{1} } & 1 \\ {\sin 2\alpha _{2} } & 1 \\ \end{array} } \\ {\begin{array}{*{20}c} \vdots \\ {\sin ^{2} \alpha _{m} } \\ \end{array} } & {\begin{array}{*{20}c} \vdots & \vdots \\ {\sin 2\alpha _{m} } & 1 \\ \end{array} } \\ \end{array} } \right]\) and using the least squares algorithm to achieve \(x\).
The coefficient of determination \({R}^{2}\) is calculated as follow.
where \({I}_{\text{f}\text{i}\text{t}}\) is the fitting function calculated by Eq. (4);\(I\) is the original Raman intensity; \(\stackrel{-}{I}\) is the average of \(I\). The coefficient of determination, which normally ranges from 0 to 1, provides a measure of how well observed outcomes are replicated by the fitting function. The microfibril orientation (β) can be easily acquired by finding the extreme values of the fitting function.
Data processing
The mathematical software MATLAB R2021a (Math Works) was applied to process the data. The original spectra were pre-processed to eliminate the spectral contaminants including baseline drifts arising from fluorescence and cosmic spikes before other data-processing approaches were implemented (Zhang et al. 2017). The principal component analysis (PCA) and clustering analysis were used to extract the spectra from plant cell walls (Zhang et al. 2015).
Results and discussion
Polarized Raman measurement at one position
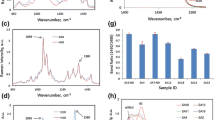
The functional groups of cell wall main components, namely cellulose, hemicelluloses and lignin, can be identified by their unique spectral pattern. The intensity of the bands may be used for the calculation of the relative content in the sampled entity. Despite some lignin peaks are observed at similar wavenumbers to cellulose and hemicelluloses, the contributions to the peaks are roughly judged by the content of each component (Agarwal and Ralph 1997). According to the variations of compositional concentration, an individual wood cell wall typically consists of cell corner (CC), compound middle lamellar (middle lamella plus primary wall, CML) and secondary wall (SW) (Côté et al. 1969).
Figure 3 shows the average spectra of different morphological regions of balsa. Typical band assignments displayed in Table 1 on the basis of previous literature (Agarwal and Ralph 1997; Wiley and Atalla 1987). Due to differences in instrument and testing conditions, the various functional groups corresponding to the wave number is slightly different from the literature, but does not affect the peaks assignments. Two evident peaks at 1601 and 1663 cm−1 are attributed to lignin. The peak at 1601 cm −1 is found as a result of aromatic ring symmetric stretching. The peak at 1663 cm −1 is the ring conjugated C=C or C=O vibration in lignin. The bands at 1096 and 1122 cm −1 are attributed to the asymmetric and symmetric stretching vibration of COC linkages of cellulose and hemicelluloses.
In addition to the different compositions, the orientation of the cellulose also affects the wood spectrum and this effect must also be understood (Gierlinger and Schwanninger 2007). The polarized Raman measurement at one position in the SW of balsa wood excluded the spectral changes due to composition. A point on the secondary wall was randomly selected as the object of polarization Raman study. As shown in Fig. 4, the intensities of almost all bands showed changes during the rotation of the half-wave plate (changing the polarized laser direction). The result is in coincidence with the analysis of Gierlinger and Sun (Gierlinger et al. 2010; Sun et al. 2016). The observed spectral changes in balsa wood were supposed to derive from the anisotropic parallel alignment of the cellulose molecules in microfibrils. The intensities of components are highly orientation-dependent and reach maximum intensity of all Raman modes when the incident and analysed polarization are aligned parallel to fibre axis and strongly suppressed when perpendicular, which is described as antenna effect (Jorio et al. 2002). This orientation judgment is obviously essential because their optimized properties always arise in their crystallographic orientation direction. Therefore, when the cellulose microfibril orientation is parallel to the polarization direction of the incident laser, the maximum Raman intensity of the characteristic cellulose peak will be obtained; when the cellulose microfibril orientation is perpendicular to the polarization direction of the incident laser, the minimum Raman intensity of the same peak will be obtained.
Changes in the fingerprint region of spectra acquired from one position of S2 layer whilst rotating the polarization direction of the incident laser in 10° steps. The polar plots show the intensity changes of 19 Raman bands. Red circles: intensity of Raman peak. Orange lines: fitting model. Blue lines (longer): highest intensity. Black lines (shorter): lowest intensity
The above theory needs to be further verified by data. In order to study the Raman peak changing with the polarization of incident laser, the intensities of 19 Raman bands listed in Table 1 were used to achieve the polar plots (red circles in Fig. 4). The concrete parameters of the models were shown in Table 2. As previous mentioned, the coefficient of determination (\({R}^{2}\)) provides a measure of how well observed outcomes are replicated by the fitting function. The closer the \({R}^{2}\) value is to 1, the higher fitting accuracy is. The \({R}^{2}\) greater than 0.9 indicates that the model can well predict the polarization behaviours of corresponding Raman modes. Otherwise, Raman modes with lower \({R}^{2}\) may not have polarization behaviours, that is, there is no clear orientation. The experimental results agree with the theoretical ones at the scanned position. the CCC ring (378 cm−1), CCO ring (436 and 456 cm−1), glycosidic CCO (496 and 522 cm−1), CH2 (973 cm−1), CO (1040 cm−1), and glycosidic COC (1096 and 1122 cm−1) have the large values of \({R}^{2}\). Most of them are attributed to cellulose and hemicelluloses. This is consistent with the clear orientation of cellulose microfibrils. The calculated β describes the direction of Raman modes of functional groups. Here, we assumed that the feature of glycosidic COC stretching is regarded as the microfibril orientation since the direction of COC can be approximated as parallel to the chain axis of cellulose (Svenningsson et al. 2019).
Polarized Raman measurement for mapping
To eliminate the particularity of one position results, the polarized investigation is performed on a region of balsa wood cell wall. The greatest difficulty of polarized Raman imaging is how to achieve high quality Raman spectra in the shortest time. The quality of Raman spectra directly affects the fitting results of the model while the testing time influences the stability of the instrument. Therefore, we had to sacrifice spectral quality to shorten the testing time. The imaging map consists of 882 pixels, i.e., polarization detection at 882 single point positions. The \({R}^{2}\) maps of 21 Raman bands are shown in Fig. 5. The images display the fitting results of the model: the clearer the shapes of the cell wall are, the better the fitting results of the model are. It is clear that the model is the best fit for functional groups of glycosidic COC (1096 cm−1). There are 692 pixels (78%) with R^2 values greater than 0.5 where the maximum is 0.9401. This result is not only consistent with the results of the polarized Raman measurement at one position, but also with the theoretical speculation. Accordingly, it is feasible to employ the polarized Raman features of 1096 cm −1ll to describe the orientation of glycosidic COC and thus determine the microfibril orientation.
As shown in Fig. 6, the variations in cellulose microfibrils orientation throughout the region are visualized in the mapping of angles β° of 1096 cm−1. The different colors are employed to distinguish the microfibrils orientation. The angles of microfibrils are between − 20° and 50° relative to laboratory coordinates and gradually change from the SW to the CML. In the region of an individual cell wall, the color of the mapping turns from blue to green indicating that the microfibrils are arranged spirally to form the cell wall. The spiral directions of two adjacent cell walls may be opposite. The microfibril angles of two adjacent cell walls is about 70° (the angles between bule region and green region). It is noted that the use of cross-section as an example in this study is to illustrate the differences in microfibril orientation in different cell wall layers. Our proposed strategy is also well applicable to the detection of microfibril orientation in radial sections of cell walls.
Conclusion
This study demonstrates a strategy for using polarized Raman spectroscopy to measure the spatial distribution of cellulose microfibrils orientation within the wood cell wall. The strategy falls into three steps: (1) sample preparation (e.g., cross-sections, radial sections); (2) polarized Raman investigation with αLVR configuration; (3) fitting of prediction model to determine the microfibril orientation. The prediction model has a solid theoretical basis to ensure the reliable analysis results. The applications of polarized Raman spectroscopy are used not only to mapping of variations in microfibrils orientation throughout a wood cell wall, but also to studying the orientation of other functional groups. Therefore, more complete understanding of organizations and interactions of cellulose, hemicellulose and lignin will then be possible.
Data availability
The datasets generated during and/or analysed during the current study are not publicly available, but are available from the corresponding author or reasonable request.
References
Agarwal UP, Ralph SA (1997) FT-Raman spectroscopy of wood: identifying contributions of lignin and carbohydrate polymers in the spectrum of black spruce (Picea mariana). Appl Spectrosc 51(11):1648–1655. https://doi.org/10.1366/0003702971939316
Agarwal UP, Ralph SA, Reiner RS, Baez C (2016) Probing crystallinity of never-dried wood cellulose with Raman spectroscopy. Cellulose 23(1):125–144. https://doi.org/10.1007/s10570-015-0788-7
Agarwal UP, Ralph SA, Reiner RS, Baez C (2018) New cellulose crystallinity estimation method that differentiates between organized and crystalline phases. Carbohyd Polym 190:262–270. https://doi.org/10.1016/j.carbpol.2018.03.003
Atalla RH, Agarwal UP (1985) Raman microprobe evidence for lignin orientation in the cell walls of native woody tissue. Science 227(4687):636–638. https://doi.org/10.1126/science.227.4687.636
Barnett JR, Bonham VA (2004) Cellulose microfibril angle in the cell wall of wood fibres. Biol Rev 79(2):461–472. https://doi.org/10.1017/S1464793103006377
Côté WA, Day A, Timell T (1969) A contribution to the ultrastructure of tension wood fibers. Wood Sci Technol 3(4):257–271. https://doi.org/10.1007/BF00352301
Donaldson L (2008) Microfibril angle: measurement, variation and relationships–a review. Iawa J 29(4):345–386. https://doi.org/10.1163/22941932-90000192
Gierlinger N, Schwanninger M (2007) The potential of Raman microscopy and Raman imaging in plant research. Spectroscopy 21(2):69–89. https://doi.org/10.1155/2007/498206
Gierlinger N, Luss S, König C, Konnerth J, Eder M, Fratzl P (2010) Cellulose microfibril orientation of Picea abies and its variability at the micron-level determined by Raman imaging. J Exp Bot 61(2):587–595. https://doi.org/10.1093/jxb/erp325
Jorio A et al (2002) Polarized resonant Raman study of isolated single-wall carbon nanotubes: symmetry selection rules, dipolar and multipolar antenna effects. Phys Rev B 65(12):121402. https://doi.org/10.1103/physrevb.65.121402
Liu X-L, Zhang X, Lin M-L, Tan P-H (2017) Different angle-resolved polarization configurations of Raman spectroscopy: a case on the basal and edge plane of two-dimensional materials. Chin Phys B 26(6):067802. https://doi.org/10.1088/1674-1056/26/6/067802
Loudon R (2001) The Raman effect in crystals. AdvPhys 50(7):813–864. https://doi.org/10.1038/122477a0
Sun L et al (2016) Non-invasive imaging of cellulose microfibril orientation within plant cell walls by polarized Raman microspectroscopy. Biotechnol Bioeng 113(1):82–90. https://doi.org/10.1002/bit.25690
Svenningsson L, Lin Y-C, Karlsson M, Martinelli A, Nordstierna L (2019) Molecular orientation distribution of regenerated cellulose fibers investigated with polarized Raman spectroscopy. Macromolecules 52(10):3918–3924. https://doi.org/10.1021/acs.macromol.9b00520
Wiley JH, Atalla RH (1987) Band assignments in the Raman spectra of celluloses. Carbohyd Res 160:113–129. https://doi.org/10.1016/0008-6215(87)80306-3
Xue Y, Qiu X, Ouyang X (2020) Insights into the effect of aggregation on lignin fluorescence and its application for microstructure analysis. Int J Biol Macromol 154:981–988. https://doi.org/10.1016/j.ijbiomac.2020.03.056
Zhang X, Ji Z, Zhou X, Ma J-F, Hu Y-H, Xu F (2015) Method for automatically identifying spectra of different wood cell wall layers in Raman imaging data set. Anal Chem 87(2):1344–1350. https://doi.org/10.1021/ac504144s
Zhang X et al (2017) Method for removing spectral contaminants to improve analysis of Raman imaging data. Sci Rep 7(1):1–10. https://doi.org/10.1038/srep39891
Acknowledgements
The authors gratefully acknowledge the financial support from the Foundation of State Key Laboratory of Biobased Material and Green Papermaking (KF201902), the National Natural Science Foundation of China (32001272), and the Programme of Introducing Talents of Discipline to Universities (Project 111, B21022).
Funding
Foundation of State Key Laboratory of Biobased Material and Green Papermaking (KF201902). National Natural Science Foundation of China (32001272). Programme of Introducing Talents of Discipline to Universities (Project 111, B21022).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have No conflict of interest exist.
Consent for publication
All authors approved the final manuscript and the submission to this journal.
Ethics approval and consent to participate
Not applicable.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, X., Li, L. & Xu, F. Polarized Raman spectroscopy for determining the orientation of cellulose microfibrils in wood cell wall. Cellulose 30, 75–85 (2023). https://doi.org/10.1007/s10570-022-04915-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10570-022-04915-w