Abstract
The modern categories of endogenous non-coding RNAs, namely circular RNAs (circRNAs), involved within the carcinogenesis and progression of various human cancers. The fundamental aim of the current investigation was the evaluation of the hsa_circ_0014130 expressions, their biological functions, and potential regulatory network in bladder cancer. The level of expression for hsa_circ_0014130 was evaluated by qRT-PCR, and its relationships to clinicopathological features and survival outcomes of cases experiencing cancer of the bladder were scrutinized. The impact of hsa_circ_0014130 expressions on biological attitudes of bladder cancer cells in vitro was investigated. The interactions between hsa_circ_0014130 and microRNA (miRNA) sponge, miRNA, and its direct targets were determined by RNA pull-down as well as luciferase reporter gene assay. The correlations of their expression were determined by Pearson’s correlation analysis. Rescue experiments were carried out to identify the biological roles of the regulation network. The expressions of hsa_circ_0014130 were markedly ameliorated in bladder cancer samples and linked with aggressive characteristics and unfavorable survival. Ectopic expression of hsa_circ_0014130 clearly enhanced the differentiation, proliferative, migratory, invasive potential of the cell in bladder cancer, and the development of tumor xenograft in vivo, while malignant biological behaviors were inhibited by hsa_circ_0014130 knockdown. The expression of hsa_circ_0014130 was tied to miR-132-3p in a negative manner with the cells and tissues of bladder cancer. hsa_circ_0014130 function as a competitive endogenous RNA for miR-132-3p to play oncogenic roles in bladder cancer cells. On the other hand, KCNJ12 was a straightforward target of miR-132-3p at the downstream, and the expressions of KCNJ12 were inversely related to that of miR-132-3p. Furthermore, a significantly positive correlation was found between hsa_circ_0014130 and KCNJ12 mRNA expression. More importantly, the oncogenic impact of hsa_circ_0014130 on bladder cancer cells was partly suppressed by ectopic expression of miR-132-3p or KCNJ12 knockdown. The underlined data revealed that hsa_circ_0014130 exerted its biological roles by regulating miR-132-3p/KCNJ12 expression. Further research revealed hsa_circ_0014130/miR-132-3p/KCNJ12 axis has participated in the Epithelial-mesenchymal transition (EMT) progress and GSK3β/AKT signaling pathway. hsa_circ_0014130 works as a sponge of miR-132-3p to advance the oncogenesis and metastasis of bladder cancer by regulation of the KCNJ12 expression. These achievements might ameliorate the comprehension of tumor pathogenesis and provide novel therapeutic targets for cancer of the bladder.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
One of the most frequent malignancies that occurred in the human urologic systems is bladder cancer with a high incidence and mortality rate in recent decades (Bray et al. 2018; Siegel et al. 2021). Although therapeutic strategies such as surgical resection, and chemoradiotherapy have been improved, the prognostic outcome of bladder cancer cases remains unsatisfactory (Antoni et al. 2017). The exploration of the molecular strategies for progress and pathogenesis of bladder cancer and evaluating novel treatment targets for early detection, prognostic evaluation, and clinical intervention is considerably required.
The modern cluster of endogenous non-coding RNAs, namely circular RNAs (circRNAs) having closed-loop skeletons and no 3′ poly(A) tails and terminal 5′ caps in the structures (Jeck and Sharpless 2014; Memczak et al. 2013). Unlike traditional linear RNAs including microRNAs (miRNAs) and long noncoding RNA (lncRNA), circRNAs had a strong resistance to RNases or RNA exonucleases, resulting in their high stability. Firstly, circRNAs were deemed as non-functional products generated through anomalous RNA splicing and did not attract notable scientific attention. However, many previous reports revealed that circRNAs considerably contribute to multiple biological procedures and the pathogenesis of various human diseases such as cancers (Chen et al. 2018; Geng et al. 2018; Guarnerio et al. 2016; Lu et al. 2019). Recently, it has been revealed that a novel circRNA, hsa_circ_0014130 expression considerably dysregulated within NSCLC specimens, and their dysregulation promoted malignant phenotypes of tumor cells (Geng et al. 2020; Wang et al. 2020; Zhang et al. 2018). Although the expressions of hsa_circ_0014130 as well as the related biological functions in bladder cancer are still unclear.
In terms of molecular mechanisms, it has been revealed that circRNAs has crucial regulatory roles by acting as RNA-binding proteins, miRNA sponges, and a template of translation into polypeptides (Chen 2016; Conn et al. 2015; Hansen et al. 2013). As another kind of non-coding RNAs, miRNAs have participated in regulating the genes expressions (at the level of transcription) through the interactions with the 3′-untranslated region (3′-UTR) of targeted mRNAs (Herranz and Cohen 2010). Reported investigations have been divulged that miRNAs contributed to the oncogenesis and metastasis of bladder cancer (Hammouz et al. 2021; Yoshino et al. 2013). For instance, a previous study demonstrated a lower expression level for miR-132 in bladder cancer samples compared with adjacent regular samples, and it played a role of tumor-suppressor in bladder cancer via TGFβ1/Smad2 signaling pathway (Wei and Lv 2019). However, the potential target genes of miR-132 in bladder cancer are still little known. In addition, as two competitive endogenous RNAs, the regulatory networks between miRNAs and circRNAs in bladder cancer need be explored and established.
Herein, the differential expressions profiles for circRNAs within bladder cancer by employing the GEO database (GSE92675 dataset) were scrutinized and initially identified the aberrant expression of hsa_circ_0014130 in bladder cancer specimens. These findings prompted us to the additional exploration of the biological performance of hsa_circ_0014130 and its regulatory networks in cancer of the bladder. Our findings demonstrated that hsa_circ_0014130 has the ability to sponge to miR-132-3p and upregulate expression of the potassium channel expression, subfamily J 12 (KCNJ12), consequently participating in the oncogenesis and metastasis for cancer of the bladder.
Materials and methods
Bladder cancer tissues and cells
By taking advantage of the database of Gene Expression Omnibus (GEO), the profile of expressions for the GSE92675 was procured (https://www.ncbi.nlm.nih.gov/geo/). The assessment of differential circRNAs expressions between cancer of the bladder and vicinal typical specimens was exerted, and the threshold was set as |log Fold Change (logFC)| > 1 and adjusted P value <0.05. In addition, matched bladder cancer and normal samples (adjoining) were acquired from 30 cases who experienced operative treatments in the Second Hospital of Tianjin Medical University between September 2019 and November 2020. Before surgery, therapies including radiotherapy and chemotherapy were not received by any patient. The informed consent was signed to notify the applying purpose of tissue samples.
Four human cell lines of bladder cancer (T24, UMUC-3, 5637 and EJ cells) and normal urothelial cell SV-HUC-1 were acquired from the Cell Bank of Chinese Academy of Sciences. The culturing of the underlined cells was carried out in a DMEM medium (GIBCO, CA, USA) comprising of FBS (10%), followed by incubation within moisturized circumstances with CO2 (5%) at 7 °C.
Plasmid construction and cell transfection
The plasmid vectors with hsa_circ_0014130 overexpression and its short interfering RNA (si-hsa_circ_0014130) were synthesized. pcDNA3.1 empty vectors and si-NC were used as the negative control. Furthermore, miR-132-3p deterrent or mimic was implemented to ectopically regulate the expressions of miR-132-3p within bladder cancer cells. The transfection progress was carried out conforming to the protocol of the producer.
qRT-PCR
The relative expressions of target genes were evaluated in the specimens related to cancer of the bladder and cell lines through qRT-PCR. Briefly, the isolation of total RNAs originated from bladder cancer cells or samples was carried out via TRIzol Reagent (TaKaRa, Japan). Extracted RNAs were reversely synthesized into cDNA through employing the kit of Prime Script RT (Takara, China). For mature miRNAs, the kit of specific miRNA synthesis (Takara, China) was implemented. The findings of the qRT-PCR assessment were shown using the 2-ΔΔ CT approach. The normalization of the miR-132-3p and hsa_circ_0014130 expressions was executed to U6 and GAPDH, respectively. The sequences of the forward (F) and reverse (R) primers were presented in Table S1.
Cell viability and colony formation
The Kit of Cell Counting (CCK-8, Beyotime Bio, China) has been utilized for the evaluation of the impacts of hsa_circ_0014130 and miR-132-3p expression on the cell viability of bladder cancer. The seeding of transfected T24 cells was carried out into a 96-well culture plate with 5000 cells/well, followed by adding a total of 10 μL CCK-8 regent into each well and then the incubation was done for 0 h, 24 h, 48 h, and 72 h, respectively. The cell viability of T24 was estimated by measuring the absorbance at 450 nm. In order to assess colony creation, the cells of bladder cancer were seeded into a plate containing 6 plates with 3000 cells/well. After 2 weeks, a light microscope was implemented for counting the number of cells.
Cell migration and transwell invasion
Seeding of T24 cells (transfected) was carried out into the upper transwell chambers with 5000 cells/well using serum-free medium. Next, cell milieu with FBS (10%) was added into the lower compartments. For the transwell invasion assessment, the upper chambers were additionally covered by 100 μg Matrigel. After incubation for 48 h, the transfected T24 cells that migrated or/and invaded into the bottom surface were subjected to 4% formaldehyde fix and 0.1% crystal violet staining. Selecting five random fields, a laser confocal microscope (Nikon, Japan) was employed to count the number of migratory as well as invasive cells.
Luciferase reporter gene assay
The luciferase plasmids containing hsa_circ_0014130 wild-type fragment (hsa_circ_0014130 WT), hsa_circ_0014130 mutant-type fragment (hsa_circ_0014130 MUT), wild-type KCNJ12 mRNA (KCNJ12 WT), and mutant-type KCNJ12 mRNA (KCNJ12 MUT) were synthesized into the luciferase vector, respectively. By employing Lipofectamine 2000 reagent (Invitrogen, USA), the co-transfection of T24 cells was carried out with the above reporter plasmids with miR-132-3p miRNA control (miRNA-NC) or mimic to determine the targeted regulatory relationship between miR-132-3p and hsa_circ_0014130 or KCNJ12 mRNA. Post 2 weeks of transfection, the luciferase activities of each research group were discerned.
RNA-pull down assay
The probe of biotinylated hsa_circ_0014130 was prepared and then transfected into bladder cancer cells. Following the incubation for 48 h, a specific lysis buffer was utilized for lysis of the transfected cells. The complex of biotin-coupled RNA was pulled-down through streptavidin-coated magnetic beads (Life Technologies, USA) (He et al. 2020). The enrichment of miR-132-3p in the bound fractions was discerned using qRT-PCR. Besides, the transfection of biotin-labeled miR-132-3p mimic or negative control was carried out into T24 cells. Following 48-h incubation, the cellular supernatants lysed by lysis buffer were harvested, and then the incubation was executed with streptavidin magnetic beads to block with yeast tRNA for 2 h. Thereafter, the RNA content was further purified. The abundance of hsa_circ_0014130 was further detected.
Western blot (WB)
Total proteins from cell and tissue samples were extracted and lysed through radio-immunoprecipitation assay (RIPA) buffer (Beyotime, China) and subsequently quantified by bicinchoninic acid method. Next, the proteins separation was carried out by SDS-PAGE (10–12%), followed by transferring onto PVDF membranes, and then incubating the membranes with various primary antibodies against epithelial-mesenchymal transition (EMT)-related markers (Twist, 1:1000, ab50887, Abcam; N-Cadherin, 1:1000, ab76011, Abcam; Vimentin, 1:1000, ab137321, Abcam; E-Cadherin, 1:1000, ab1416, Abcam; MMP-7,1:1000, ab205525, Abcam; β-actin, 1:1000, ab8226, Abcam) under 4 °C (for 24 h), and incubated with the second antibody (IgG-HRP, 1:1000, ab6721, Abcam) for 1 h.
Tumor xenograft model
In total, the injection of 40 μL cell suspensions transfected with hsa_circ_0014130 or si-hsa_circ_0014130 overexpression vector was conducted into the back of nude mice (subcutaneously) at the dose of 1 × 107 cells/ml. One month later, the euthanasia process for all mice was fulfilled, and the xenograft of tumors were isolated and weighted. The volume of the tumor was determined as the length × width2 × 0.5.
Immunohistochemistry
The sections were dewaxed by xylene and dehydrated by decreasing gradient alcohol. Following the retrieval of the antigen, 3% hydrogen peroxide (H2O2) was employed for blocking the activity of endogenous peroxidase, and 10% serum was used for blocking non-specific binding sites. Next, the incubation of tissue sections was carried out with primary antibody against loricrin (1:500, ab176322, Abcam), involucrin (1:500, ab68, Abcam), filaggrin (1:200, ab218395, Abcam), and keratin K1(1:500, ab185628, Abcam) at 4 °C for 24 h, respectively, and then incubated by utilizing secondary antibody (labeled with horseradish peroxidase). Subsequently, they were stained by hematoxylin, dehydrated by xylene solution, and examined under a light microscope.
Statistical analysis
Each experimental procedure was carried out thricely. The obtained results were indicated as mean ± SD. Various tests, namely paired Student’s t test, one-way ANOVA, or Student’s t test, were employed for comparing the variations between research groups if the outcomes were consistent with the normal distribution. The curves of Kaplan-Meier and the assessment of log-rank were employed for the evaluation of the relationships between the expression of hsa_circ_0014130 or KCNJ12 and the long-term survival of bladder cancer cases. In addition, the relation between the expressions of hsa_circ_0014130 and miR-132-3p, miR-132-3p and KCNJ12 mRNA expressions, hsa_circ_0014130 and KCNJ12 mRNA expression were analyzed through Pearson’s coefficient. A statistical investigation was conducted via SPSS 21.0 version computer program (IBM Corp, USA). A p value <0.05 was regarded as statistically considerable.
Results
The expression of hsa_circ_0014130 in bladder cancer specimens and its impact on the long-term survival of patients
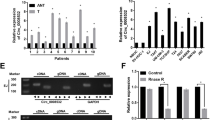
Among differentially expressed circRNAs identified from the GSE92675 dataset, the expressions of hsa_circ_0014130 were evidently dysregulated in bladder cancer samples (Fig. S1 and S2). Therefore, the expressions of hsa_circ_0014130 in 30 pairs of bladder cancer specimens were further identified through qRT PCR assessment. As presented in Fig. 1A, bladder cancer samples had a more notable expression of hsa_circ_0014130 than adjacent specimens (P < 0.001). By analyzing the link between clinicopathological traits of bladder cancer cases and the expression of hsa_circ_0014130, it was noted that the expression of hsa_circ_0014130 was considerably linked with lymph node metastasis (LNM) (Fig. 1B, P < 0.01) and TNM III-IV stage (Fig. 1C, P < 0.01). All bladder cancer patients were split into the lower (n = 15) and elevated expression (n = 15) group according to the median of hsa_circ_0014130 expression. The Kaplan-Meier curves illuminated that the expression of hsa_circ_0014130 was a prognostic marker for the cases of cancer of the bladder, and the related considerable expressions were closely tied to worse survival (P < 0.05) (Fig. 1D).
hsa_circ_0014130 expressions in bladder cancer specimens were detected (A). The link between the expressions of hsa_circ_0014130 and LNM (B) as well as the TNM stage (C) of bladder cancer patients were further explored. Survival curves of Kaplan-Meier for bladder cancer cases were stratified by hsa_circ_0014130 expression (D). hsa_circ_0014130 expression in four bladder cancer cell lines (E). hsa_circ_0014130 expression in bladder cancer cells could not be affected by RNase R treatment (F). The schematic diagram indicating that hsa_circ_0014130 was originated from the exons 7 of PIP5K1A gene (G). **P < 0.01, ***P < 0.001
Comparable with the above outcomes, elevated expression of hsa_circ_0014130 was evaluated in bladder cancer cells in comparison to normal human urothelial cells (P < 0.001) (Fig. 1E). The assay of RNase R treatment revealed that linear GAPDH mRNA could be eliminated by RNase R, but it had no impact on the expression of hsa_circ_0014130 (Fig. 1F). These findings further validated the circular nature of hsa_circ_0014130, suggesting that it had resistance to RNase R. According to the annotation of circBase, hsa_circ_0014130 was found to be originated from the exons 7 of the PIP5K1A gene, which is located at chr1:151206672–151,212,515 (Fig. 1G).
hsa_circ_0014130 promotes malignant phenotypes of bladder cancer cells in vitro
The biological roles of hsa_circ_0014130 in the oncogenesis and metastasis of cancer of the bladder were investigated by transfecting with hsa_circ_0014130 overexpression vector and its specific siRNAs into T24 cells. Figure 2A showed the efficiency of ectopic overexpression and knockdown of hsa_circ_0014130 within T24 cells (P < 0.001). CCK-8 and the assay of colony creation divulged that ectopic expressions of hsa_circ_0014130 substantially enhanced the cell viability and growth of bladder cancer, while its expression silencing inhibited this biological behavior (P < 0.001) (Fig. 2B and C). On the other hand, our results suggested that the number of migratory and invasive cells was evidently increased by hsa_circ_0014130 overexpression and decreased by downregulation of hsa_circ_0014130 (P < 0.01) (Fig. 2D and E).
The effectiveness of ectopic overexpression and knockdown of hsa_circ_0014130 were validated through qRT-PCR (A). The influences of hsa_circ_0014130 expressions on biological characteristics of the cells of bladder cancer in vitro were evaluated by the assay of CCK-8 (B), colony creation (C), migration (D), and the assay of transwell invasion (E). The impacts of hsa_circ_0014130 expression on the expression of cell differentiation-related markers, i.e., loricrin, involucrin, filaggrin, and keratin K1 (F). **P < 0.01, ***P < 0.001
We also discovered the impact of hsa_circ_0014130 expressions on cell differentiation of bladder cancer by detecting differentiation-related markers. The obtained data demonstrated that the ectopic expression of hsa_circ_0014130 considerably elevated the expression of markers (linked with cell differentiation), i.e., loricrin, involucrin, filaggrin, and keratin K1 in bladder cancer cells (Fig. 2F). In contrast, the reduced expression of cell differentiation-related markers was observed in T24 cells transfected with short interfering RNA for hsa_circ_0014130 (Fig. 2F).
hsa_circ_0014130 binds to miR-132-3p as a miRNA sponge
The CircInteractome database was employed to identify the potential target of miRNA for hsa_circ_0014130, and the result of bioinformatics analysis revealed a potential interacting site between miR-132-3p and hsa_circ_0014130 (Fig. 3A and supplementary data 1). Then, bladder cancer cells were co-transfected with hsa_circ_0014130-WT or hsa_circ_0014130-MUT with miR-132-3p mimic. The reporter gene assay of luciferase demonstrated that an elevated expression of miR-132-3p obviously lowered the luciferase performance of hsa_circ_0014130-WT (P < 0.001), but no remarkable alteration was found in the hsa_circ_0014130-MUT vector (Fig. 3B). For more elucidation of the binding between miR-132-3p and hsa_circ_0014130 in bladder cancer cells, the probe of biotin-labeled hsa_circ_0014130 was designed to conduct an RNA pull-down assessment. We noted that the miR-132-3p expression was evidently enriched after purifying the RNAs interacting with hsa_circ_0014130 (P < 0.001) (Fig. 3C). On the other hand, the biotin-labeled miR-132-3p probe was transfected into bladder cancer cells. As expected, the pull-down assessment exhibited that the enrichment of hsa_circ_0014130 was considerably ameliorated compared to the negative control (P < 0.001) (Fig. 3D).
Bioinformatics analysis revealed that hsa_circ_0014130 had a potential site of binding for miR-132-3p (A). The binding between hsa_circ_0014130 and miR-132-3p in bladder cancer cells was validated with the aid of the reporter gene assay of luciferase (B), qRT-PCR assay (C), RNA pull-down assay (D and E), respectively. The expression levels of miR-132-3p in bladder cancer samples (F) and its correlation to hsa_circ_0014130 expressions were determined through the analysis of Pearson’s correlation (G). ***P < 0.001
According to the findings of this exploration, the ectopic expression of hsa_circ_0014130 considerably reduced the miR-132-3p expression in the cells of bladder cancer, although the knockdown of hsa_circ_0014130 resulted in an increased level of miR-132-3p expression (P < 0.001), as depicted in Fig. 3E. These data supported the binding of miR-132-3p to hsa_circ_0014130 in bladder cancer cells.
According to the achievements, we subsequently detected the miR-132-3p expression in bladder cancer specimens using qRT-PCR analysis. Our results proposed that bladder cancer samples had a decreased expression of miR-132-3p than non-tumor samples (P < 0.001) (Fig. 3F). The Pearson’s relationship analysis demonstrated that the hsa_circ_0014130 expression was inversely relevant to that of miR-132-3p in bladder cancer specimens (r = − 0.368, P < 0.05) (Fig. 3G).
KCNJ12 is a downstream target of miR-132-3p
To explore the downstream targets of miR-132-3p in bladder cancer cells, online database such as TargetScan and miRDB were used. Herein, it has been proposed that the 3′-UTR of KCNJ12 mRNA had a putative site of binding for miR-132-3p (Fig. 4A). For confirming the aforesaid interaction, luciferase reporter gene assay was conducted, and we found that the 3′-UTR of KCNJ12 wild type could be specifically bind by miR-132-3p (Fig. 4B). Furthermore, qRT-PCR displayed that mRNA expression of KCNJ12 in T24 cells was downregulated by transfecting with the mimic of miR-132-3p (P < 0.001), while was upregulated through the inhibitor of miR-132-3p (P < 0.001), as depicted in Fig. 4C. The biotin-labeled miR-132-3p probe further identify its interaction to KCNJ12 in bladder cancer cells (P < 0.001), as depicted in Fig. 4D. The underlined data illustrated that KCNJ12 was a straightforward target of miR-132-3p at the downstream.
Bioinformatics analysis revealed that the 3′-UTR of KCNJ12 mRNA had a putative binding site of miR-132-3p (A). The interaction between KCNJ12 mRNA and miR-132-3p in the cells of bladder cancer was validated by luciferase reporter gene assessment (B), qRT-PCR assay (C), RNA pull-down assessment (D), respectively. The expression levels of KCNJ12 mRNA (E) and protein (F) in bladder cancer samples. The association between KCNJ12 mRNA expression and LNM (G) and TNM stage (H) of bladder cancer patients were further explored. Survival curves of Kaplan-Meier for bladder cancer cases were stratified by the mRNA expression of KCNJ12 (I). The link between miR-132-3p and KCNJ12 mRNA expression was evaluated (J). The link between hsa_circ_0014130 and KCNJ12 mRNA expression was analyzed (K). **P < 0.01, ***P < 0.001
Subsequently, qRT-PCR and WB were exerted for determining the expression of KCNJ12 mRNA and protein in the samples related to the cancer of the bladder. KCNJ12 was markedly overexpressed in bladder cancer samples (P < 0.001) (Fig. 4E and F). In particular, bladder cancer patients with LNM and/or TNM stages III–IV had a greater level of expression for KCNJ12 (P < 0.01) (Fig. 4G and H). According to the median of KCNJ12 expression, 30 bladder cancer patients were further split into low (n = 15) and high expression (n = 15) group. An elevated expression of KCNJ12 was considerably linked with poorer survival in bladder cancer patients (P < 0.05) (Fig. 4I). Interestingly, there existed a negative link between miR-132-3p and KCNJ12 mRNA expressions in bladder cancer samples (r = − 0.766, P < 0.001), as depicted in Fig. 4J. Moreover, we also investigated the link between hsa_circ_0014130 and KCNJ12 mRNA expression, considering that they shared an inverse correlation to miR-132-3p. The underlined data revealed that the expression of hsa_circ_0014130 was linked with that of KCNJ12 in bladder cancer samples in a positive manner (r = 0.516, P < 0.01), as represented in Fig. 4K.
hsa_circ_0014130 promotes traits of bladder cancer cells in vitro via regulating miR-132-3p/KCNJ12 axis
It was hypothesized that hsa_circ_0014130 exerted the related biological functions in the cells of bladder cancer with the aid of mediating the miR-132-3p/KCNJ12 axis. For this purpose, T24 cells were transfected with the vector of hsa_circ_0014130, hsa_circ_0014130 + miR-NC, hsa_circ_0014130 + miR-132-3p mimic, hsa_circ_0014130 + sh-NC, and hsa_circ_0014130 + sh-KCNJ12, respectively. The data proposed that ectopic expression of hsa_circ_0014130 augmented the expression of KCNJ12 at the level of protein and mRNA, but the underlined effects were partially eliminated through transfecting with miR-132-3p mimic or siRNA for KCNJ12 (P < 0.001) (Fig. 5A and B). In terms of biological function, the cell proliferation of bladder cancer mediated through the overexpression of hsa_circ_0014130 could be partially abrogated by miR-132-3p mimic or KCNJ12 silencing (P < 0.001) (Fig. 5C and D). Furthermore, the promoting influences of hsa_circ_0014130 overexpression on invasion and migration capability of bladder cancer cells can be partially suppressed through elevated expression of miR-132-3p or KCNJ12 knockdown (P < 0.001) (Fig. 5E and F). The similar findings were also observed in the studies on the expression of cell differentiation-associated markers (P < 0.001) (Fig. 5G).
An elevated expression of KCNJ12 mRNA (A) and protein (B) mediated by hsa_circ_0014130 overexpression were partially eliminated by miR-132-3p mimic or si-KCNJ12. The promoting impact of hsa_circ_0014130 overexpression on cell growth (C and D), migration (E), invasion (F), and differentiation (G) were partially suppressed by miR-132-3p mimic or si-KCNJ12, respectively. ***P < 0.001 vs control; ##P < 0.01, ###P < 0.001 vs hsa_circ_0014130 + mimic control; &P < 0.05, &&P < 0.01, &&&P < 0.001 vs hsa_circ_0014130 + sh-NC
hsa_circ_0014130 promotes the growth of tumor xenograft in nude mice
To further investigate whether hsa_circ_0014130 function as an oncogene to play a similar role in vivo, tumor xenograft models were established in nude mice. Figure 6A–C indicated that the volume and weight of tumor xenograft and the expression of cell differentiation-related markers were markedly increased after hsa_circ_0014130 overexpression (P < 0.001). In contrast, bladder cancer cells transfected with siRNA for hsa_circ_0014130 resulted in a reduction in tumor weight, volume, and the expression cell differentiation-related markers (P < 0.001). However, we did not observe similar findings in KCNJ12−/− nude mice. In the context of KCNJ12 knockout, ectopic expression of hsa_circ_0014130 demonstrated no impact on the growth of tumor xenograft (Fig. 7A–C).
hsa_circ_0014130/miR-132-3p/KCNJ12 axis promotes the expression of EMT-associated markers and activates GSK3β/AKT signaling cascades
To evaluate whether the impact of hsa_circ_0014130/miR-132-3p/KCNJ12 axis on bladder cancer progression were associated with EMT process, we evaluated the expression of EMT-associated markers in bladder cancer cells. The obtained outcomes divulged that the increased expressions of hsa_circ_0014130 clearly elevated the expression of N-Cadherin, Twist, Vimentin, and MMP-7, as depicted in Fig. 8A. By contrast, the increased expression of these EMT-related markers mediated by hsa_circ_0014130 overexpression were neutralized by KCNJ12 knockdown (P < 0.001) (Fig. 8A). The similar findings were detected in the expression of p-GSK3β and p-AKT, suggesting that the oncogenic roles of hsa_circ_0014130/miR-132-3p/KCNJ12 axis could be implicated in GSK3β/AKT signaling pathway (P < 0.001)(Fig. 8B). Additionally, we also conducted the WB to evaluate the expressions of E-Cadherin and GSK3β/AKT pathway-related proteins in bladder cancer tissues. Based on Fig. 8C, the expression of E-Cadherin was reduced, and phosphorylation levels of GSK3β and AKT protein were evidently increased in patients with elevated hsa_circ_0014130 expressions compared with low expression (Fig. 8C).
The influences of hsa_circ_0014130 and KCNJ12 expression on EMT-related markers (A) and GSK3β/AKT signaling pathway in bladder cancer cells (B). ***P < 0.001 vs vector; #P < 0.05, ##P < 0.01, ###P < 0.001 vs hsa_circ_0014130 + sh-NC. The expression of E-Cadherin and GSK3β/AKT pathway-related proteins in bladder cancer samples (C)
Discussion
Reported investigations have shown that circRNAs are contributing to the tumorigenesis and development of several cancers in humans (Geng et al. 2018; Guarnerio et al. 2016). Herein, it was reported the abnormal expression of hsa_circ_0014130 in bladder cancer specimens and its biological performances in this malignancy. The obtained achievements demonstrated that increased hsa_circ_0014130 expression was substantially linked with LNM, advanced stage of TNM, and unfavorable prognosis of bladder cancer patients. An enhanced expression of hsa_circ_0014130 considerably accelerated the cell growth, migration, invasion, and differentiation for cancer of the bladder in vitro, while malignant biological behaviors were inhibited by hsa_circ_0014130 knockdown.
The dysregulation of hsa_circ_0014130 expression in human cancer samples has been reported by already published articles (Wang et al. 2020; Zhang et al. 2018). Recently, Geng et al. revealed that the expression of hsa_circ_0014130 was elevated in NSCLC samples, and its elevated expression was a predictor of bad survival of these patients (Geng et al. 2020). Using small interfering RNA targeted to hsa_circ_0014130, the biological aggressiveness of lung cancer cells was effectively suppressed (Geng et al. 2020). Our and these achievements consistently proposed that hsa_circ_0014130 can be a useful therapeutic target and prognostic marker of cancer cases. It has been well-established that circRNAs have higher biological stability than traditional linear RNAs, making them an attractive candidate for the diagnosis and prognostic evaluation (Suzuki and Tsukahara 2014; Wilusz 2018). In future research, we plan to identify the prognostic and diagnostic values for hsa_circ_0014130 in peripheral blood samples of bladder cancer cases.
Previous explorations have been eliminated that circRNAs behave as competitive endogenous RNAs for regulation of the expressions of mRNA through sponging to miRNAs (Hansen et al. 2013; Yang et al. 2021). Herein, miR-132-3p was initially evaluated as a putative target of hsa_circ_0014130 using online bioinformatics tools. Dual reporter of luciferase and RNA pull-down assessment then validated the interactions between hsa_circ_0014130 and miR-132-3p in bladder cancer cells. As a suppressor of a tumor, the aberrant expressions of miR-132-3p also were implicated in various human cancers, i.e., lung adenocarcinoma, bladder, breast, and colorectal cancers (Li et al. 2019; Liu et al. 2019; Su et al. 2020; Zhang et al. 2019). A recent report has shown that circDOCK1 executed its oncogenic roles through sponging to miR-132-3p and modulating Sox5 pathway (Liu et al. 2019). In the current exploration, our achievements illuminated that malignant characteristics of bladder cancer cells mediated by hsa_circ_0014130 overexpression can be partly reversed by upregulation of miR-132-3p. The underlined results further supported the effect of anti-cancer for miR-132-3p in the tumorigenesis and metastasis of bladder cancer.
To deeper comprehending the fundamental strategy of anti-cancer performance of miR-132-3p, we explored its potential target genes. The result of bioinformatics analysis showed that the 3′-UTR of KCNJ12 mRNA had a putative site of binding for miR-132-3p. Therefore, we executed a luciferase reporter gene assessment to confirm this hypothesis and observed that KCNJ12 was a straightforward downstream target of miR-132-3p. KCNJ12 is one of the main control proteins of K+ channels, which regulate cell excitability and implicate in multiple pathophysiologic processes (Houtman et al. 2012; Hugnot et al. 1997). Previous explorations demonstrated that KCNJ12 had an important value of research on anti-tumor treatment, and the expression loss of KCNJ12 resulted in cell cycle arrest (Lee et al. 2013; Lee et al. 2010). Recently, Khalilipour et al. found that specific mutations of KCNJ12 were related to a high risk of esophageal squamous cell carcinoma (Khalilipour et al. 2018), suggesting that KCNJ12 might contribute to tumor development. However, the expression level of KCNJ12 and its specific biological performance in bladder cancer was not fully understood. Herein, we first reported that KCNJ12 expression was markedly elevated in bladder cancer specimens and its considerable expression was linked with LNM and progressed tumor stage. Moreover, bladder cancer cases with elevated expression of KCNJ12 possessed a poorer prognosis than those with decreased expression, suggesting that KCNJ12 was a novel prognostic biomarker for bladder cancer cases. More importantly, knockdown of KCNJ12 partially suppressed the promoting influences of hsa_circ_0014130 on cell growth, invasion, migration, and differentiation of bladder cancer. These data proposed that KCNJ12 could function as an oncogene in tumors development and progression. EMT was a crucial step toward cancer progression and metastasis (Yang et al. 2020). Herein, it has been revealed that the expression of markers (linked with EMT), such as N-Cadherin, Vimentin, and MMP-7, was increased by hsa_circ_0014130 overexpression and was partially suppressed by KCNJ12 knockdown. The oncogenic tasks of hsa_circ_0014130/miR-132-3p/KCNJ12 axis in cell migration and invasion for cancer of the bladder might be implicated with EMT process.
To conclude, the underlined outcomes illuminated that hsa_circ_0014130 was decidedly overexpressed in bladder cancer specimens and related to aggressive characteristics and unfavorable prognosis of patients. hsa_circ_0014130 behave as a miR-132-3p sponge to perform oncogenic tasks in the oncogenesis and metastasis for cancer of the bladder by regulating KCNJ12 expression. These achievements might augment the comprehending of tumor pathogenesis and provide novel therapeutic targets for bladder cancer patients.
References
Antoni S, Ferlay J, Soerjomataram I, Znaor A, Jemal A, Bray F. Bladder cancer incidence and mortality: a global overview and recent trends. Eur Urol. 2017;71(1):96–108. https://doi.org/10.1016/j.eururo.2016.06.010.
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. https://doi.org/10.3322/caac.21492.
Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol. 2016;17(4):205–11. https://doi.org/10.1038/nrm.2015.32.
Chen X, Chen RX, Wei WS, Li YH, Feng ZH, Tan L, et al. PRMT5 circular RNA promotes metastasis of urothelial carcinoma of the bladder through sponging miR-30c to induce epithelial-mesenchymal transition. Clin Cancer Res. 2018;24(24):6319–30. https://doi.org/10.1158/1078-0432.ccr-18-1270.
Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, et al. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160(6):1125–34. https://doi.org/10.1016/j.cell.2015.02.014.
Geng Y, Bao Y, Zhang W, Deng L, Su D, Zheng H. Circular RNA hsa_circ_0014130 inhibits apoptosis in non-small cell lung cancer by sponging miR-136-5p and upregulating BCL2. Mol Cancer Res. 2020;18(5):748–56. https://doi.org/10.1158/1541-7786.mcr-19-0998.
Geng Y, Jiang J, Wu C. Function and clinical significance of circRNAs in solid tumors. J Hematol Oncol. 2018;11(1):98. https://doi.org/10.1186/s13045-018-0643-z.
Guarnerio J, Bezzi M, Jeong JC, Paffenholz SV, Berry K, Naldini MM, et al. Oncogenic role of fusion-circRNAs derived from cancer-associated chromosomal translocations. Cell. 2016;165(2):289–302. https://doi.org/10.1016/j.cell.2016.03.020.
Hammouz RY, Kołat D, Kałuzińska Ż, Płuciennik E, Bednarek AK. MicroRNAs: their role in metastasis, angiogenesis, and the potential for biomarker utility in bladder carcinomas. Cancers (Basel). 2021;13(4). https://doi.org/10.3390/cancers13040891.
Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–8. https://doi.org/10.1038/nature11993.
He Q, Yan D, Dong W, Bi J, Huang L, Yang M, et al. circRNA circFUT8 upregulates Krüpple-like factor 10 to inhibit the metastasis of bladder cancer via sponging miR-570-3p. Mol Ther Oncolytics. 2020;16:172–87. https://doi.org/10.1016/j.omto.2019.12.014.
Herranz H, Cohen SM. MicroRNAs and gene regulatory networks: managing the impact of noise in biological systems. Genes Dev. 2010;24(13):1339–44. https://doi.org/10.1101/gad.1937010.
Houtman MJ, Takanari H, Kok BG, van Eck M, Montagne DR, Vos MA, et al. Experimental mapping of the canine KCNJ2 and KCNJ12 gene structures and functional analysis of the canine K(IR)2.2 ion channel. Front Physiol, 3, 9. 2012. https://doi.org/10.3389/fphys.2012.00009.
Hugnot JP, Pedeutour F, Le Calvez C, Grosgeorge J, Passage E, Fontes M, Lazdunski M. The human inward rectifying K+ channel Kir 2.2 (KCNJ12) gene: gene structure, assignment to chromosome 17p11.1, and identification of a simple tandem repeat polymorphism. Genomics. 1997;39(1):113–6. https://doi.org/10.1006/geno.1996.4450.
Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32(5):453–61. https://doi.org/10.1038/nbt.2890.
Khalilipour N, Baranova A, Jebelli A, Heravi-Moussavi A, Bruskin S, Abbaszadegan MR. Familial esophageal squamous cell carcinoma with damaging rare/germline mutations in KCNJ12/KCNJ18 and GPRIN2 genes. Cancer Genet. 2018;221:46–52. https://doi.org/10.1016/j.cancergen.2017.11.011.
Lee I, Lee SJ, Kang TM, Kang WK, Park C. Unconventional role of the inwardly rectifying potassium channel Kir2.2 as a constitutive activator of RelA in cancer. Cancer Res. 2013;73(3):1056–62. https://doi.org/10.1158/0008-5472.can-12-2498.
Lee I, Park C, Kang WK. Knockdown of inwardly rectifying potassium channel Kir2.2 suppresses tumorigenesis by inducing reactive oxygen species-mediated cellular senescence. Mol Cancer Ther. 2010;9(11):2951–9. https://doi.org/10.1158/1535-7163.mct-10-0511.
Li S, Xu JJ, Zhang QY. MicroRNA-132-3p inhibits tumor malignant progression by regulating lysosomal-associated protein transmembrane 4 beta in breast cancer. Cancer Sci. 2019;110(10):3098–109. https://doi.org/10.1111/cas.14164.
Liu P, Li X, Guo X, Chen J, Li C, Chen M, et al. Circular RNA DOCK1 promotes bladder carcinoma progression via modulating circDOCK1/hsa-miR-132-3p/Sox5 signalling pathway. Cell Prolif. 2019;52(4):e12614. https://doi.org/10.1111/cpr.12614.
Lu Q, Liu T, Feng H, Yang R, Zhao X, Chen W, et al. Circular RNA circSLC8A1 acts as a sponge of miR-130b/miR-494 in suppressing bladder cancer progression via regulating PTEN. Mol Cancer. 2019;18(1):111. https://doi.org/10.1186/s12943-019-1040-0.
Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–8. https://doi.org/10.1038/nature11928.
Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin. 2021;71(1):7–33. https://doi.org/10.3322/caac.21654.
Su Y, Shetty A, Jiang F. Integrated analysis of miRNAs and DNA methylation identifies miR-132-3p as a tumor suppressor in lung adenocarcinoma. Thorac Cancer. 2020;11(8):2112–24. https://doi.org/10.1111/1759-7714.13497.
Suzuki H, Tsukahara T. A view of pre-mRNA splicing from RNase R resistant RNAs. Int J Mol Sci. 2014;15(6):9331–42. https://doi.org/10.3390/ijms15069331.
Wang M, Shi J, Jiang H, Xu K, Huang Z. Circ_0014130 participates in the proliferation and apoptosis of nonsmall cell lung cancer cells via the miR-142-5p/IGF-1 axis. Cancer Biother Radiopharm. 2020;35(3):233–40. https://doi.org/10.1089/cbr.2019.2965.
Wei XC, Lv ZH. MicroRNA-132 inhibits migration, invasion and epithelial-mesenchymal transition via TGFβ1/Smad2 signaling pathway in human bladder cancer. Onco Targets Ther. 2019;12:5937–45. https://doi.org/10.2147/ott.s201731.
Wilusz JE. A 360° view of circular RNAs: from biogenesis to functions. Wiley Interdiscip Rev RNA. 2018;9(4):e1478. https://doi.org/10.1002/wrna.1478.
Yang J, Antin P, Berx G, Blanpain C, Brabletz T, Bronner M, et al. Guidelines and definitions for research on epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2020;21(6):341–52. https://doi.org/10.1038/s41580-020-0237-9.
Yang X, Ye T, Liu H, Lv P, Duan C, Wu X, et al. Expression profiles, biological functions and clinical significance of circRNAs in bladder cancer. Mol Cancer. 2021;20(1):4. https://doi.org/10.1186/s12943-020-01300-8.
Yoshino H, Seki N, Itesako T, Chiyomaru T, Nakagawa M, Enokida H. Aberrant expression of microRNAs in bladder cancer. Nat Rev Urol. 2013;10(7):396–404. https://doi.org/10.1038/nrurol.2013.113.
Zhang M, Li Y, Wang H, Yu W, Lin S, Guo J. LncRNA SNHG5 affects cell proliferation, metastasis and migration of colorectal cancer through regulating miR-132-3p/CREB5. Cancer Biol Ther. 2019;20(4):524–36. https://doi.org/10.1080/15384047.2018.1537579.
Zhang S, Zeng X, Ding T, Guo L, Li Y, Ou S, Yuan H. Microarray profile of circular RNAs identifies hsa_circ_0014130 as a new circular RNA biomarker in non-small cell lung cancer. Sci Rep. 2018;8(1):2878. https://doi.org/10.1038/s41598-018-21300-5.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Graphical highlights
1. hsa_circ_0014130 was markedly overexpressed in bladder cancer samples and related to aggressive biological characteristics and poor survival.
2. hsa_circ_0014130 function as a miR-132-3p sponge to promote cell proliferation, migration, invasion, and differentiation of bladder cancer in vitro.
3. KCNJ12 was a direct target of miR-132-3p at the downstream, and its silencing partially abrogated the oncogenic roles mediated by the overexpression of hsa_circ_0014130 in bladder cancer cells.
4. hsa_circ_0014130/miR-132-3p/KCNJ12 axis was involved in the development and progression of bladder cancer, and it might be a novel therapeutic target for bladder cancer.
Rights and permissions
About this article
Cite this article
Li, G., Guo, By., Wang, Hd. et al. CircRNA hsa_circ_0014130 function as a miR-132-3p sponge for playing oncogenic roles in bladder cancer via upregulating KCNJ12 expression. Cell Biol Toxicol 38, 1079–1096 (2022). https://doi.org/10.1007/s10565-021-09668-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10565-021-09668-z