Abstract
Exposure to environmental and occupational contaminants leads to lung cancer. 3-Nitrobenzanthrone (3-nitro-7H-benz[de]anthracen-7-one, 3-NBA) is a potential carcinogen in ambient air or diesel particulate matter. Studies have revealed that short-term exposure to 3-NBA induces cell death, reactive oxygen species activation, and DNA adduct formation and damage. However, details of the mechanism by which chronic exposure to 3-NBA influences lung carcinogenesis remain largely unknown. In this study, human lung epithelial BEAS-2B cells were continuously exposed to 0–10-μM 3-NBA for 6 months. NanoString analysis was conducted to evaluate gene expression in the cells, revealing that 3-NBA–mediated transformation results in a distinct gene expression signature including carbon cancer metabolism, metastasis, and angiogenesis. Alterations in tumor-promoting genes such as EREG (epiregulin), SOX9, E-cadherin, TWIST, and IL-6 were involved in epithelial cell aggressiveness. Kaplan–Meier plotter analyses indicated that increased EREG and IL-6 expressions in early-stage lung cancer cells are correlated with poor survival. In vivo xenografts on 3-NBA–transformed cells exhibited prominent tumor formation and metastasis. EREG knockout cells exposed to 3-NBA for a short period exhibited high apoptosis and low colony formation. By contrast, overexpression of EREG in 3-NBA–transformed cells markedly activated the PI3K/AKT and MEK/ERK signaling pathways, resulting in tumorigenicity. Furthermore, elevated IL-6 and EREG expressions synergistically led to STAT3 signaling activation, resulting in clonogenic cell survival and migration. Taken together, chronic exposure of human lung epithelial cells to 3-NBA leads to malignant transformation, in which the EREG signaling pathway plays a pivotal mediating role.
Graphical abstract

• Short-term exposure of lung epithelial cells to 3-NBA can lead to ROS production and cell apoptosis.
• Long-term chronic exposure to 3-NBA upregulates the levels of tumor-promoting genes such as EREG and IL-6.
• Increased EREG expression in 3-NBA–transformed cells markedly contributes to tumorigenesis through PI3K/AKT and MEK/ERK activation and synergistically enhances the IL-6/STAT3 signaling pathway, which promotes tumorigenicity.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Lung cancer has been commonly associated with exposure to environmental and occupational pollutants (Field and Withers 2012). The high number of lung cancer deaths among individuals who have never smoked indicates that risk factors other than tobacco smoking substantially contribute to lung cancer mortality (Casal-Mourino et al. 2019). Diesel engine exhaust (DEE) is a major source of urban pollution, and its composition is complex owing to its gas and particulate phases (Steiner et al. 2016). Toxic chemicals adsorbed onto diesel exhaust particles (DEPs) include polycyclic aromatic hydrocarbons (PAHs) and nitro-PAHs, which exert carcinogenic properties through intracellular metabolic activation by binding to DNA and inducing mutations (Long et al. 2016; White et al. 2017). Case-control studies of 11 populations with smoking-adjusted analysis have revealed an increased risk of lung cancer after exposure to DEE (Olsson et al. 2011). Moreover, animal studies have revealed that long-term (3 months) exposure to low (100 μg/m3) or high (3 mg/m3) concentrations of DEP for 1 h/day 5 days/week increased airway inflammation, pulmonary fibrosis, and goblet cell hyperplasia (Kim et al. 2016). These results indicate that chronic exposure to DEPs may promote airway remodeling and angiogenesis. Additionally, DEP extracts instilled intratracheal lung carcinomas and sarcomas in animals (Kunitake et al. 1986; Grimmer et al. 1987).
The nitro-PAH 3-nitrobenzanthrone (3-nitro-7H-benz[de]anthracen-7-one, 3-NBA) is a potential carcinogen typically present in diesel emissions, gasoline exhaust particles, and atmospheric particles (Arlt 2005; Santos et al. 2019) and has been reported to possess high genotoxic and mutagenic potential (Nagy et al. 2006, 2007; Bieler et al. 2007). Notably, 3-aminobenzanthrone was identified as the predominant metabolite of 3-NBA in the urine of DEE-exposed workers (Seidel et al. 2002). In addition, after 3-NBA is metabolically activated, the final electrophilic metabolite can bind to nucleophilic sites in biomolecules such as DNA (Arlt et al. 2005; Stiborova et al. 2010). Thus, nucleophilic attacks may occur at the nitrogen and carbon rings of DNA (Takamura-Enya et al. 2007). Intratracheal instillation of 3-NBA (total dose = 2.5 mg) for 18 months caused tumor formation in rats; specifically, 3-NBA induced squamous cell carcinoma in 11 of 16 rats (68.7%) and lung adenocarcinoma in one rat (Nagy et al. 2005). Evidently, 3-NBA causes premutagenic lesions in DNA and may initiate tumor progression in target tissues. In the MutaMouse model, 3-NBA also causes DNA adducts in lung tissue when administered through gavage (Arlt et al. 2008). In addition, 3-NBA induces cell death, the production of reactive oxygen species (ROS), and DNA adduct formation and lesion damage in several types of cells and tissues (Arlt et al. 2008; Arlt et al. 2001; Hansen et al. 2007; Stiborova et al. 2012). These data suggest that 3-NBA is a potential lung carcinogen.
Increasing evidence suggests that the 3-NBA present in ambient air and diesel particulate matter (PM) is a critical carcinogenic compound. In fact, DEPs contain a heterogeneous mixture of transition metals, PAHs, and nitro-PAHs, including 3-NBA. An individual’s daily breath intake of 3-NBA contained in DEPs may be as high as 90 pg/day (Arlt 2005). However, the qualitative and quantitative compositions of DEE vary according to fuel, engine, and emission system. Moreover, 3-NBA exposure concentrations may vary between the general population and specific high-risk groups exposed to engine exhaust, domestic coal-burning stoves, rainwater, and ambient air PM. Accordingly, the detailed mechanism of carcinogenicity mediated by consistent, long-term exposure to single-compound 3-NBA remains unclear.
To determine the cellular and molecular processes involved in carcinogenesis, lung epithelial BEAS-2B cells were continuously exposed to 3-NBA in a culture over an extended period. Analysis of NanoString PanCancer gene expression profiling revealed that 3-NBA–mediated cell transformation significantly altered several tumor-promoting genes such as EREG. EREG is an agonist of EGFR that homodimerizes EGFR, thereby activating EGFR downstream signaling cascades such as the MEK/ERK pathway and subsequently promoting cancer cell survival and proliferation (Shelly et al. 1998). Signaling of the EREG/EGFR pathway regulates tumor development processes, including apoptosis resistance, cell proliferation, metastasis, and angiogenesis (Sunaga and Kaira 2015). Notably, EREG expression is typically low in nontumor cells but is elevated in several human tumors such as lung cancer (Sunaga et al. 2013). We hypothesized that EREG may play a key role in cell survival, and we further prepared BEAS-2B EREG-depleted cells by using CRISPR/Cas9 technology. EREG knockout can markedly increase more apoptosis and reduce clonogenic cell survival upon stimulation with 3-NBA. In the current study, chronic 3-NBA exposure induced malignant transformation through the altered expression of tumor-promoting genes that mediated clonogenic cell survival, anchorage-independent cell growth, migration, and in vitro angiogenesis. In addition, EREG-mediated downstream signaling paths including PI3K/AKT, MEK/ERK, and STAT3 may contribute to tumorigenicity in 3-NBA–transformed cells.
Materials and methods
Cell culture
Human lung epithelial cells (BEAS-2B cells) were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA). The cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS; Invitrogen, Thermo Fisher Scientific, Carlsbad, CA, USA) in a 5% CO2 humidified incubator at 37°C. 3-NBA (Carbosynth, Berkshire, UK) was suspended and diluted to appropriate concentrations in the culture medium. In total, 1 × 106 BEAS-2B cells were seeded onto a 10-cm dish and treated with 0–10-μM 3-NBA per passage. This process was sustained for 6 months (2 passages/week, BEAS-2B 3-NBA-6M). Continuous culture-matched BEAS-2B cells were treated with dimethyl sulfoxide (DMSO) as controls (BEAS-2B control-6M).
Cell proliferation assay
In total, 1 × 105 BEAS-2B parental and transformed 3-NBA-6M or control-6M cells were seeded on 6-cm dishes. Growth rates were then examined with trypan blue exclusion, and cells were quantitated on days 1–4 by using the Countess cell counter (Invitrogen). The results are presented as total cell numbers.
Transfection and RNA interference
In total, 5 × 106 BEAS-2B 3-NBA-6M or control-6M cells were transiently transfected with 500-nM small interfering RNAs targeting EREG and IL-6 (Dharmacon, Lafayette, CO, USA) or a scramble control according to the manufacturer’s instructions. The samples were then transferred into an electroporation cuvette by using a Nucleofector system; the transfections were performed according to the manufacturer’s instructions. After transfection, the cells were immediately transferred into prewarmed complete DMEM medium and cultured in a regular culture condition.
Cell apoptosis assay
Apoptosis analysis was performed through Annexin V-FITC/propidium iodide (PI) staining (BD Biosciences, San Jose, California, USA) with a detection kit according to the manufacturer’s instructions. In brief, 3-NBA–treated cells were harvested, washed twice with cold phosphate-buffered saline (PBS), and then resuspended in binding buffer. The cell suspension (105 cells) was mixed with equal volumes (5 μL) of Annexin V-FITC and PI at room temperature without exposure to light for 15 min. Subsequently, the percentage of apoptotic cells was measured using a BD LSRFortessa cell analyzer.
NanoString gene expression analysis
Gene expression assay was performed with the NanoString PanCancer Pathway Panel by using 50 ng of total RNA according to the manufacturer’s instructions. The data were then processed and analyzed using nSolver analysis software 4.0 (NanoString Technologies, WA, USA). The geometric mean of housekeeping genes was calculated to normalize all samples. The fold change (FC) of differentially expressed gene expression between 3-NBA-6M and control-6M cells was obtained. Deregulated KEGG pathways were identified using STRING (http://string-db.org).
Immunoblot analysis
Total cell lysates (40 μg/lane) were added to each lane of 10% SDS-PAGE gel and analyzed by blotting with antibodies specific to p53 (Santa Cruz, Dallas, TX, USA, sc-126), SOX9 ([SRY (sex determining region Y)-box 9]; ABclonal, Woburn, MA, USA, A2479), TWIST1 (Santa Cruz, sc-81417), TWIST2 (ABclonal, A5599), SANI2 (Proteintech, Rosemont, IL, USA, 12129-1-AP), GREM1 (ABclonal, A11595), THBS (ABclonal, A2125), E-cadherin (Thermo, 13-1700), STAT3 (Cell Signaling, Danvers, MA, USA, 9131), p-STAT3 (Cell Signaling, 9139), ERK (Thermo, MA5-15605), p-ERK (Cell Signaling, 5683), AKT (Proteintech, 10176-2-AP), p-AKT (Cell Signaling, 9271), MET (Cell Signaling, 3127), and EREG (R&D, Minneapolis, MN, USA, AF1195).
Soft agar colony formation assays
In total, 10,000 cells per well were seeded in 12-well plates for 3 weeks to achieve layer agar cultures according to the method described in our previous publication (Wu et al. 2017). Thereafter, the cells were resuspended in 10% FBS in DMEM with 0.33% agar; the complete medium in the upper layer was replaced once per week. The colonies were stained with crystal violet (0.01%, w/v) after 3 weeks.
Clonogenic assays
EREG knockout or control cells (250 cells/well) were seeded in 6-well plates, and 10-μM 3-NBA was replenished every 3 days for 2 weeks. Induction of colony formation was also measured in 3-NBA-6M or control-6M cells with recombinant human IL-6 (rIL-6) treatment in the presence or absence of indicated inhibitors. Monolayers were then fixed and stained with 0.5% crystal violet, washed, and dried. After the plates were photographed, the crystal violet was solubilized with 20% acetic acid, and the intensity was spectrophotometrically quantified at 490 nm.
Migration and invasion assay
The BEAS-2B–transformed and control cells (2 × 105) were resuspended in 100 μL of serum-free DMEM before being placed in the upper chamber of 24-well transwell plates (Costar, St Lowell, MA, USA); 1 mL of 20% FBS medium was used as a chemoattractant in the lower chamber. For invasion assays, the upper chambers were coated with 100 μL of diluted Matrigel and were left to gel at 37°C for 4 h. BEAS-2B–transformed and control cells were preincubated in serum-free medium overnight. Subsequently, the cells were suspended in 100 μL of serum-free medium before being placed in the upper Matrigel-coated chamber; the lower well was placed in a 20% FBS medium. After 24 h of incubation, the migration or invasion ability of the cells was measured as described previously (Wu et al. 2017).
Endothelial cell tube formation assay
The Matrigel matrix was placed on ice overnight before tube formation analysis. Next, 100 μL of Matrigel was transferred to one well in a 48-well plate, which was placed in regular culture condition overnight. Primary human umbilical vein endothelial cells (HUVECs) derived from PromoCell (Heidelberg, Germany) were cultured in EGM-2 medium, and 3 × 104 cells/100 μL were mixed with 80 μg of conditioned medium from BEAS-2B 3NBA-6M or control-6M cells from each well. After 16 h of incubation, the absolute number of tube formations in five randomly selected fields per well was measured using light microscopy.
Enzyme-linked immunosorbent assay
The IL-6 protein levels in the supernatants were determined using commercial enzyme-linked immunosorbent assay (ELISA) kits (eBioscience, San Diego, CA, USA) according to the manufacturer’s instructions. ELISA kits were also used to confirm secretory EREG protein levels (Abnova, Taipei, Taiwan, KA5094).
Generation of EREG knockout BEAS-2B cells through CRISPR/Cas9-mediated genome editing
Human EREG knockout BEAS-2B cells were generated using CRISPR/Cas9 technology. Two sgRNAs targeting the third or seventh coding exon were separately cloned into pAll-Cas9. We purchased pPpuro plasmid from the National RNAi Core Facility (Academia Sinica, Taipei, Taiwan). The sgRNA targeting sites 5′-ATGGAATCACAGTTGTACTGAGG-3′ and 5′-GACAGAAGACAATCCACGTGTGG-3′ were used for sgRNA#1 and sgRNA#2, respectively. Human BEAS-2B cells were transfected with two sgRNA plasmids by using the 4D-Nucleofector system according to the manufacturer’s instructions (Lonza, Morristown, NJ, USA). After 2 days, the transfected cells were treated with puromycin (2 μg/mL) for 1 week. The live cells were then diluted in a 96-well plate to isolate single-cell clones. The depletion of EREG in the EREG knockout cells was confirmed through immunoblot analysis and genomic DNA sequencing.
Xenograft assays of tumorigenicity in vivo
A total of 20 female BALB/cAnN.Cg-Foxn1nu/CrlNarl nude mice aged 6 weeks were obtained from the National Laboratory Animal Center (Taipei, Taiwan) and housed in a pathogen-free facility. To evaluate solid tumors generated in the nude mice, 2 × 105 BEAS-2B 3-NBA-6M (10 μM)–transformed (3-NBA–T) or control cells in 100-μL PBS were subcutaneously inoculated (n = 5 nude mice/group), and tumor size was determined as described previously (Wu et al. 2017). The animals were sacrificed after the 6-week tumor growth experiment. The invasive ability of the 3-NBA–T cells was further determined by intravenously injecting transformed or control cells (1 × 106) into five nude mice/group. After 7 weeks, all animals involved in invasion experiments were sacrificed. We examined the tumor foci in the lung tissues of the nude mice and determined EREG and IL-6 expressions by using immunohistochemistry with EREG (Abcam, ab175118) and IL-6 (Bioss, Woburn, MA, USA, bs-0781R) antibodies. Animal care and experiment protocols were approved by the Institutional Animal Care and Use Committee of Taipei Medical University (Approval No. LAC-2020-0061).
Kaplan–Meier plotter database
The Kaplan–Meier (KM) survival plotter database (http://kmplot.com/analysis/), which includes data on 1925 patients with lung cancer, was used to evaluate the prognostic values of the 3-NBA–mediated genes. KM plots were obtained for each gene symbol in the database. The high- and low-expression groups of patients with lung cancer were classified according to optimal gene expression cutoff values, and 10-year overall survival analyses were conducted. The number of patients, upper quartile or median survival months, and hazard ratios with 95% confidence intervals and log rank p values were extracted from the database.
Statistical analysis
Data were presented as means with standard deviations and statistically analyzed using GraphPad Prism 8.0 (GraphPad Software, San Diego, CA, USA). Between-group differences were analyzed using the Student’s t test. To compare the results obtained from multiple treatments, one-way analysis of variance and Tukey’s honestly significant difference post hoc test were conducted.
Results
Differential gene expression in human lung epithelial cells after long-term exposure to 3-NBA
The chemical structure of 3-NBA is presented in Supplementary Material 1. BEAS-2B cells exposed to 1–160-μM 3-NBA for 24 h revealed a dose-dependent 10.43–65.42% reduction in cell viability and a 15.10–110.28% increase in intracellular ROS when compared with DMSO-exposed control cells (Supplementary Material 1). Moreover, 1- or 10-μM 3-NBA treatment resulted in a 1.89–5.12% increase in cell apoptosis for 24 h and repressed cell proliferation by 20.28–30.38% for 4 days. These results indicate that treatment with 1- or 10-μM 3-NBA can induce cellular ROS formation and apoptotic cell death within a short period.
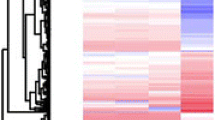
The malignant transformation potential of BEAS-2B cells in vitro was assessed after long-term exposure to 3-NBA. As presented in Fig. 1a, subconfluent cell cultures continuously exposed to 3-NBA (0.1–10 μM) and DMSO control for 6 months were designated as BEAS-2B 3-NBA-6M and control-6M, respectively. To identify a specific gene alteration pattern caused by 1- or 10-μM 3-NBA, gene expression profiling was performed using the NanoString PanCancer Panel. The gene expression profile of BEAS-2B 3-NBA-6M (1 or 10 μM) cells differed from that of the control-6M cells (Fig. 1b). The most differentially expressed genes in each gene set were identified; the global significance score indicates the extent of differential expression. The heatmap represents the scores of the different gene sets between BEAS-2B cells treated with 1- or 10-μM 3-NBA and the control cells treated with DMSO (Fig. 1c). The pathway scores condense the gene expression profile of each sample assessed in later experiments. The scores were plotted to compare pathway differences between 1- and 10-μM 3-NBA (Fig. 1d). The average scores for pathways related to carbon cancer metabolism, epithelial-to-mesenchymal transition (EMT) to metastasis, angiogenesis regulation, and sprouting angiogenesis increased in a 3-NBA dose-dependent manner. The top 20 up- and downregulated genes in BEAS-2B 3-NBA-6M (10 μM) cells and the relative gene sets of the cancer progression pathways are presented in Fig. 1e.
Differential gene expression after long-term exposure of BEAS-2B cells to 3-NBA. a Schematic representation of BEAS-2B cells continuously exposed to 0–10-μM 3-NBA for 6 months (BEAS-2B 3-NBA-6M cells). b mRNA quantified using the PanCancer Progression Panel through NanoString analysis with BEAS-2B samples exposed to 3-NBA and DMSO control for an extended period. Results are depicted as a volcano plot indicating the differential gene expression of BEAS-2B cells treated with 1- or 10-M 3-NBA for 6 months compared with the BEAS-2B DMSO control. c Heatmap displaying differential gene expression between BEAS-2B cells treated with DMSO control and those treated with 1- or 10-μM 3-NBA. The most differentially expressed genes in each gene set were identified, and the extent of differential expression in each gene set was summarized using a global significance score. The heatmap displays each sample’s global significance score. Orange denotes gene sets whose genes exhibited higher differential expression with the covariate, and blue denotes gene sets with lower differential expression. d Pathway scores plotted to compare variation between 0-, 1-, and 10-μM 3-NBA-6M. The lines denote each pathway’s average score among the group conditions. e Top 20 upregulated and downregulated genes in the 10 μM 3-NBA-6M group; p values and log2 FC on the left panel correspond to the cancer progression pathway gene sets. False discovery rate was controlled using the Benjamini–Yekutieli procedure, and adjusted p values were generated
3-NBA–mediated cell transformation promotes proliferation
To determine whether 3-NBA can transform lung epithelial cells into the cancer progression phenotype, NanoString gene expression analysis was performed by applying the nCounter PanCancer Progression Pathways Panel to RNA from 3-NBA–transformed cells. The panel contains 770 genes involved in several key cancer progression steps such as cell proliferation, extracellular matrix remodeling, EMT, and metastasis. Carbon cancer metabolism gene sets such as FGFR2 and MET were upregulated, but key tumor suppressor genes such as p53 were downregulated. The p53 protein was downregulated in BEAS-2B 3-NBA-6M cells compared with the control-6M cells (Supplementary Material 2).
To examine the proliferation alteration in BEAS-2B 3-NBA-6M cells, we analyzed the proliferation gene set signature (Fig. 2). Figure 2a indicates that the genes in 3-NBA–transformed cells were significantly altered. The analysis of the proliferation pathway gene set revealed differentially expressed genes with log2 FC > 1.5, including EREG, IL-6, SOX9, THBS1, and CD24, in BEAS-2B cells transformed through exposure to 10-μM 3-NBA. Notably, the most differentially expressed gene, EREG, was significantly upregulated by log2 FCs of 3.00 and 6.88 in 1- and 10-μM 3-NBA–transformed cells, respectively. To evaluate the potential functional interrelationships within the proliferation gene set, the STRING database (Szklarczyk et al. 2017) was used to determine protein–protein interactions after 3-NBA transformation. The results revealed that the upregulated genes affected after 3-NBA transformation were mainly in the EREG/AKT, IL-6/SOX9, and MET protein networks. As expected, 1- or 10-μM 3-NBA–transformed BEAS-2B cell proliferation was enhanced by 33.76–48.47% compared with control cells after treatment for 4 days (Fig. 2b).
Cell proliferation increased in lung epithelial cells exposed to 3-NBA for an extended period. a Differentially expressed genes with p ≤ 0.01 and log2 FC > 1 or log2 FC < −1 in BEAS-2B cells treated with 10-μM 3-NBA for 6 months when compared with the BEAS-2B DMSO control (0-μM 3-NBA). The analysis of proliferation-related genes in BEAS-2B 3-NBA-6M cells treated with 1- or 10-μM 3-NBA is depicted as a volcano plot. The inter protein–protein interaction of the genes upregulated (log2 FC > 0.5) under long-term 10-μM 3-NBA treatment during cell proliferation was obtained through STRING network analysis. The network yielded up to three clusters by using k-means clustering and had a high confidence of interaction score (0.700). b BEAS-2B 3-NBA-6M (1–10 μM)–transformed cells exhibited increased cell proliferation when compared with control cells after treatment for 4 days. c 3-NBA-transformed cells grown in plates with soft agar cultures. Representative images of colonies (scale bar, 400 μm) are displayed. The 3-NBA–mediated transformation significantly increased colony numbers in BEAS-2B 3-NBA-6M cells when compared with the DMSO control. d Expression levels of EREG, SOX9, and MET proteins examined through immunoblotting. Actin served as a loading control, and protein expression was normalized to actin. The relative values of proteins are expressed as the FC over control. Data are presented as the mean ± standard deviation of three independent experiments. (*p < 0.05 and **p < 0.01)
Anchorage-independent growth in soft agar in vitro is an indication of cellular transformation and is closely associated with tumorigenesis in vivo (Cox and Der 1994). Data from the clonogenic soft agar assay revealed 2.43- to 2.77-fold increases in colony numbers in the BEAS-2B 3-NBA–transformed group compared with the controls, indicating malignancy (Fig. 2c). To determine whether the changes in the gene expression patterns were consistent with those in protein levels, we investigated several principal components involved in cell proliferation. The levels of EREG, AKT, SOX9, and MET were increased in the BEAS-2B 3-NBA group treated with 10-μM 3-NBA (Fig. 2d). Notably, an evident dose-dependent increase (1.83- and 2.69-fold) in EREG proteins was observed in cells transformed with 1–10-μM 3-NBA.
Long-term exposure to 3-NBA induces human lung epithelial cell aggressiveness
Active tumor cell migration is a crucial prerequisite for tumor cell invasion and metastasis. To determine whether 3-NBA–mediated transformation can affect EMT phenotype, we conducted a NanoString gene expression analysis, revealing that 10-μM 3-NBA–mediated transformation upregulated EMT and EMT to metastasis gene sets such as SOX9 (log2 FC = 1.76) and SANI2 (log2 FC = 1.35) but downregulated CDH1 (E-cadherin) (log2 FC = −4.42) during EMT (Fig. 3a and b). To examine the functional role of 3-NBA–mediated transformation in cell migration, we performed a transwell migration assay, which indicated that migration ability increased 2.12- to 2.34-fold in cells transformed with 1- or 10-μM 3-NBA (Fig. 3c). Furthermore, compared with control cells, 1- or 10-μM 3-NBA–transformed BEAS-2B cells exhibited 2.59- to 3.56-fold enhancements in invasion ability (Fig. 3d). We further examined several key regulators of EMT in 3-NBA–transformed cells. Western blot assays revealed markedly reduced protein levels of E-cadherin (0.32-fold) and increased levels of TWIST1 and TWIST2 (1.91- and 2.56-fold, respectively) in BEAS-2B cells transformed with 10-μM 3-NBA (Fig. 3e).
3-NBA–transformed lung epithelial cells exhibited increased cell migration and invasion. a The EMT-related differentially expressed genes with p ≤ 0.01 and log2 FC > 1 or log2 FC < −1 in BEAS-2B cells treated with 10-μM 3-NBA for 6 months when compared with the DMSO control cells (0-μM 3-NBA). The analysis of proliferation-related genes in BEAS-2B 3-NBA-6M cells treated with 1- or 10-μM 3-NBA is depicted as a volcano plot. The protein network of the EMT-related genes upregulated after long-term 10-μM 3-NBA treatment during cell proliferation was obtained through STRING network analysis. b The differentially expressed genes in EMT and metastasis-related genes with p ≤ 0.01 are listed and depicted as a volcano plot. The protein network displays upregulated genes after 10-μM 3-NBA treatment. c BEAS-2B 3-NBA-6M (1–10 μM)–transformed cells exhibited increased cell migration compared with DMSO control cells. d BEAS-2B 3-NBA-6M (1–10 μM)–transformed cells exhibited increased cell invasion compared with control cells. e Expression levels of TWIST1, TWIST2, p-AKT, and AKT proteins examined through immunoblotting and normalized to actin. The relative expressions of proteins are indicated as the FC over control. Data are presented as the mean ± standard deviation of three independent experiments. (*p < 0.05 and **p < 0.01)
Angiogenesis is critical to cancer development and is characterized by abnormal tumor vascularization. Therefore, we conducted NanoString gene expression analysis to examine angiogenesis regulation in 3-NBA–transformed cells. Differentially expressed genes from the angiogenesis or sprouting angiogenesis processes were observed (Fig. 4a and b). To determine whether 3-NBA–transformed cells induced angiogenesis in vitro, we performed tube formation assays, revealing that 3-NBA–mediated transformation markedly enhanced tube formation (1.77- to 2.04-fold), particularly at concentrations of 1 and 10 μM (Fig. 4c). The IL-6/STAT3 axis may facilitate angiogenesis in several cancers. High levels of systemic IL-6 expression are correlated with poor prognosis in patients with non-small-cell lung cancer (NSCLC) (Silva et al. 2017). The gene expression levels of IL-6 increased 3.09- to 4.62-fold in 3-NBA–transformed cells (1 or 10 μM; Fig. 4d). Moreover, secretory IL-6 protein levels were markedly elevated (1.58- to 2.15-fold; Fig. 4e). However, NanoString analysis indicated that the upregulation of THBS1 and GREM1 in 3-NBA–transformed cells did not significantly alter protein levels (Fig. 4f).
3-NBA–transformed lung epithelial cells exhibited increased in vitro angiogenesis. a Angiogenesis-related differentially expressed genes with p ≤ 0.01 and log2 FC > 0.5 or log2 FC < −0.5 in BEAS-2B cells treated with 10-μM 3-NBA for 6 months compared with the DMSO control cells. The analysis of angiogenesis-related genes in BEAS 3-NBA-6M cells treated with 1- or 10-μM 3-NBA is depicted as a volcano plot. The protein network of the EMT-related genes upregulated after long-term 10-μM 3-NBA treatment during cell proliferation was obtained through STRING network analysis. b Differentially expressed genes in sprouting angiogenesis with p ≤ 0.01 and log2 FC > 0.5 or log2 FC < −0.5. All differentially expressed genes are depicted as a volcano plot. The protein network indicates the upregulated genes in cells transformed after 10-μM 3-NBA treatment. c BEAS-2B 3-NBA-6M (1–10 μM)–transformed cells exhibited increased tube formation compared with DMSO control cells. In vitro angiogenesis assays were conducted. HUVECs were grown in 3-NBA–transformed cell-derived conditioned medium (CM). The number of closed tubes was counted in five random fields from each well, and relative tube formation was then calculated. d IL-6 expression measured using quantitative real-time polymerase chain reaction. e Secretory IL-6 protein was elevated in BEAS-2B 3-NBA-6M (1–10 μM) cells according to ELISA assays. f GREM1 and THBS1 protein expression levels measured through immunoblotting. No significant difference was observed between the treated and control cells. The relative expressions of proteins are indicated as the FC over control. Data are presented as the mean ± standard deviation of three independent experiments. (*p < 0.05 and **p < 0.01)
Collectively, these data suggest that chronic exposure to 3-NBA may mediate tumor-promoting gene alteration, including cell proliferation, independent cell growth, migration, invasion, and tube formation, thereby inducing epithelial cell aggressiveness.
3-NBA–mediated gene expression is correlated with poorer overall lung cancer survival
We used KM plotter with the transcriptomic data of 1925 NSCLC specimens from the lung cancer gene expression dataset (Gyorffy et al. 2013) to evaluate the prognostic significance of the biomarkers in the genes that were markedly mediated by 3-NBA. The 10-year overall survival analysis suggested that most upregulated genes in the 3-NBA transformation network, including IL-6, SOX9, and SANI2, were highly expressed in patients with poor lung cancer prognosis (Fig. 5a). By contrast, the levels of several downregulated genes such as CDH1, SYNE1, and TCF4 were lower, with significant differences in patients with poor lung cancer poor prognosis (Fig. 5b). Furthermore, survival analyses were performed according to subtypes of clinical lung cancer characteristics, revealing that 3-NBA markedly induced EREG gene correlation with lung cancer. Notably, in an analysis of patients with lung cancer, female patients, those who never smoked, and those with adenocarcinoma exhibited a high correlation of EREG expression with poor overall survival (Fig. 5c). Moreover, poor survival was observed in patients with lung cancer and high EREG expression in the following subtypes: small tumor size (stage T1, ≤3 cm), no regional lymph node or ipsilateral mediastinal lymph node spreading (N0 or N2 status), and well-differentiated lung cancer cells (grade I). Patients with stage 1A (T1N0M0) lung cancer and high EREG expression had poor survival (low expression, 48.0 months; high expression, 25.0 months; p = 0.023; Fig. 5d). The survival analysis demonstrated that high IL-6 expression was associated with unfavorable prognosis in most subtypes of clinical characteristics (except stages T2–T4; Fig. 5e). Similarly, in patients with stage 1A lung cancer, survival was lower in patients with high IL-6 expression (30.0 months) than in those with low IL-6 expression (91.0 months; p < 0.001; Fig. 5f). Collectively, 3-NBA–transformed cells demonstrated alterations in several tumor-promoting genes such as EREG and IL-6, and their expression in early-stage lung cancer was correlated with shorter survival.
Analysis of association between 3-NBA–mediated gene expression and survival in patients with lung cancer. a, b Prognostic hazard ratios of the top 20 genes that were upregulated or downregulated genes through 3-NBA mediation obtained using KM plotter with published lung cancer microarray datasets. c Overall survival for EREG (Affymetrix ID: 205767_at) evaluated using KM analysis with transcriptomic data of 1925 patients with lung cancer. Clinical characteristics of the datasets were included in the survival analysis. d Overall survival for EREG according to data of patients with stage 1A lung cancer. e Survival analysis for IL-6 (Affymetrix ID: 205207_at) according to clinical characteristics of patients with lung cancer. Forest plots indicate hazard ratios and 95% confidence intervals. f Overall survival for IL-6 according to data of patients with stage 1A lung cancer
3-NBA–transformed cells mediate malignant properties in vivo
On the basis of the malignant phenotypes caused by 3-NBA in vitro, we inoculated xenograft mice with BEAS2B 3-NBA-6M (10 μM)–transformed (3-NBA–T) cells to examine tumorigenicity in vivo. Nude mice were inoculated with 3-NBA–T and control cells through subcutaneous injection. No tumor formation was observed in the mice inoculated with BEAS-2B control cells after 6 weeks, but all five mice in the subcutaneous xenograft mouse model injected with 3-NBA–T cells exhibited tumor formation (average volume > 500 mm3; Fig. 6a and b). To determine the potential metastasis ability in vivo, nude mice received tail-vein injections of 3-NBA–T or control cells. Inoculation of 3-NBA–T cells resulted in higher metastasis capability compared with the control cells (5.68-fold; Fig. 6c). Immunostaining revealed that the lungs of mice receiving 3-NBA–T contained EREG and IL-6 overexpressing micrometastases (Fig. 6c). These findings indicate that 3-NBA–T cells induce tumor formation and metastasis in vivo.
Tumorigenicity of 3-NBA–T human lung epithelial cells in nude mice. a Tumor formation in nude mice and tumor xenografts 6 weeks after subcutaneous inoculation with 2 × 105 BEAS-2B 3-NBA–T cells (10-μM 3-NBA-6M transformed) or control cells. b Tumor growth after injection with BEAS-2B control or 3-NBA–T cells. Tumor volume was measured every 7 days until the end of the experiment. Images display tumors from nude mice inoculated with 3-NBA–T cells. None of the nude mice injected with BEAS-2B control cells formed tumors. c Immunohistochemical staining of lung sections for BEAS-2B control or 3-NBA–T cells with metastasis (n = 5/group). All analyses were conducted 7 weeks after tail-vein injection of 1 × 106 3-NBA–T or control cells. The metastasis index (fold, tumor density in control or 3-NBA–T cells per cm2) in the lung is presented on the right panel
3-NBA induces lung epithelial cell malignancy through the EREG signaling pathway
The present results indicate that 3-NBA–mediated cell malignancy transformation increases clonogenic cell survival, in vitro angiogenesis, and migration. Moreover, 3-NBA–T cells contributed to tumorigenesis in the mouse xenograft model. These results suggest that EREG and IL-6 are crucial for inducing the tumorigenic properties of 3-NBA–transformed cells. To determine whether EREG plays a critical role in cell transformation, EREG expression was stably knocked out in the parental (BEAS-2B) cells. After 10-μM 3-NBA treatment, the EREG knockout cells exhibited higher apoptosis (1.71-fold) and formed substantially fewer (0.46-fold) and smaller colonies when compared with knockout control cells (Fig. 7a and b). To determine the signaling pathway involved in colony formation, BEAS-2B 3-NBA-6M (10 μM)–transformed cells were treated with kinase or STAT3 inhibitors. Treatments with PI3K and MEK inhibitors caused reduced colony formation in the 3-NBA–transformed cells (reduction from 2.33-fold to 0.51- and 0.29-fold, respectively; Fig. 7c). However, treatment with STAT3 inhibitors partially reduced clonogenic cell survival (30.0%, reduction from 2.33-fold to 1.63-fold). Moreover, recombinant IL-6 stimulation enhanced colony formation ability by 28.8%, and this was attenuated to 10.4% through the suppression of STAT3 activation. These results imply that 3-NBA–transformed cells may markedly promote cell survival through the PI3K/AKT and MEK/ERK signaling pathways. To clarify the signaling pathway involved in migration, we treated 3-NBA–transformed cells with PI3K and MEK inhibitors, which significantly disrupted migration in the 3-NBA–transformed cells (reduction from 2.50-fold to 0.59- and 0.41-fold, respectively; Fig. 7d). Similarly, treatment with STAT3 inhibitors partially blocked cell migration (35.6%, reduction from 2.50-fold to 1.61-fold). IL-6 stimulation increased migration by 26.4%, and this was attenuated to 8.8% by the STAT3 inhibitors.
EREG-mediated clonogenic cell survival and migration in 3-NBA–transformed cells. a Knockout of EREG in parental BEAS-2B cells significantly increased cell apoptosis after treatment with 3-NBA for 24 h. b The 3-NBA–mediated colony formation was diminished by EREG knockout, suggesting involvement of EREG in colony formation. c) Treatment with rIL-6 (10 ng/mL) increased colony formation capabilities of 3-NBA-6M (10-μM 3-NBA–transformed) and control-6M cells. Induction of colony formation in 3-NBA-6M cells with IL-6 stimulation was mostly abrogated after treatment with Ly294002 (PI3K inhibitor, 10 μM) or U0126 (ERK1/2 inhibitor, 10 μM) when compared with control cells. However, 3-NBA–transformed cells with or without IL-6 stimulation still exhibited partly increased colony formation after S3I-201 (STAT3 activity inhibitor, 100 μM) treatment when compared with control cells. d Transwell migration assay of 3-NBA-6M cells after treatment with inhibitors and rIL-6 for 16 h. The 3-NBA transformation increased the migration ability that was abrogated by Ly294002, U0126, or S3I-201 treatment when compared with the control cells. e Cytosolic and secreted EREG protein levels examined using immunoblot and ELISA assays, respectively. Cytosolic or secreted EREG protein levels were not significantly altered in BEAS-2B 3-NBA-6M or control cells after IL-6 knockdown. The levels of p-STAT3, pAKT, and pERK were measured through immunoblotting. f EREG knockdown in BEAS-2B 3-NBA-6M cells determined through immunoblotting and ELISA analysis. The levels of p-STAT3, pAKT, pERK, SOX9, and TWIST2 were reduced in EREG knockdown cells. EREG knockdown in BEAS-2B 3-NBA-6M cells abrogated transformation-mediated clonogenic cell survival and migration. Data are presented as the mean ± standard deviation of three independent experiments. (*p < 0.05; **p < 0.01; ns, not significant)
Next, we investigated the effects of IL-6/STAT3 signaling on EREG expression. The levels of cytosolic or secreted EREG proteins were not significantly altered in BEAS-2B 3-NBA-6M (10 μM) or control-6M cells after IL-6 knockdown (Fig. 7e). In addition, the signaling pathways involved in STAT3, AKT, and ERK activation were only partially diminished in the BEAS-2B-6M IL-6 knockdown cells, suggesting that elevated EREG expression mediated these signaling pathways in 3-NBA–transformed cells. To elucidate whether EREG plays a crucial role in tumorigenic properties, EREG knockdown assays were conducted with BEAS-2B 3-NBA-6M (10 μM) cells. Western blot and ELISA assays indicated efficient reduction of EREG expression through knockdown (Fig. 7f). Moreover, secretory IL-6 protein levels were reduced in EREG knockdown cells. The STAT3, AKT, and ERK signaling pathways were markedly repressed in the 3-NBA-6M EREG knockdown cells. Similarly, EREG knockdown in 3-NBA–transformed cells abrogated colony formation and migration. Notably, the expression levels of proliferation- and migration-related proteins such as SOX9 and TWIST2 were suppressed in 3-NBA–transformed cells after EREG knockdown.
Collectively, these findings indicate that 3-NBA–transformed cells induce EREG overexpression, thereby activating the PI3K/AKT and MEK/ERK signaling pathways and resulting in tumorigenicity (Fig. 8). Furthermore, elevated IL-6 and EREG expressions can synergistically lead to STAT3 signaling activation, promoting clonogenic cell survival and migration.
Schematic of increased EREG expression in 3-NBA–transformed cells, leading to tumorigenicity. The short-term exposure of lung epithelial cells to 3-NBA can lead to ROS production and cell apoptosis. However, long-term exposure to 3-NBA resulted in distinct alterations in the expressions of genes such as E-cadherin, SOX9, TWIST, IL-6, and EREG, which are involved in carbon cancer metabolism, metastasis, and angiogenesis. The EREG ligand can bind to the EGFR and ErbB4 receptors, thereby activating the downstream signaling pathways. The 3-NBA–transformed cells exhibited marked EREG and IL-6 overexpressions, which activated the STAT3, PI3K/AKT, and MEK/ERK signaling pathways and contributed to tumorigenicity. Depletion of EREG in 3-NBA-T cells abrogated IL-6 expression and reduced colony formation and migration. Furthermore, IL-6 stimulated the downstream STAT3 signaling pathway, enhancing clonogenic cell survival and migration. However, STAT3 signaling blockade partially suppressed colony formation and migration capabilities. Taken together, increased EREG expression in 3-NBA–transformed cells markedly contributed to tumorigenesis through PI3K/AKT and MEK/ERK activation and synergistically enhanced IL-6 signaling pathways, thus promoting tumorigenicity.
Discussion
As indicated, 3-NBA is a potent inducer of BEAS-2B cell apoptosis through the DNA damage response (Oya et al. 2011). The metabolic activation of nitro-PAHs often increases ROS production. In human lung cancer A549 cells, short-term exposure of 3-NBA significantly induced superoxide levels and reduced total antioxidant capacity as well as HMOX1 and TXNRD1 expressions (Libalova et al. 2018). Our results reveal that chronic exposure to 3-NBA can cause malignant transformation in human lung epithelial cells because of increased cellular proliferation, colony formation, and cell migration. Moreover, 3-NBA–T cells induced tumor formation and metastasis in the xenograft mouse model. Lung tumors exhibited high mutation rates in oncogenes such as KRAS and in emerging tumor suppressors such as p53 (Gibbons et al. 2014). Findings in both primary tumors and cell lines, including NSCLCs and SCLCs, revealed that p53 genome locations are frequently deleted or mutated (Gibbons et al. 2014). Evidence has revealed that the p53 protein confers epigenetic fidelity and that the depletion of p53 gene functions increases epigenetic status alterations in cancer cells (Levine et al. 2016). The translesion DNA synthesis of dGC8-N-ABA catalyzed by hPolκ is error-prone, which may be the mechanism by which 3-NBA promotes tumorigenesis (Phi et al. 2019). The present results also reveal a reduction of p53 protein levels in 3-NBA–transformed cells. The genetic and signaling mechanisms in the malignant cell transformation process induced by long-term 3-NBA exposure were further investigated.
In the current study, NanoString analysis was conducted using the PanCancer Progression Pathways Panel to analyze RNA expression in the 3-NBA–transformed cells and to subsequently identify the novel EREG-mediated network. The gene signature scores in the cancer progression pathways involved with carbon cancer metabolism, EMT to metastasis, regulation of angiogenesis, and sprouting angiogenesis were increased in 3-NBA–transformed cells. Notably, the critical tumor suppressor gene p53 was also downregulated in the transformed cells, potentially promoting cell proliferation and clonogenic survival. Advanced data mining and STRING network analysis revealed that the identified gene products IL-6, SOX9, AKT1, TWIST2, and MET interacted with EREG, a potential therapeutic target associated with lung cancer malignancy (Sunaga and Kaira 2015). Moreover, reduced E-cadherin expression was observed as a key event during EMT in the 3-NBA–transformed cells. Studies have revealed that upregulated EREG expression is crucial to cell growth in hTERT-mediated fibroblast immortalization and transformation (Lindvall et al. 2003). Moreover, EREG expression levels were inversely correlated with EREG methylation in lung cancer (Qu et al. 2016). EREG overexpression in lung cancer may activate the EGFR pathway to promote malignant progression (Sunaga and Kaira 2015). The present results reveal that EREG expression was significantly upregulated in 3-NBA–transformed lung epithelial cells, promoting clonogenic colony formation and migration. Similarly, EREG overexpression may predominantly contribute to tumorigenesis in 3-NBA–transformed cells through the EGFR downstream PI3K/AKT and MEK/ERK signaling pathways.
EREG overexpression may be a predictive functional marker triggering head and neck squamous cell carcinoma oncogenic transformation through activation of the EREG–EGFR–C–Myc axis signaling pathway (Liu et al. 2020). Moreover, EREG deficiency reduced lung tumor formation in a primary two-stage mouse model; tumor promotion was initiated upon exposure to 3-methylcholanthrene, and tumor progression was subsequently promoted after multiple exposures to butylated hydroxytoluene (Bauer et al. 2017). EGFR was constitutively activated in metastatic lung subtypes of salivary adenoid cystic carcinoma (SACC) cells and may have been induced by EREG autocrine expression (Liu et al. 2016). EREG-activated EGFR stabilizes Snail and Slug, thus promoting SACC cell metastatic capability. NADPH oxidase 4/AKT and IL-6/STAT3 signaling pathways positively regulate each other, thereby enhancing the proliferation and survival of NSCLC cells (Li et al. 2015). In the present study, EREG knockout in parental epithelial cells exhibited more apoptosis and reduced colony formation after short-term 3-NBA treatment. EREG expression knockdown abrogated colony formation and migration, and the levels of SOX9, TWIST2, p-STAT3, p-AKT, and p-ERK were reduced in 3-NBA–transformed cells.
Studies have associated ambient air pollutants with lung cancer incidence and mortality (Hamra et al. 2014; Fischer et al. 2015). Several risk factors, such as female sex, environmental factors, hormones, genetic factors, and viruses, may cause lung cancer development in never-smokers (Sun et al. 2007). Notably, 3-NBA is a crucial carcinogen present in atmospheric and diesel exhaust PM (Arlt et al. 2005; Santos et al. 2019). Patients with lung cancer (particularly early-stage adenocarcinomas) presented short survival because of exposure to high ambient air pollutants such as NO2, PM2.5, and PM10 (Eckel et al. 2016). The present study revealed that EREG was highly expressed in 3-NBA–transformed cells and associated with cancer progression. Regarding the prognostic significance of EREG expression in lung cancer, an NSCLC biopsy specimen immunohistochemical staining analysis indicated that EREG-positive tumors were correlated with poor clinical outcomes (Zhang et al. 2008). In addition, the expression level of EREG is significantly higher in lung adenocarcinoma than in squamous cell carcinoma (Sunaga et al. 2013). Moreover, EREG contributes to aggressive tumor phenotypes in KRAS-mutant lung adenocarcinoma and may serve as a prognostic marker in NSCLC (Sunaga et al. 2013). In the present study, survival analyses based on the subtypes of female sex, never-smoker status, and adenocarcinoma in patients with lung cancer revealed a negative correlation between EREG and outcomes. Furthermore, high EREG and IL-6 expressions in early-stage lung cancer are correlated with poor survival. However, in most nonsmokers, lung cancer is more common in women and is often diagnosed at more advanced stages, the predominant histologic type of which is adenocarcinoma (Casal-Mourino et al. 2019). Whether specific EREG overexpression is correlated with driver mutations such as those in EGFR, ALK, and ROS1 in nonsmokers remains undetermined. Additionally, IL-6 is a pleotropic and complex cytokine that might play an anti-inflammatory or pro-inflammatory role in response to tissue damage through classical and trans-signaling pathways, respectively (Scheller et al. 2011). IL-6 may play an antioxidant role in pulmonary homeostasis, particularly in protecting alveolar epithelial cells from ROS-mediated cell death (Kida et al. 2005). Chronic signaling of IL-6 is correlated with tumor development, possibly acting intrinsically on cancer cells through various mediators to increase cell survival, proliferation, and metastatic dissemination (Fisher et al. 2014). Furthermore, IL-6/STAT3 signaling was revealed to play a critical role in lung premetastatic niche formation and immunosuppression (Jing et al. 2020). The present study revealed that elevated EREG signaling can activate IL-6 expression, thereby synergistically contributing to cell survival capabilities and migration through the STAT3 pathway.
In vitro cell transformation assays can be used as alternative tools to evaluate human tumorigenesis induced by potent environmental carcinogens (Steinberg 2017). The air–liquid interface (ALI) culture of these assays may cause morphological and functional differentiation in epithelial cells, thereby creating a more realistic environment for exposure (Chen and Schoen 2019). ALI culture experiments (He et al. 2021) have revealed that only the human cancer cell line Calu-3, and not BEAS-2B, can sustain a monolayer structure and a strong tight junction for 2 weeks. Additionally, three-dimensional (3D) cell culture (MucilAir) experiments revealed the effects of short-term (24-h) and long-term (7- and 28-day) exposure to 0.1- or 1-μM 3-NBA (Cervena et al. 2019). Exposure to 0.1-μM 3-NBA for 11 days caused a marked reduction in junction tightness (i.e., transepithelial electrical resistance, TEER), whereas 1-μM 3-NBA exposure for 9 days caused only a slight reduction in TEER. Furthermore, CYP1A1 and HMOX1 expression levels were upregulated after 7 days of 1-μM 3-NBA exposure. However, the mRNA levels of several genes did not significantly change after 28 days of 3-NBA exposure at any concentration, possibly because functioning cells and tight junctions affected 3-NBA entry into the lower cell layers. Therefore, further examination is required to determine environmentally relevant doses or higher concentrations of 3-NBA for long-term exposure (>1 month). Nevertheless, MucilAir is a useful 3D culture including ciliated, goblet, and basal human cells mimicking the heterogeneity of the lung environment for in vitro study of disease progression. However, commercially available systems are often limited in terms of flexibility (cell or genetic manipulation) and customization. In the present study, the effects of malignant cell transformation were mainly determined using BEAS-2B cells with long-term exposure to 0.1–10-μM 3-NBA, which may be higher than typical exposure concentrations. Oxidative stress and metabolomic profiles were investigated in human urothelial cancer cells exposed to a 0.0003-μM 3-NBA, which is a relevant environmental dose, and reaching 80 μM after 24 h of exposure (Pink et al. 2017). These results showed elevated levels of various antioxidants at low doses of 3-NBA (0.0003 and 0.01 μM). However, additional metabolic pathways were increased to maintain cellular homeostasis at 0.04–10-μM 3-NBA, and prominent toxic effects were observed at 80 μM. Treatments with higher doses of 3-NBA (10 and 80 μM) may change energy metabolism through the pentose phosphate pathway. The present data indicated that short-term (24-h) exposure to 3-NBA (1–160 μM) reduced cell viability and increased ROS production in a dose-dependent manner. Moreover, several tumor-promoting functions were dose-dependently altered in 1- and 10-μM 3-NBA–transformed cells, possibly because long-term exposure to higher doses of 3-NBA markedly increased cell survival and caused malignant transformation through critical expression alterations of genes such as EREG.
The current results clearly indicate that chronic exposure to 3-NBA mediated lung epithelial cell transformation. Cancer aggressiveness, measured in terms of EMT, is regulated by various signaling cascades, which the PI3K/AKT and STAT3 pathways significantly activate during lung cancer progression (Liu et al. 2017; Wang et al. 2016). Our results identified the EREG regulatory cascade as a novel pathway associated with 3-NBA–mediated lung epithelial cell transformation through the activation signaling cascade involving the PI3K/AKT, MEK/ERK, and STAT3 pathways. We also revealed that E-cadherin reduction and the induction of SNAI2 and TWIST1/2 expression may confer migration and invasion capabilities in 3-NBA–transformed cells. Our findings are consistent with those of relevant studies, strongly supporting the critical role of the EREG regulatory cascade in the molecular mechanism underlying cancer development. Additionally, studies revealed that numerous stem cell regulatory proteins and intracellular signaling pathways can regulate the cancer stem cells (CSCs) that contribute to tumor malignancies (Yang et al. 2020). Notably, EREG blockade through anti-EREG antibodies efficiently represses cancer development, whereas colon cancer enriches CSCs (Kobayashi et al. 2012). SOX9 is a transcription factor of the SOX family that acts a key regulator in lung cancer progression and patient survival (Zhou et al. 2012; Matheu et al. 2012). Studies have indicated that long-term exposure to the single-walled carbon nanotubes (SWCNTs) of BEASE-2B cells results in a highly invasive and stem cell–like subpopulation (Luanpitpong et al. 2014). Moreover, SOX9 increases CSC-like and metastatic properties in SWCNT-exposed cells (Voronkova et al. 2017). In the current study, SOX9 was also highly expressed in 3-NBA–transformed cells. However, whether 3-NBA mediates stemness properties in tumorigenesis remains undetermined.
Recently, BEAS-2B and human mesenchymal stem cell lines (hMSC1) were revealed to share the same expression profiles as human umbilical cord–derived MSC (hMSC) surface markers and exhibited similar osteogenic and adipogenic differentiation potentials (Han et al. 2020). However, BEAS-2B cells did not display expression of mesenchymal proteins such as α-SMA, collagen I, and fibronectin, which are abundantly expressed in hMSCs. Additionally, BEAS-2B did not promote type-2 macrophage (M2) polarization, whereas hMSC1 induced a dramatic increase in the proportion of M2 macrophages. Furthermore, BEAS-2B cells exhibited a gene expression pattern similar to that of primary human bronchial epithelial cells (Courcot et al. 2012). In addition, the EMT process of BEAS-2B cells mediated by TGF-β1 caused morphological changes and reduced E-cadherin (Kamitani et al. 2011). The CSC-like and metastatic properties were also observed in transformed BEAS-2B cells (Voronkova et al. 2017). In addition, BEAS-2B exposed to FBS induced sustained but reversible EMT (Malm et al. 2018). Notably, EREG is required for fibroblast transformation, which induces EMT in a manner dependent on JAK2, STAT3, and IL-6 in oral squamous cell carcinoma (OSCC) cells (Wang et al. 2019). In addition, overexpression of EREG in patients with OSCC was correlated with tumor invasion and shorter overall survival. Similarly, increased EREG expression in 3-NBA–transformed cells enhanced migration and the synergistically activated IL-6/STAT3 signaling pathway. Moreover, we observed that 3-NBA–transformed cells, which cause high levels of EREG and IL-6 expressions, induced tumor metastasis in vivo. Patients with lung cancer and high levels of EREG and IL-6 expressions had poor survival. Notably, primary human airway epithelia cocultured with lung fibroblasts may secrete ErbB ligands including EREG, thus stimulating airway epithelial differentiation (Vermeer et al. 2006). On the basis of these findings, EREG overexpression confers downstream signaling pathway activation, possibly promoting tumor development malignant cell transformation. However, this study could not sufficiently replicate the in vivo phenotype. The EREG signaling pathways and other mediated signaling pathways must be further analyzed in a more appropriate human microenvironment by using 3D primary cell or lung tissue cultures correlated with exposure to 3-NBA at various concentrations.
On the basis of the tumor-promoting potential of 3-NBA, we focused on predefined PanCancer genes in 3-NBA–transformed cells, revealing that significant changes in cancer progression analysis have not been realized. However, the results of NanoString gene expression analysis did not rule out other potential targets or pathways in 3-NBA–transformed cells that could be analyzed through a transcriptome microarray or RNA sequencing. Even so, the NanoString platform possesses several advantages, including high sensitivity and reproducibility, low possibility of cDNA introduction bias, and suitability for detecting low-expression RNA in clinical applications (Veldman-Jones et al. 2015). The intake of 3-NBA may be variable in individuals exposed to environmental contaminants. By characterizing the chemical composition of environmental pollutants, the direct effects of cancer progression in certain patient groups exposed to specific compounds (e.g., 3-NBA) can be further verified and correlated. Notably, low-quantity or poor-quality RNA derived from lung biopsy and formalin-fixed paraffin-embedded tissue can be examined through NanoString analysis. In future longitudinal studies, comprehensive analyses of genomic, transcriptomic, and proteomic profiling in these human tissue specimens could clarify the detailed characteristics of tumor microenvironments.
In conclusion, we demonstrated the 3-NBA–mediated transformation of normal lung epithelial cells preceding cancer cell progression through increased proliferation, anchorage-independent cell growth, migration, and in vitro angiogenesis. Long-term exposure to 3-NBA may promote lung cancer progression through EREG signaling activation.
References
Arlt VM. 3-Nitrobenzanthrone, a potential human cancer hazard in diesel exhaust and urban air pollution: a review of the evidence. Mutagenesis. 2005;20(6):399–410. https://doi.org/10.1093/mutage/gei057.
Arlt VM, Bieler CA, Mier W, Wiessler M, Schmeiser HH. DNA adduct formation by the ubiquitous environmental contaminant 3-nitrobenzanthrone in rats determined by (32)P-postlabeling. Int J Cancer. 2001;93(3):450–4.
Arlt VM, Gingerich J, Schmeiser HH, Phillips DH, Douglas GR, White PA. Genotoxicity of 3-nitrobenzanthrone and 3-aminobenzanthrone in MutaMouse and lung epithelial cells derived from MutaMouse. Mutagenesis. 2008;23(6):483–90. https://doi.org/10.1093/mutage/gen037.
Arlt VM, Stiborova M, Henderson CJ, Osborne MR, Bieler CA, Frei E, et al. Environmental pollutant and potent mutagen 3-nitrobenzanthrone forms DNA adducts after reduction by NAD(P)H:quinone oxidoreductase and conjugation by acetyltransferases and sulfotransferases in human hepatic cytosols. Cancer Res. 2005;65(7):2644–52. https://doi.org/10.1158/0008-5472.CAN-04-3544.
Bauer AK, Velmurugan K, Xiong KN, Alexander CM, Xiong J, Brooks R. Epiregulin is required for lung tumor promotion in a murine two-stage carcinogenesis model. Mol Carcinog. 2017;56(1):94–105. https://doi.org/10.1002/mc.22475.
Bieler CA, Cornelius MG, Stiborova M, Arlt VM, Wiessler M, Phillips DH, et al. Formation and persistence of DNA adducts formed by the carcinogenic air pollutant 3-nitrobenzanthrone in target and non-target organs after intratracheal instillation in rats. Carcinogenesis. 2007;28(5):1117–21. https://doi.org/10.1093/carcin/bgl219.
Casal-Mourino A, Valdes L, Barros-Dios JM, Ruano-Ravina A. Lung cancer survival among never smokers. Cancer Lett. 2019;451:142–9. https://doi.org/10.1016/j.canlet.2019.02.047.
Cervena T, Vrbova K, Rossnerova A, Topinka J, Rossner P Jr. Short-term and long-term exposure of the MucilAir model to polycyclic aromatic hydrocarbons. Altern Lab Anim. 2019;47(1):9–18. https://doi.org/10.1177/0261192919841484.
Chen S, Schoen J. Air-liquid interface cell culture: from airway epithelium to the female reproductive tract. Reprod Domest Anim. 2019;54(Suppl 3):38–45. https://doi.org/10.1111/rda.13481.
Courcot E, Leclerc J, Lafitte JJ, Mensier E, Jaillard S, Gosset P, et al. Xenobiotic metabolism and disposition in human lung cell models: comparison with in vivo expression profiles. Drug Metab Dispos. 2012;40(10):1953–65. https://doi.org/10.1124/dmd.112.046896.
Cox AD, Der CJ. Biological assays for cellular transformation. Methods Enzymol. 1994;238:277–94. https://doi.org/10.1016/0076-6879(94)38026-0.
Eckel SP, Cockburn M, Shu YH, Deng H, Lurmann FW, Liu L, et al. Air pollution affects lung cancer survival. Thorax. 2016;71(10):891–8. https://doi.org/10.1136/thoraxjnl-2015-207927.
Field RW, Withers BL. Occupational and environmental causes of lung cancer. Clin Chest Med. 2012;33(4):681–703. https://doi.org/10.1016/j.ccm.2012.07.001.
Fischer PH, Marra M, Ameling CB, Hoek G, Beelen R, de Hoogh K, et al. Air pollution and mortality in seven million adults: the Dutch Environmental Longitudinal Study (DUELS). Environ Health Perspect. 2015;123(7):697–704. https://doi.org/10.1289/ehp.1408254.
Fisher DT, Appenheimer MM, Evans SS. The two faces of IL-6 in the tumor microenvironment. Semin Immunol. 2014;26(1):38–47. https://doi.org/10.1016/j.smim.2014.01.008.
Gibbons DL, Byers LA, Kurie JM. Smoking, p53 mutation, and lung cancer. Mol Cancer Res. 2014;12(1):3–13. https://doi.org/10.1158/1541-7786.MCR-13-0539.
Grimmer G, Brune H, Deutsch-Wenzel R, Dettbarn G, Jacob J, Naujack KW, et al. Contribution of polycyclic aromatic hydrocarbons and nitro-derivatives to the carcinogenic impact of diesel engine exhaust condensate evaluated by implantation into the lungs of rats. Cancer Lett. 1987;37(2):173–80. https://doi.org/10.1016/0304-3835(87)90160-1.
Gyorffy B, Surowiak P, Budczies J, Lanczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One. 2013;8(12):e82241. https://doi.org/10.1371/journal.pone.0082241.
Hamra GB, Guha N, Cohen A, Laden F, Raaschou-Nielsen O, Samet JM, et al. Outdoor particulate matter exposure and lung cancer: a systematic review and meta-analysis. Environ Health Perspect. 2014;122(9):906–11. https://doi.org/10.1289/ehp.1408092201289/ehp/1408092.
Han X, Na T, Wu T, Yuan BZ. Human lung epithelial BEAS-2B cells exhibit characteristics of mesenchymal stem cells. PLoS One. 2020;15(1):e0227174. https://doi.org/10.1371/journal.pone.0227174.
Hansen T, Seidel A, Borlak J. The environmental carcinogen 3-nitrobenzanthrone and its main metabolite 3-aminobenzanthrone enhance formation of reactive oxygen intermediates in human A549 lung epithelial cells. Toxicol Appl Pharmacol. 2007;221(2):222–34. https://doi.org/10.1016/j.taap.2007.03.003.
He RW, Braakhuis HM, Vandebriel RJ, Staal YCM, Gremmer ER, Fokkens PHB, et al. Optimization of an air-liquid interface in vitro cell co-culture model to estimate the hazard of aerosol exposures. J Aerosol Sci. 2021;153:105703. https://doi.org/10.1016/j.jaerosci.2020.105703.
Jing B, Wang T, Sun B, Xu J, Xu D, Liao Y, et al. IL6/STAT3 signaling orchestrates premetastatic niche formation and immunosuppressive traits in lung. Cancer Res. 2020;80(4):784–97. https://doi.org/10.1158/0008-5472.CAN-19-2013.
Kamitani S, Yamauchi Y, Kawasaki S, Takami K, Takizawa H, Nagase T, et al. Simultaneous stimulation with TGF-beta1 and TNF-alpha induces epithelial mesenchymal transition in bronchial epithelial cells. Int Arch Allergy Immunol. 2011;155(2):119–28. https://doi.org/10.1159/000318854.
Kida H, Yoshida M, Hoshino S, Inoue K, Yano Y, Yanagita M, et al. Protective effect of IL-6 on alveolar epithelial cell death induced by hydrogen peroxide. Am J Phys Lung Cell Mol Phys. 2005;288(2):L342–9. https://doi.org/10.1152/ajplung.00016.2004.
Kim BG, Lee PH, Lee SH, Kim YE, Shin MY, Kang Y, et al. Long-term effects of diesel exhaust particles on airway inflammation and remodeling in a mouse model. Allergy, Asthma Immunol Res. 2016;8(3):246–56. https://doi.org/10.4168/aair.2016.8.3.246.
Kobayashi S, Yamada-Okabe H, Suzuki M, Natori O, Kato A, Matsubara K, et al. LGR5-positive colon cancer stem cells interconvert with drug-resistant LGR5-negative cells and are capable of tumor reconstitution. Stem Cells. 2012;30(12):2631–44. https://doi.org/10.1002/stem.1257.
Kunitake E, Shimamura K, Katayama H, Takemoto K, Yamamoto A, Hisanaga A, et al. Studies concerning carcinogenesis of diesel particulate extracts following intratracheal instillation, subcutaneous injection, or skin application. Dev Toxicol Environ Sci. 1986;13:235–52.
Levine AJ, Puzio-Kuter AM, Chan CS, Hainaut P. The role of the p53 protein in stem-cell biology and epigenetic regulation. Cold Spring Harb Perspect Med. 2016;6(9). https://doi.org/10.1101/cshperspect.a026153.
Li J, Lan T, Zhang C, Zeng C, Hou J, Yang Z, et al. Reciprocal activation between IL-6/STAT3 and NOX4/Akt signalings promotes proliferation and survival of non-small cell lung cancer cells. Oncotarget. 2015;6(2):1031–48. https://doi.org/10.18632/oncotarget.2671.
Libalova H, Milcova A, Cervena T, Vrbova K, Rossnerova A, Novakova Z, et al. Kinetics of ROS generation induced by polycyclic aromatic hydrocarbons and organic extracts from ambient air particulate matter in model human lung cell lines. Mutat Res Genet Toxicol Environ Mutagen. 2018;827:50–8. https://doi.org/10.1016/j.mrgentox.2018.01.006.
Lindvall C, Hou M, Komurasaki T, Zheng C, Henriksson M, Sedivy JM, et al. Molecular characterization of human telomerase reverse transcriptase-immortalized human fibroblasts by gene expression profiling: activation of the epiregulin gene. Cancer Res. 2003;63(8):1743–7.
Liu S, Wang Y, Han Y, Xia W, Zhang L, Xu S, et al. EREG-driven oncogenesis of head and neck squamous cell carcinoma exhibits higher sensitivity to erlotinib therapy. Theranostics. 2020;10(23):10589–605. https://doi.org/10.7150/thno.47176.
Liu S, Ye D, Xu D, Liao Y, Zhang L, Liu L, et al. Autocrine epiregulin activates EGFR pathway for lung metastasis via EMT in salivary adenoid cystic carcinoma. Oncotarget. 2016;7(18):25251–63. https://doi.org/10.18632/oncotarget.7940.
Liu XL, Zhang XT, Meng J, Zhang HF, Zhao Y, Li C, et al. ING5 knockdown enhances migration and invasion of lung cancer cells by inducing EMT via EGFR/PI3K/Akt and IL-6/STAT3 signaling pathways. Oncotarget. 2017;8(33):54265–76. https://doi.org/10.18632/oncotarget.17346.
Long AS, Lemieux CL, Arlt VM, White PA. Tissue-specific in vivo genetic toxicity of nine polycyclic aromatic hydrocarbons assessed using the MutaMouse transgenic rodent assay. Toxicol Appl Pharmacol. 2016;290:31–42. https://doi.org/10.1016/j.taap.2015.11.010.
Luanpitpong S, Wang L, Castranova V, Rojanasakul Y. Induction of stem-like cells with malignant properties by chronic exposure of human lung epithelial cells to single-walled carbon nanotubes. Part Fibre Toxicol. 2014;11:22. https://doi.org/10.1186/1743-8977-11-22.
Malm SW, Amouzougan EA, Klimecki WT. Fetal bovine serum induces sustained, but reversible, epithelial-mesenchymal transition in the BEAS-2B cell line. Toxicol in Vitro. 2018;50:383–90. https://doi.org/10.1016/j.tiv.2018.04.008.
Matheu A, Collado M, Wise C, Manterola L, Cekaite L, Tye AJ, et al. Oncogenicity of the developmental transcription factor Sox9. Cancer Res. 2012;72(5):1301–15. https://doi.org/10.1158/0008-5472.CAN-11-3660.
Nagy E, Adachi S, Takamura-Enya T, Zeisig M, Moller L. DNA damage and acute toxicity caused by the urban air pollutant 3-nitrobenzanthrone in rats: characterization of DNA adducts in eight different tissues and organs with synthesized standards. Environ Mol Mutagen. 2006;47(7):541–52. https://doi.org/10.1002/em.20227.
Nagy E, Adachi S, Takamura-Enya T, Zeisig M, Moller L. DNA adduct formation and oxidative stress from the carcinogenic urban air pollutant 3-nitrobenzanthrone and its isomer 2-nitrobenzanthrone, in vitro and in vivo. Mutagenesis. 2007;22(2):135–45. https://doi.org/10.1093/mutage/gel067.
Nagy E, Zeisig M, Kawamura K, Hisamatsu Y, Sugeta A, Adachi S, et al. DNA adduct and tumor formations in rats after intratracheal administration of the urban air pollutant 3-nitrobenzanthrone. Carcinogenesis. 2005;26(10):1821–8. https://doi.org/10.1093/carcin/bgi141.
Olsson AC, Gustavsson P, Kromhout H, Peters S, Vermeulen R, Bruske I, et al. Exposure to diesel motor exhaust and lung cancer risk in a pooled analysis from case-control studies in Europe and Canada. Am J Respir Crit Care Med. 2011;183(7):941–8. https://doi.org/10.1164/rccm.201006-0940OC.
Oya E, Ovrevik J, Arlt VM, Nagy E, Phillips DH, Holme JA. DNA damage and DNA damage response in human bronchial epithelial BEAS-2B cells following exposure to 2-nitrobenzanthrone and 3-nitrobenzanthrone: role in apoptosis. Mutagenesis. 2011;26(6):697–708. https://doi.org/10.1093/mutage/ger035.
Phi KK, Smith MC, Tokarsky EJ, Suo Z. Kinetic investigation of translesion synthesis across a 3-nitrobenzanthrone-derived DNA lesion catalyzed by human DNA polymerase kappa. Chem Res Toxicol. 2019;32(8):1699–706. https://doi.org/10.1021/acs.chemrestox.9b00219.
Pink M, Verma N, Zerries A, Schmitz-Spanke S. Dose-dependent response to 3-nitrobenzanthrone exposure in human urothelial cancer cells. Chem Res Toxicol. 2017;30(10):1855–64. https://doi.org/10.1021/acs.chemrestox.7b00174.
Qu X, Sandmann T, Frierson H Jr, Fu L, Fuentes E, Walter K, et al. Integrated genomic analysis of colorectal cancer progression reveals activation of EGFR through demethylation of the EREG promoter. Oncogene. 2016;35(50):6403–15. https://doi.org/10.1038/onc.2016.170.
Santos AG, da Rocha GO, de Andrade JB. Occurrence of the potent mutagens 2-nitrobenzanthrone and 3-nitrobenzanthrone in fine airborne particles. Sci Rep. 2019;9(1):1. https://doi.org/10.1038/s41598-018-37186-2.
Scheller J, Chalaris A, Schmidt-Arras D, Rose-John S. The pro- and anti-inflammatory properties of the cytokine interleukin-6. Biochim Biophys Acta. 2011;1813(5):878–88. https://doi.org/10.1016/j.bbamcr.2011.01.034.
Seidel A, Dahmann D, Krekeler H, Jacob J. Biomonitoring of polycyclic aromatic compounds in the urine of mining workers occupationally exposed to diesel exhaust. Int J Hyg Environ Health. 2002;204(5-6):333–8. https://doi.org/10.1078/1438-4639-00116.
Shelly M, Pinkas-Kramarski R, Guarino BC, Waterman H, Wang LM, Lyass L, et al. Epiregulin is a potent pan-ErbB ligand that preferentially activates heterodimeric receptor complexes. J Biol Chem. 1998;273(17):10496–505. https://doi.org/10.1074/jbc.273.17.10496.
Silva EM, Mariano VS, Pastrez PRA, Pinto MC, Castro AG, Syrjanen KJ, et al. High systemic IL-6 is associated with worse prognosis in patients with non-small cell lung cancer. PLoS One. 2017;12(7):e0181125. https://doi.org/10.1371/journal.pone.0181125.
Steinberg P. In vitro-in vivo carcinogenicity. Adv Biochem Eng Biotechnol. 2017;157:81–96. https://doi.org/10.1007/10_2015_5013.
Steiner S, Bisig C, Petri-Fink A, Rothen-Rutishauser B. Diesel exhaust: current knowledge of adverse effects and underlying cellular mechanisms. Arch Toxicol. 2016;90(7):1541–53. https://doi.org/10.1007/s00204-016-1736-5.
Stiborova M, Cechova T, Borek-Dohalska L, Moserova M, Frei E, Schmeiser HH, et al. Activation and detoxification metabolism of urban air pollutants 2-nitrobenzanthrone and carcinogenic 3-nitrobenzanthrone by rat and mouse hepatic microsomes. Neuro Endocrinol Lett. 2012;33(Suppl 3):8–15.
Stiborova M, Martinek V, Svobodova M, Sistkova J, Dvorak Z, Ulrichova J, et al. Mechanisms of the different DNA adduct forming potentials of the urban air pollutants 2-nitrobenzanthrone and carcinogenic 3-nitrobenzanthrone. Chem Res Toxicol. 2010;23(7):1192–201. https://doi.org/10.1021/tx100052d.
Sun S, Schiller JH, Gazdar AF. Lung cancer in never smokers--a different disease. Nat Rev Cancer. 2007;7(10):778–90. https://doi.org/10.1038/nrc2190.
Sunaga N, Kaira K. Epiregulin as a therapeutic target in non-small-cell lung cancer. Lung Cancer (Auckl). 2015;6:91–8. https://doi.org/10.2147/LCTT.S60427.
Sunaga N, Kaira K, Imai H, Shimizu K, Nakano T, Shames DS, et al. Oncogenic KRAS-induced epiregulin overexpression contributes to aggressive phenotype and is a promising therapeutic target in non-small-cell lung cancer. Oncogene. 2013;32(34):4034–42. https://doi.org/10.1038/onc.2012.402.
Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45(D1):D362–8. https://doi.org/10.1093/nar/gkw937.
Takamura-Enya T, Kawanishi M, Yagi T, Hisamatsu Y. Structural identification of DNA adducts derived from 3-nitrobenzanthrone, a potent carcinogen present in the atmosphere. Chem Asian J. 2007;2(9):1174–85. https://doi.org/10.1002/asia.200700061.
Veldman-Jones MH, Brant R, Rooney C, Geh C, Emery H, Harbron CG, et al. Evaluating robustness and sensitivity of the NanoString Technologies nCounter Platform to enable multiplexed gene expression analysis of clinical samples. Cancer Res. 2015;75(13):2587–93. https://doi.org/10.1158/0008-5472.CAN-15-0262.
Vermeer PD, Panko L, Karp P, Lee JH, Zabner J. Differentiation of human airway epithelia is dependent on erbB2. Am J Phys Lung Cell Mol Phys. 2006;291(2):L175–80. https://doi.org/10.1152/ajplung.00547.2005.
Voronkova MA, Luanpitpong S, Rojanasakul LW, Castranova V, Dinu CZ, Riedel H, et al. SOX9 regulates cancer stem-like properties and metastatic potential of single-walled carbon nanotube-exposed cells. Sci Rep. 2017;7(1):11653. https://doi.org/10.1038/s41598-017-12037-8.
Wang X, Wang Y, Xiao G, Wang J, Zu L, Hao M, et al. Hypermethylated in cancer 1(HIC1) suppresses non-small cell lung cancer progression by targeting interleukin-6/Stat3 pathway. Oncotarget. 2016;7(21):30350–64. https://doi.org/10.18632/oncotarget.8734.
Wang Y, Jing Y, Ding L, Zhang X, Song Y, Chen S, et al. Epiregulin reprograms cancer-associated fibroblasts and facilitates oral squamous cell carcinoma invasion via JAK2-STAT3 pathway. J Exp Clin Cancer Res. 2019;38(1):274. https://doi.org/10.1186/s13046-019-1277-x.
White PA, Douglas GR, Phillips DH, Arlt VM. Quantitative relationships between lacZ mutant frequency and DNA adduct frequency in MutaMouse tissues and cultured cells exposed to 3-nitrobenzanthrone. Mutagenesis. 2017;32(2):299–312. https://doi.org/10.1093/mutage/gew067.
Wu SM, Lin SL, Lee KY, Chuang HC, Feng PH, Cheng WL, et al. Hepatoma cell functions modulated by NEK2 are associated with liver cancer progression. Int J Cancer. 2017;140(7):1581–96. https://doi.org/10.1002/ijc.30559.
Yang L, Shi P, Zhao G, Xu J, Peng W, Zhang J, et al. Targeting cancer stem cell pathways for cancer therapy. Signal Transduct Target Ther. 2020;5(1):8. https://doi.org/10.1038/s41392-020-0110-5.
Zhang J, Iwanaga K, Choi KC, Wislez M, Raso MG, Wei W, et al. Intratumoral epiregulin is a marker of advanced disease in non-small cell lung cancer patients and confers invasive properties on EGFR-mutant cells. Cancer Prev Res (Phila). 2008;1(3):201–7. https://doi.org/10.1158/1940-6207.CAPR-08-0014.
Zhou CH, Ye LP, Ye SX, Li Y, Zhang XY, Xu XY, et al. Clinical significance of SOX9 in human non-small cell lung cancer progression and overall patient survival. J Exp Clin Cancer Res. 2012;31:18. https://doi.org/10.1186/1756-9966-31-18.
Acknowledgements
We thank the CRISPR Gene Targeting Core Lab at Taipei Medical University for providing technical support. This manuscript was edited by Wallace Academic Editing.
Availability of data and material
The datasets used or analyzed are available from the corresponding author upon reasonable request.
Code availability
Deregulated KEGG pathways were identified using STRING software (http://string-db.org). Differential genes were analyzed (www.kmplot.com) using Kaplan–Meier plotter with published lung cancer microarray datasets.
Funding
This work was supported by grants from the Ministry of Science and Technology of the Republic of China (MOST: 108-2314-B-038-111-MY3 and 108-2314-B-038-063-MY3), Ministry of Education of the Republic of China (DP2-109-21121-01-T-01), Taipei Medical University and Shuang Ho Hospital (109TMU-SHH-09), Taipei Medical University (TMU107-AE1-B04), and Shuang Ho Hospital (108-FRP-04 and 109-FRP-03).
Author information
Authors and Affiliations
Contributions
KYC and CHT designed the study and wrote the manuscript. PHF and WLS drafted the manuscript and interpreted the data. SCH, CWL, NVH, CSL, and WTL collected the primary data and prepared the figures. TTC and YHT contributed to the literature review and performed statistical analysis. KYL and SMW supervised and reviewed the entire project and manuscript preparation. All authors have read and approved the manuscript.
Ethics declarations
Ethics approval
All protocols for animal experiments were approved by the Taipei Medical University Institutional Animal Care and Use Committee.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflicts of interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
(DOCX 3717 kb)
Rights and permissions
About this article
Cite this article
Chen, KY., Tseng, CH., Feng, PH. et al. 3-Nitrobenzanthrone promotes malignant transformation in human lung epithelial cells through the epiregulin-signaling pathway. Cell Biol Toxicol 38, 865–887 (2022). https://doi.org/10.1007/s10565-021-09612-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10565-021-09612-1