Abstract
Objectives
LncRNAs (long noncoding RNAs) have been reported to critically regulate colorectal cancer (CRC). We prospectively investigated effects and mechanisms of lncRNA LINC00858 on regulation of CRC progression.
Methods
Expression of LINC00858 and its target were analyzed by quantitative real-time polymerase chain reaction and in situ hybridization. MTT and bromodeoxyuridine/5-bromo-2′-deoxyuridine (BrdU) staining to assess cell proliferation ability. Flow cytometry, wound healing, and transwell assays were conducted to evaluate cell apoptosis, migration, and invasion, respectively. Interaction between LINC00858 and its target was confirmed by luciferase activity assay and RNA immunoprecipitation. Subcutaneous xenotransplanted tumor model was established and employed to detect tumorigenic functions of LINC00858, and further evaluated by qRT-PCR, western blot, immunohistochemistry, and hematoxylin and eosin staining.
Results
With a predicted poor prognosis, LINC00858 was upregulated in CRC patients. LINC00858 knockdown suppressed cell proliferation, invasion, and migration abilities, meanwhile induced cell apoptosis. Moreover, LINC00858 could target and inhibit the miR-4766-5p expression, thus promoting CRC progression. miR-4766-5p further suppressed serine/threonine kinase PAK2. Interestingly, interference of LINC00858 suppressed tumorigenic ability of CRC in vivo by downregulating PAK2.
Conclusions
LINC00858 promoted CRC progression by sponging miR-4766 to upregulate PAK2, shedding lights on LINC00858 as a potential therapeutic target candidate in CRC treatment from bench to clinic.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Ranked as the third leading cause of cancer-related death worldwide (Hashim et al. 2016), colorectal cancer (CRC) is assigned with stages I, II, III, and IV due to TNM (tumor-node-metastasis) classification (Chapuis et al. 1986). Current therapeutic strategies targeting CRC, such as surgery, radiation, chemotherapeutic agents, and target therapy, have been progressed tremendously. However, the overall survival rate of CRC patients has not been ameliorated owing to local recurrence and distant metastasis (Lee and Chu 2018). Investigations of the genetic and epigenetic molecular alterations in CRC are urgent and crucial for early diagnosis, prognosis, and survival rate for patients.
Noncoding RNAs (ncRNAs) are the large proportion of transcripts in human genome, consisting of short and long ncRNAs (Consortium et al. 2007). Regarded as mediators of gene expression (Kornienko et al. 2013), lncRNAs participate in various pathological processes (Fatica and Bozzoni 2014). For example, lncRNAs participate in tumor progression (Han et al. 2017). LINC01296 was shown to be a potential prognostic biomarker in CRC (Qiu and Yan 2015). LncRNAs AFAP1-AS1 (Han et al. 2016) and SLCO4A1-AS1 (Yu et al. 2018) promote tumor metastasis of CRC. LINC00858 exhibited effects on development of osteosarcoma (Gu et al. 2018) and non-small cell lung cancer (Zhu et al. 2017b). Recently, upregulation of LINC00858 level was validated in CRC (Yamada et al. 2018), and effects of LINC00858 on CRC progression have also been investigated (Sha et al. 2019). However, the detailed molecular mechanisms of LINC00858 in CRC remain largely unclear.
LncRNAs mediate gene expression in various manners, including hybridization to specific pre-mRNAs to alternatively splice mRNAs (Wilusz et al. 2009), sponge and silence microRNA (miRNA) expression (Ebert and Sharp 2010), or bind with proteins and thus leading to dysregulation of protein activity and cellular localization (Geisler and Coller 2013). Downstream targets of LINC00858 in osteosarcoma (Gu et al. 2018) and non-small cell lung cancer (Zhu et al. 2017b) are identified as miR-139 and miR-422a. However, downstream targets of LINC00858 in CRC have not yet been assessed. LncRNA-PRLB interacts with miR-4766-5p, and regulates breast cancer progression (Liang et al. 2018). Whether LINC00858/miR-4766-5p axis participates in regulation process of CRC progression requires further investigation. Here, we not only evaluated the influence of LINC00858 on CRC progression, but also explored the underlying mechanisms, with the aim to clarify the CRC pathologic process at molecular and cellular level.
Materials and methods
Patients
This study was approved by Ethics Committee on Drug Clinical Trials in Affiliated Hospital of Guizhou Medical University (Approval No. 2018-123-01). Fifty paired CRC and adjacent normal tissues were collected from 50 patients with written informed consents in Affiliated Hospital of Guizhou Medical University. Patients aged from 52 to 80 years old were diagnosed via pathology examination, and their clinicopathological characteristics were shown in Table 1. Patients, diagnosed or received diagnosis care outside the Affiliated Hospital of Guizhou Medical University, were excluded from the study. Patients were returned for follow-up visit with an interval of 2 months in 5 years. During follow-up, 5 patients died of CRC.
ISH (in situ hybridization)
Five micrometers paraffined CRC or adjacent normal tissues were dewaxed and rehydrated, and then digested with 20 μg/mL proteinase K. Four percent paraformaldehyde-fixed samples were hybridized overnight with 8 ng/μL specific antisense oligonucleotide DNA probe at 55 °C, which (5′-cacttcattgggtgttctaa-3′) was synthesized by Invitrogen (Carlsbad, CA, USA). Samples were then incubated with horseradish peroxidase (HRP, Sigma Aldrich, St. Louis, MO, USA). Hybridization signal was amplified with diaminobenzidine (DAB, Sigma Aldrich), and images were taken by fluorescent microscope (DP12 SZX7, Olympus Inc., Japan).
Cell culture and transfection
CRC cell lines (T84, HCT 116, SW480 and HT-29) and NCM460 were acquired from American Type Culture Collection (Manassas, VA, USA), and cultured in RPMI 1640 medium (Lonza, Basel, Switzerland) at 37 °C constant temperature incubator with 5% CO2.
shRNAs (1#: 5′-GCTAAGACCTAATAGCCAATA-3′, 2#: 5′-GCCATCCACTTCAAAGTATTC-3′) for LINC00858 knockdown were subcloned into pLKO.1 (Biosettia, San Diego, CA, USA). pLKO-1# LINC00858 (sh1#-LINC00858), pLKO-2# LINC00858 (sh2#-LINC00858), or pLKO scramble (Scramble) were then co-transfected with psPAX2 and pMD2.G using Lipofectamine 2000 into HEK-293T cells for lentivirus production. HCT 116 or SW480 cells were infected with lentiviruses under 8 mg/mL polybrene and ViraPower™ Packaging Mix (Thermo Fisher, Waltham, MA, USA). Lastly, under the treatment with 5 μg/mL puromycin for 7 days, the stable cell lines were got. Cells were transfected with 40 nM miR-4766-5p mimics, inhibitor, or their negative controls (NC mimics, NC-inh) (GenePharmam Suzhou, China) using Lipofectamine 2000.
Fluorescence in situ hybridization (FISH)
Fixed and permeabilized HCT 116 cells were hybridized with 40 nM fluorescence-conjugated LINC00858 probes (Invitrogen), and then photographed using laser scanning confocal microscopy (Carl Zeiss, Jena, Germany).
Cell proliferation assay
After seeding 2 × 103 HCT 116 or SW480 cells per well in 96-well plates for 24, 48, 72, or 96 h, cells were mixed with 20 μL 5 mg/mL MTT (Sigma Aldrich) for 4 h. The solution were removed, and the cells were incubated with 150 μL DMSO. Four hundred fifty nanometers absorbance was measured.
For BrdU (5-bromo-2′-deoxyuridine) staining, 5 × 103 HCT 116 or SW480 cells per well were incubated with 100 μL 10 μM BrdU labeling solution (Sigma Aldrich). Fixed and permeabilized cells were incubated with 1 × Apollo® reaction cocktail (RiboBio, China), and then incubated overnight with a monoclonal rat anti-BrdU antibody (1:100, Abcam, Cambridge, MA, USA), and then incubated with fluorescently labeled secondary antibody (1:1000, Alexa Fluor 488 goat anti-rat IgG; Abcam). Cells were visualized using a fluorescence microscope (Nikon, Japan) after staining nuclei with Hoechst 33342 solution.
Apoptosis assay
1 × 106 HCT 116 or SW480 cells were resuspended in 100 μL binding buffer (KeyGEN BioTech, Jiangning, Nanjing, China) containing 5 μL propidium iodide (100 μg/mL) with 1 U/mL ribonuclease (Abcam, Cambridge, MA, USA) in dark. The cells were then incubated with additional 5 μL of FITC conjugated annexin V and then evaluated by FACS flow cytometer (Attune, Life Technologies, Darmstadt, Germany).
Wound healing assay
5 × 105 HCT 116 or SW480 cells per well were seeded, and a linear scratch wound was generated with a sterile pipette tip. After removing the suspended cells, wound was imaged and calculated the distance 24 h post-scratch.
Transwell assay
Upper chamber of well (Corning, Tewksbury, MA, USA) with 0.1 mL Matrigel-coated membrane (50 μg/mL; BD Biosciences, Bedford, MA, USA) contained 2 × 104 HCT 116 or SW480 cells in 200 μL serum-free RPMI 1640 medium. Lower chamber contained 400 μL RPMI 1640 medium with 20% FBS. Twenty-four hours later, cells in the lower chamber were stained with 1% crystal violet, and cell numbers were counted under a microscope.
Immunofluorescence
Fixed and permeabilized HCT 116 or SW480 cells were firstly blocked with 5% BSA, and then incubated overnight with the following primary antibodies: anti-E-cadherin anti-N-cadherin (Abcam). Lastly, cells were incubated with fluorescein isothiocyanate (FITC)-conjugated secondary antibody, and photographed after staining with DAPI.
Dual luciferase reporter assay
Wild-type or mutant 3′-UTR of LINC00858 or PAK2 were synthesized and then subcloned into pmirGLO (Promega, Madison, WI, USA). 3 × 104 HEK-293T cells per well were seeded and then co-transfected pmirGLO-wt-LINC00858, pmirGLO-mut-LINC00858, pmirGLO-wt-PAK2, or pmirGLO-mut-PAK2 with miR-4766-5p mimic or miR-NC. Forty-eight hours post-transfection, luciferase activities were performed via Lucifer Reporter Assay System (Promega).
RNA immunoprecipitation (RIP)
HCT 116 or SW480 cells were collected, and lysed using a Magna RIP Kit (EMD Millipore, Billerica, MA). Cell lysates were incubated with protein G Sepharose beads coated with anti-AGO2 antibody (Abcam). Anti-IgG antibody was used as a negative control. RNAs were isolated for qRT-PCR analysis.
qRT-PCR (quantitative real-time polymerase chain reaction)
RNAs and miRNAs were isolated with Trizol (Invitrogen) and miRcute miRNA isolation kit (Tiangen, Beijing, China), respectively. cDNAs were then obtained, and qRT-PCR analyses were performed using SYBR Green Master (Roche, Mannheim, Germany). GAPDH and U6 were applied as endogenous controls. The primer used in this study was shown as below.
Primers table
ID | Sequence (5′-3′) |
|---|---|
GAPDH_F | ACCACAGTCCATGCCATCAC |
GAPDH_R | TCCACCACCCTGTTGCTGTA |
LINC00858_F | CCCAGCTCCTTACACACGTT |
LINC00858_R | TTCAGAGGCCTGCATCACTG |
miR-4766-5p_F | CAGACATACTTTATCATCCCTT |
miR-4766-5p_R | ACAATGCCACCTCCTCC |
PAK2_F | TGGTCGGAACGCCATACTG |
PAK2_R | TTCTGGGGTTCCATTAGTTGC |
U6_F | CTCGCTTCGGCAGCACA |
U6_R | AACGCTTCACGAATTTGCGT |
Western blot
Thirty micrograms protein samples were separated by SDS-PAGE, and then transferred onto PVDF membrane. Membrane was incubated overnight with primary antibodies, including anti-CDK2, p21 antibodies (1:1500, Abcam), Bcl-2, Bax and cleaved caspase-3 (1:2000, Abcam), MMP2, MMP9 (1:2500, Abcam), PAK2, GAPDH (1:3000, Abcam) after blocking with 5% BSA. The immunoreactivities of membranes were determined via enhanced chemiluminescence (KeyGen, Nanjing, China) after incubation with HRP labeled secondary antibody (1:5000; Abcam).
Mouse xenograft assay
All studies involving animals were approved by Experimental Animal Ethics Committee of Guizhou Medical University and conducted in accordance with the guidelines set out by Experimental Animal Ethics Committee of Guizhou Medical University.
Twenty female BALB/c nude mice with 6-week-old were purchased from ARS/Sprague Dawley Division (Madison, WI, USA), and then separated into two groups. One hundred microliters 1 × 108 SW480 cells expressing scrambled shRNA or stable LINC00858 knockdown via pLKO-1# LINC00858 were subcutaneously injected in the right flank of nude mice. Tumors, as well as the volume, were measured every 7 days. Five weeks post-transplantation, tumor tissues were isolated and weighted after the mice were sacrificed.
Immunohistochemistry
Paraffined CRC tissues were dewaxed and rehydrated, and then incised into 4 μm thick sections. Sections were incubated in 3% H2O2, and then immersed for antigen retrieval. After incubation in 4% dry milk and 0.3% goat serum, tissue sections were incubated with anti-PAK2, anti-E-cadherin, or Ki-67 (Abcam) antibody with 10% rabbit serum overnight. After incubation with HRP goat anti-rabbit IgG secondary antibody, tissues were examined under light microscope upon hematoxylin staining.
Statistical analysis
Data were expressed as mean ± SEM. Statistical analyses were conducted via one-way analysis of variance using GraphPad Prism software. Kaplan–Meier method was demonstrated to plot survival curves. Overall survival analysis of patients was determined via log-rank test. P < 0.05 was regarded as statistically significant (*P < 0.05; **P < 0.01; ***P < 0.005).
Results
LncRNA LINC00858 was induced in CRC
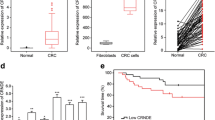
The lncRNA expression level LINC00858 in human CRC tissues was investigated via qRT-PCR. LINC00858 was elevated in CRC tumor tissues (Tumor) (Fig. 1a). In situ hybridization (ISH) further confirmed the augment of LINC00858 in CRC tissues (Fig. 1b). Moreover, median expression level of LINC00858 in CRC patients was considered as a cut-off, patients were then divided into two groups: high LINC00858 expression group (fold change ≥ 2.5, N = 26) and low LINC00858 expression group (N = 24). Kaplan-Meier survival analysis illustrated high LINC00858 expression was related to short overall survival (OS) (*P = 0.0351) (Fig. 1c). Further refinement analysis suggested high LINC00858 expression was dramatically associated with tumor size (**P = 0.010), distance metastasis (***P = 0.005), and TNM stage (*P = 0.020) (Table 1) among the 50 patients. Moreover, other clinical features, including age (P = 0.233) and gender (P = 0.982), showed no significant correlation with LINC00858 expression (Table 1). Therefore, LINC00858 upregulation was related to poor prognosis of CRC, suggesting a potential role of LINC00858 functioning as a prognostic biomarker for CRC treatment. Consistent with its level in tissues, LINC00858 was also increased in CRC cell lines (Fig. 1d). HCT 116 and SW480 cells exhibited higher LINC00858 expression were selected for following experiments. RNA fluorescence in situ hybridization (RNA-FISH) showed LINC00858 was localized in the cytoplasm in HCT 116 and SW480 cells (Fig. 1e).
LncRNA LINC00858 was induced in CRC. a The expression of lncRNA LINC00858 in CRC tissues and adjacent normal tissues detected by qRT-PCR (N = 50). ** represents tumor vs. non-tumor tissues, P < 0.01. b In situ hybridization (ISH) analysis of LINC00858 expression in CRC tissues and non-tumor tissues. c OS analysis of CRC patients with high or low LINC00858 expression levels. d LINC00858 levels in CRC cell lines (T84, HCT 116, SW480, and HT-29) and normal human colon mucosal epithelial cell line (NCM460) were detected by qRT-PCR. ** represents CRC cell lines vs. NCM460, P < 0.01. e Subcellular localization of LINC00858 in HCT 116 and SW480 cells via RNA-FISH
LINC00858 knockdown suppressed CRC proliferation and induced cell apoptosis
HCT 116 and SW480 cells were transfected with pLKO-1# LINC00858 (sh1#-LINC00858) or pLKO-2# LINC00858 (sh2#-LINC00858), and the downstream gene expression was confirmed by qRT-PCR in Fig. 2a. Moreover, MTT (Fig. 2b) and BrdU labeling (Fig. 2c) assays indicated that LINC00858 knockdown suppressed cell proliferation. Furthermore, flow cytometry suggested LINC00858 knockdown would induce apoptotic cell death (Fig. 2d). Apoptosis biomarkers were investigated using western blot, and results revealed CDK2 and Bcl-2 were decreased upon LINC00858 knockdown while p21, Bax, and cleaved caspase-3 were increased (Fig. 2e). Taken together, data obtained from this section suggested LINC00858 might be contributed to abnormal cell proliferation of CRC.
LINC00858 knockdown suppressed CRC proliferation and induced cell apoptosis. a Transfection efficiency of sh1#-LINC00858 or sh2#-LINC00858 in HCT 116 and SW480 cell lines detected by qRT-PCR. ** represents sh1#-LINC00858 or sh2#-LINC00858 vs. Scramble, P < 0.01. b Effects of sh-LINC00858 on cell viability of HCT 116 and SW480 cells detected by MTT. ** represents sh1#-LINC00858 or sh2#-LINC00858 vs. Scramble, P < 0.01. c Effects of sh-LINC00858 on cell proliferation of HCT 116 and SW480 cells detected by BrDU staining. The number of BrdU positive cells were counted. ** represents sh1#-LINC00858 or sh2#-LINC00858 vs. Scramble, P < 0.01. d Effects of sh-LINC00858 on cell apoptosis of HCT 116 and SW480 cells detected by flow cytometry. The number of apoptosis cells were counted. *, ** represents sh1#-LINC00858 or sh2#-LINC00858 vs. Scramble, P < 0.05, P < 0.01. e Effects of sh-LINC00858 on protein expression of CDK2, p21, Bcl-2, Bax, and cleaved caspase-3 in HCT 116 and SW480 cells detected by western blot. *, ** represents sh1#-LINC00858 or sh2#-LINC00858 vs. Scramble, P < 0.05, P < 0.01
LINC00858 knockdown suppressed CRC cell migration and invasion
Wound healing (Fig. 3a) and transwell (Fig. 3b) assays showed that LINC00858 knockdown inhibited cell migration and invasion rate, respectively. Immunofluorescence revealed the upregulation of E-cadherin and downregulation of N-cadherin upon sh1#-LINC00858 and sh2#-LINC00858 expression (Fig. 3c), illustrating the suppression effects of LINC00858 knockdown on cell migration and invasion. Matrix metalloproteinases (MMP2 and MMP9) were decreased by sh-LINC00858 using western bot (Fig. 3d).
LINC00858 knockdown suppressed CRC cell migration and invasion. a Effects of sh-LINC00858 on cell migration of HCT 116 and SW480 cells detected by wound healing assay. *, ** represents sh1#-LINC00858 or sh2#-LINC00858 vs. Scramble, P < 0.05, P < 0.01. b Effects of sh-LINC00858 on cell invasion of HCT 116 and SW480 cells detected by transwell assay. ** represents sh1#-LINC00858 or sh2#-LINC00858 vs. Scramble, P < 0.01. c Effects of sh-LINC00858 on protein expression of E-cadherin and N-cadherin in HCT 116 and SW480 cells detected by immunofluorescence. d Effects of sh-LINC00858 on protein expression of MMP2 and MMP9 in HCT 116 and SW480 cells detected by western blot. ** represents sh1#-LINC00858 or sh2#-LINC00858 vs. Scramble, P < 0.01
LINC00858 directly bound to miR-4766-5p
Given FISH assay showed LINC00858 expressed in the cytoplasm, we subsequently explore the downstream targets of LINC00858. The potential binding target of LINC00858 was predicted, and the transfection efficiency of miR-4766-5p mimics was assessed via qRT-PCR (Fig. 4a,b). miR-47766-5p mimics inhibited luciferase activity of pmirGLO-wt-LINC00858, which did not change pmirGLO-mut-LINC00858 (Fig. 4c). RNA immunoprecipitation (RIP) assay furtherly confirmed the binding between LINC00858 and miR-4766-5p, LINC00858 and miR-4766-5p were both enriched in Ago2-containing beads (Fig. 4d). Moreover, miR-4766-5p expression was decreased upon sh1#-LINC00858 or sh2#-LINC00858 (Fig. 4e), indicating the directly inhibition ability of LINC00858 on miR-4766-5p. Collectively, LINC00858 inhibited miR-4766-5p expression by directly binding. Decreased miR-4766-5p was observed in CRC tumor tissues (Fig. 4f), showing negative correlation between LINC00858 and miR-4766-5p (Fig. 4g).
LINC00858 directly bound to miR-4766-5p. a Transfection efficiency of miR-4766-5p mimics in HCT 116 and SW480 cells. ** represents miR-4766-5p mimics vs. NC mimics, P < 0.01. b The predicted wild-type (WT) binding site of miR-4766-5p in LINC00858. Mutant (MUT) with disrupted binding site of miR-4766-5p in LINC00858 was also shown. c Effects of miR-4766-5p mimics on luciferase activity of reporter gene with wild-type or mutant LINC00858 detected by luciferase reporter assay. ** represents miR-4766-5p mimics vs. NC mimics, P < 0.01. d RNA immunoprecipitation (RIP) assay were performed using the AGO2 antibody, and qRT-PCR was used to detected the enrichment of miR-4766-5p and LINC00858 in HCT 116 and SW480 cells. *, ** represents Anti-Ago2 or input vs. Anti-IgG, P < 0.05, P < 0.01. e Effects of sh-LINC00858 on miR-4766-5p expression in HCT 116 and SW480 cells. ** represents sh1#-LINC00858 or sh2#-LINC00858 vs. Scramble, P < 0.01. f miR-4766-5p levels in CRC tissues and adjacent normal tissues detected by qRT-PCR (N = 60). ** represents tumor vs. non-tumor tissues, P < 0.01. g Negative correlation between LINC00858 and miR-4766-5p in CRC tissues
PAK2 was a direct target of miR-4766-5p
Similarly, PAK2 was predicted as a miR-4766-5p binding target via Targetscan, miRDB, and miRTarBase (Fig. 5a). The predicted binding site was shown in Fig. 5b, and then binding ability was verified via dual luciferase reporter assay (Fig. 5c). Then HCT 116 and SW480 cells were transfected with miR-4766-5p mimics or inhibitor, which was confirmed via qRT-PCR (Fig. 5d). miR-4766-5p mimics decreased protein expression of PAK2 (Fig. 5e), similarly miR-4766-5p inhibitor could restore PAK2 level (Fig. 5e). Interestingly, immunohistochemistry showed PAK2 was upregulated in CRC tumor tissues (Fig. 5f). Bivariate correlation analysis showed positive correlation between PAK2 and LINC00858 in context of CRC, while there was a negative correlation with miR-4766-5p (Fig. 5g). The elevated PAK2 in CRC tissues was also confirmed by western blot analysis (Fig. 5h).
PAK2 was a direct target of miR-4766-5p. a Predicted binding targets for miR-4766-5p binding target via Targetscan, miRDB, and miRTarBase. b The predicted wild-type (WT) binding site of miR-4766-5p in PAK2. Mutant (MUT) with disrupted binding site of miR-4766-5p in PAK2 was also shown. c Effects of miR-4766-5p mimics on luciferase activity of reporter gene with wild-type or mutant PAK2 detected by luciferase reporter assay. ** represents miR-4766-5p mimics vs. NC mimics, P < 0.01. d Transfection efficiency of miR-4766-5p inhibitor in HCT 116 and SW480 cells. ** represents miR-4766-5p inhibitor vs. NC-inh, P < 0.01. e Effects of miR-4766-5p mimics or inhibitor on the protein expression of PAK2 in HCT 116 and SW480 cells. ** represents miR-4766-5p inhibitor vs. NC-inh or miR-4766-5p mimics vs. NC mimics, P < 0.01. f Immunohistochemistry of human CRC tissues and non-tumor tissues. g Positive correlation between PAK2 and LINC00858 and negative correlation between PAK2 and miR-4766-5p in CRC. h Western blot analysis of PAK2 in human CRC tissues and non-tumor tissues. ** represents tumor vs. non-tumor tissues, P < 0.01
LINC00858 knockdown suppressed CRC progression via sponging miR-4766-5p
Cells were then co-transfected with sh1#-LINC00858 and miR-4766-5p inhibitor. Firstly, MTT (Fig. 6a) and BrdU labeling (Fig. 6b) assays indicated that inhibition of cell proliferation upon sh1#-LINC00858 could be promoted by miR-4766-5p inhibitor. In addition, miR-4766-5p inhibitor reversed promotion effects of sh1#-LINC00858 on cell apoptosis (Fig. 6c). The reduced migration (Fig. 6d) and invasion (Fig. 6e) abilities upon sh1#-LINC00858 could also be reversed by miR-4766-5p inhibitor. Secondly, immunofluorescence showed increase of E-cadherin and decrease of N-cadherin upon sh1#-LINC00858 could be furtherly restored by miR-4766-5p inhibitor (Fig. 6f). Western blot analysis showed the decreased levels of PAK2, CDK2, Bcl-2, MMP2, and MMP9, and the increased p21, Bax, and cleaved caspase-3 upon sh1#-LINC00858 could be reversed by miR-4766-5p inhibitor, further demonstrating functions of LINC00858 in sponging miR-4766 to promote cell progression of CRC.
LINC00858 knockdown suppressed CRC progression via sponging miR-4766-5p. a The inhibited cell viability by sh1#-LINC00858 was promoted by additional transfection with miR-4766-5p inhibitor in HCT 116 and SW480 cells detected by MTT assay. *, ** represents sh1#-LINC00858 + NC-inh vs. Scramble + NC-inh or sh1#-LINC00858 + miR-4766-5p inh, P < 0.05, P < 0.01. b The inhibited cell proliferation by sh1#-LINC00858 was promoted by additional transfection with miR-4766-5p inhibitor in HCT 116 and SW480 cells detected by BrDU staining assay. ** represents sh1#-LINC00858 + NC-inh vs. Scramble + NC-inh or sh1#-LINC00858 + miR-4766-5p inh, P < 0.01. c The promoted cell apoptosis by sh1#-LINC00858 was inhibited by additional transfection with miR-4766-5p inhibitor in HCT 116 and SW480 cells detected by flow cytometry. ** represents sh1#-LINC00858 + NC-inh vs. Scramble + NC-inh or sh1#-LINC00858 + miR-4766-5p inh, P < 0.01. d The inhibited cell migration by sh1#-LINC00858 was promoted by additional transfection with miR-4766-5p inhibitor in HCT 116 and SW480 cells detected by wound healing assay. ** represents sh1#-LINC00858 + NC-inh vs. Scramble + NC-inh or sh1#-LINC00858 + miR-4766-5p inh, P < 0.01. e The inhibited cell invasion by sh1#-LINC00858 was promoted by additional transfection with miR-4766-5p inhibitor in HCT 116 and SW480 cells detected by transwell assay. ** represents sh1#-LINC00858 + NC-inh vs. Scramble + NC-inh or sh1#-LINC00858 + miR-4766-5p inh, P < 0.01. f The increase of E-cadherin and decrease of N-cadherin in HCT 116 and SW480 cells transfected with sh1#-LINC00858 were reversed by cotransfection with sh1#-LINC00858 and miR-4766-5p inhibitor detected by immunofluorescence. g Protein expression of PAK2, CDK2, p21, Bcl-2, Bax, cleaved caspase-3, MMP2 and MMP9 in HCT 116 and SW480 cells transfected with sh1#-LINC00858 were reversed by cotransfection with sh1#-LINC00858 and miR-4766-5p inhibitor detected by western blot. ** represents sh1#-LINC00858 + NC-inh vs. Scramble + NC-inh or sh1#-LINC00858 + miR-4766-5p inh, P < 0.01
LINC00858 knockdown suppressed in vivo CRC tumor growth
We then inoculated SW480 cells expressing sh1#-LINC00858 into nude mice. Firstly, the transfection efficiency was confirmed (Fig. 7a), as shown by a downregulation of LINC00858 and upregulation of miR-4766-5p. Moreover, intratumoral injection of sh1#-LINC00858 suppressed tumor growth (Fig. 7b), as shown by a reduced tumor volume and weight (Fig. 7c). Furthermore, immunohistochemistry demonstrated decrease of PAK2 and Ki67, increase of E-cadherin in xenograft tumor tissues injected with sh1#-LINC00858 (Fig. 7d). Finally, we investigated the effect of LINC00858 on tumor metastasis in vivo, and discovered that LINC00858 knockdown dramatically reduced the metastatic nodules in the lung (Figs. 7e and 8), confirming the suppression effect of LINC00858 knockdown on xenograft tumor growth.
LINC00858 knockdown suppressed in vivo CRC tumor growth. a Transfection efficiency of sh1#-LINC00858 and the expression of miR-4766-5p in xenograft tumor mice detected by qRT-PCR. ** represents sh1#-LINC00858 vs. Scramble, P < 0.01. b Effects of sh1#-LINC00858 on tumor growth in xenograft tumor mice. c Effects of sh1#-LINC00858 on the tumor volume and weight. ** represents sh1#-LINC00858 vs. Scramble, P < 0.01. d Immunohistochemistry staining was used to determine expression of PAK2, E-cadherin, and Ki-67 affected by sh1#-LINC00858. Black bar: 50 μm. e LINC00858 knockdown remarkably reduced the metastatic nodules in the lung. The liver sections were shown via H&E staining. Scale bar, 50 μm. ** represents sh1#-LINC00858 vs. Scramble, P < 0.01
Discussion
Recently, studies have demonstrated functional roles of lncRNAs as promoter or inhibitor of cancer-critical genes in CRC (Zhu et al. 2017a), which mediated the regulation process of cell progression (Smolle et al. 2014). Due to the increasing mortality and incidence of CRC worldwide, especially in China (Li et al. 2014), lncRNAs became a research hotspot due to its potential role as prognosis biomarker in CRC. lncRNAs may not only contribute to early diagnosis, but also would improve the individualized treatment of CRC patients (Yang et al. 2017). Accumulating evidence have reported LINC00858 participates in carcinogenesis and progression of various types of tumors. We hypothesized that LINC00858 dysregulation in CRC might be a molecular therapeutic target in CRC (Yamada et al. 2018). However, the functional role and clinical relevance of LINC00858 in CRC is far from clear. Therefore, we investigated potential mechanisms of LINC00858 in tumorigenesis of CRC.
We discovered that high expression level of LINC00858, occurred in CRC tissues, was closely related to some clinical parameters and predicted a poor prognosis in CRC patients, which is in consistent with previous studies in other cancers including osteosarcoma (Gu et al. 2018) and non-small cell lung cancer (Zhu et al. 2017b). Similar to Sha et al (Sha et al. 2019)’s work, high expression of LINC00858 was related to poor OS of CRC patients with more frequently occurred in patients with TNM stages III and IV, suggesting LINC00858 might be a poor prognosis indicator of CRC. However, due to the small sample size of our current clinical analysis (N = 50), the significant correlation between expression levels of LINC00858 with histological grades and lymph nodes metastasis found in research before (Sha et al. 2019) was not reported in this study. A larger patient cohort is required to strengthen the clinical significance of LINC00858 in CRC patients for future investigations.
Similar to the clinical results, in vitro loss-of-function assays using LINC00858 shRNAs showed LINC00858 knockdown not only inhibited CRC progression, but also induced cell apoptosis, which is consistent with Sha et al (Sha et al. 2019)’s study. Moreover, our in vivo subcutaneous xenotransplanted tumor model for the first time revealed that interference of LINC00858 could suppress CRC growth in vivo, indicating the potential clinical application of LINC00858 in CRC treatment.
Although Sha et al. (Sha et al. 2019) have reported the oncogenic role of LINC00858 on CRC depended on the promotion of cell apoptosis, its functional targets remain unclear. In this study, our results showed to regulate cell apoptosis, CDK2, p21, Bcl-2, Bax, and cleaved caspase-3 were found to be involved in regulation of LINC00858 on CRC. Generally, CDK2 (cyclin-dependent kinase 2), as the mediator of cell cycle transition, has been found to be upregulated in CRC (Diaz-Moralli et al. 2013). CDK2 inhibitor exhibits anti-proliferative effect on CRC (Shi et al. 2015). Moreover, as a cyclin-dependent kinases inhibitors, p21 suppressed cell growth as an antitumor agent (Pasz-Walczak et al. 2001). Bax and cleaved caspase-3 promoted CRC apoptosis, and Bcl-2 inhibited apoptosis by suppression of Bax (Khodapasand et al. 2015; Noble et al. 2013).We showed that LINC00858 knockdown increased p21, Bax, and cleaved caspase-3 and decreased CDK2 and Bcl-2 levels, and thus promoting CRC cell apoptosis. Flow cytometry also confirmed the pro-apoptotic effects of LINC00858 knockdown. Meanwhile, the effects of LINC00858 on CRC cell cycle should be further investigated.
In addition to promotion of cell apoptosis, the oncogenic role of LINC00858 in CRC was also dependent on the promotion of epithelial to mesenchymal transition (EMT) (Sha et al. 2019). EMT is essential for the development of metastasis, which contributes to the unfavorable prognosis in cancer (Pino et al. 2010). Reduced E-cadherin level accompanied by elevated mesenchymal markers (N-cadherin or vimentin) led to disassembly of cell adherens junctions and cell invasion (Buhrmann et al. 2019). Sha et al (2019) only reported LINC00858 knockdown decreased N-cadherin and vimentin, we reported that LINC00858 knockdown increased E-cadherin and decreased N-cadherin, demonstrating the anti-migration and invasion abilities of LINC00858 knockdown on CRC. Moreover, MMP2 and MMP9, biomarkers significantly associated with invasive cancer stages of CRC (Araujo Jr. et al. 2015), were downregulated upon LINC00858 knockdown in the present study, which is beneficial for the inhibition of CRC metastasis (Damodharan et al. 2011). EMT also participate in mediation of cancer stemness (Fender et al. 2015). lncRNA UICLM induced EMT and promoted stemness properties of CRC for the metastasis (Chen et al. 2017). Considering that LINC00858 was associated with EMT of CRC, the impact of LINC00858 on stemness properties of CRC also required further investigation.
As well known, lncRNAs depends on its binding targets (miRNAs and proteins) to perform biological functions. Sha et al. (2019) reported that miR-22-3p was a target of LINC00858, which was associated with regulation of CRC cell progression. By dual luciferase reporter assay and RIP, miR-4766-5p was validated as a binding target of LINC00858. Other than miR-22-3p/YWHAZ axis reported before (Sha et al. 2019), our results for the first time confirmed LINC00858 could promote CRC progression by sponging miR-4766-5p to upregulate PAK2. PAKs (p21-activated kinases), as commonly oncogenic kinases, have been considered as promoter of CRC progression (Ye and Field 2012). PAKs could promote cell cycle progression via Erk, Akt, and Wnt signaling pathways, trigger apoptosis via nuclear factor κB (NF-κB) pathway, as well as activate invasion and metastasis via various pathways (Senapedis et al. 2016; Carter et al. 2004). However, different from other PAKs, PAK2 inhibits cell cycle progression, performs anti-apoptotic and anti-metastatic in CRC (Lv et al. 2018). Loss-of-function assays showed that LINC00858 knockdown decreased PAK2 expression, and thus suppressing cell progression of CRC. However, other isoforms of PAKs in CRC have yet not been evaluated.
In summary, this study reported lncRNA LINC00858 knockdown inhibited CRC progression and induced cell apoptosis, via miR-4766-5p-mediated PAK2. The research regarding LINC00858/miR-4766-5p/PAK2 regulatory axis shed lights on LINC00858 as a potential therapeutic candidate targeting tumor progression for CRC treatment from bench to clinic.
References
Araujo RF Jr, Lira GA, Vilaca JA, Guedes HG, Leitao MC, Lucena HF, et al. Prognostic and diagnostic implications of MMP-2, MMP-9, and VEGF-alpha expressions in colorectal cancer. Pathol Res Pract. 2015;211(1):71–7. https://doi.org/10.1016/j.prp.2014.09.007.
Buhrmann C, Yazdi M, Popper B, Kunnumakkara AB, Aggarwal BB, Shakibaei M. Induction of the epithelial-to-mesenchymal transition of human colorectal cancer by human TNF-beta (lymphotoxin) and its reversal by resveratrol. Nutrients. 2019;11(3). https://doi.org/10.3390/nu11030704.
Carter JH, Douglass LE, Deddens JA, Colligan BM, Bhatt TR, Pemberton JO, et al. Pak-1 expression increases with progression of colorectal carcinomas to metastasis. Clin Cancer Res. 2004;10(10):3448–56. https://doi.org/10.1158/1078-0432.CCR-03-0210.
Chapuis PH, Dent OF, Newland RC, Bokey EL, Pheils MT. An evaluation of the American Joint Committee (pTNM) staging method for cancer of the colon and rectum. Dis Colon Rectum. 1986;29(1):6–10.
Chen DL, Lu YX, Zhang JX, Wei XL, Wang F, Zeng ZL, et al. Long non-coding RNA UICLM promotes colorectal cancer liver metastasis by acting as a ceRNA for microRNA-215 to regulate ZEB2 expression. Theranostics. 2017;7(19):4836–49. https://doi.org/10.7150/thno.20942.
Consortium EP, Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447(7146):799–816. https://doi.org/10.1038/nature05874.
Damodharan U, Ganesan R, Radhakrishnan UC. Expression of MMP2 and MMP9 (gelatinases A and B) in human colon cancer cells. Appl Biochem Biotechnol. 2011;165(5–6):1245–52. https://doi.org/10.1007/s12010-011-9342-8.
Diaz-Moralli S, Tarrado-Castellarnau M, Miranda A, Cascante M. Targeting cell cycle regulation in cancer therapy. Pharmacol Ther. 2013;138(2):255–71. https://doi.org/10.1016/j.pharmthera.2013.01.011.
Ebert MS, Sharp PA. Emerging roles for natural microRNA sponges. Curr Biol. 2010;20(19):R858–61. https://doi.org/10.1016/j.cub.2010.08.052.
Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15(1):7–21. https://doi.org/10.1038/nrg3606.
Fender AW, Nutter JM, Fitzgerald TL, Bertrand FE, Sigounas G. Notch-1 promotes stemness and epithelial to mesenchymal transition in colorectal cancer. J Cell Biochem. 2015;116(11):2517–27. https://doi.org/10.1002/jcb.25196.
Geisler S, Coller J. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Biol. 2013;14(11):699–712. https://doi.org/10.1038/nrm3679.
Gu Z, Hou Z, Zheng L, Wang X, Wu L, Zhang C. Long noncoding RNA LINC00858 promotes osteosarcoma through regulating miR-139-CDK14 axis. Biochem Biophys Res Commun. 2018;503(2):1134–40. https://doi.org/10.1016/j.bbrc.2018.06.131.
Han X, Wang L, Ning Y, Li S, Wang Z. Long non-coding RNA AFAP1-AS1 facilitates tumor growth and promotes metastasis in colorectal cancer. Biol Res. 2016;49(1):36. https://doi.org/10.1186/s40659-016-0094-3.
Han F, Wang C, Wang Y, Zhang L. Long noncoding RNA ATB promotes osteosarcoma cell proliferation, migration and invasion by suppressing miR-200s. Am J Cancer Res. 2017;7(4):770–83.
Hashim D, Boffetta P, La Vecchia C, Rota M, Bertuccio P, Malvezzi M, et al. The global decrease in cancer mortality: trends and disparities. Ann Oncol. 2016;27(5):926–33. https://doi.org/10.1093/annonc/mdw027.
Khodapasand E, Jafarzadeh N, Farrokhi F, Kamalidehghan B, Houshmand M. Is Bax/Bcl-2 ratio considered as a prognostic marker with age and tumor location in colorectal cancer? Iran Biomed J. 2015;19(2):69–75.
Kornienko AE, Guenzl PM, Barlow DP, Pauler FM. Gene regulation by the act of long non-coding RNA transcription. BMC Biol. 2013;11:59. https://doi.org/10.1186/1741-7007-11-59.
Lee JJ, Chu E. The adjuvant treatment of stage III colon cancer: might less be more? Oncology. 2018;32(9):437–42 44.
Li X, Hu F, Wang Y, Yao X, Zhang Z, Wang F, et al. CpG island methylator phenotype and prognosis of colorectal cancer in Northeast China. Biomed Res Int. 2014;2014:236361. https://doi.org/10.1155/2014/236361.
Liang Y, Song X, Li Y, Sang Y, Zhang N, Zhang H, et al. A novel long non-coding RNA-PRLB acts as a tumor promoter through regulating miR-4766-5p/SIRT1 axis in breast cancer. Cell Death Dis. 2018;9(5):563. https://doi.org/10.1038/s41419-018-0582-1.
Lv C, Wang H, Tong Y, Yin H, Wang D, Yan Z, et al. The function of BTG3 in colorectal cancer cells and its possible signaling pathway. J Cancer Res Clin Oncol. 2018;144(2):295–308. https://doi.org/10.1007/s00432-017-2561-9.
Noble P, Vyas M, Al-Attar A, Durrant S, Scholefield J, Durrant L. High levels of cleaved caspase-3 in colorectal tumour stroma predict good survival. Br J Cancer. 2013;108(10):2097–105. https://doi.org/10.1038/bjc.2013.166.
Pasz-Walczak G, Kordek R, Faflik M. P21 (WAF1) expression in colorectal cancer: correlation with P53 and cyclin D1 expression, clinicopathological parameters and prognosis. Pathol Res Pract. 2001;197(10):683–9. https://doi.org/10.1078/0344-0338-00146.
Pino MS, Kikuchi H, Zeng M, Herraiz MT, Sperduti I, Berger D, et al. Epithelial to mesenchymal transition is impaired in colon cancer cells with microsatellite instability. Gastroenterology. 2010;138(4):1406–17. https://doi.org/10.1053/j.gastro.2009.12.010.
Qiu JJ, Yan JBJTB. Long non-coding RNA LINC01296 is a potential prognostic biomarker in patients with colorectal cancer. 2015;36(9):7175–83.
Senapedis W, Crochiere M, Baloglu E, Landesman Y. Therapeutic potential of targeting PAK signaling. Anti Cancer Agents Med Chem. 2016;16(1):75–88.
Sha Q-K, Chen L, Xi J-Z, Song H. Long non-coding RNA LINC00858 promotes cells proliferation, migration and invasion by acting as a ceRNA of miR-22-3p in colorectal cancer. Artif Cells Nanomed Biotechnol. 2019;47(1):1057–66. https://doi.org/10.1080/21691401.2018.1544143.
Shi XN, Li H, Yao H, Liu X, Li L, Leung KS, et al. Adapalene inhibits the activity of cyclin-dependent kinase 2 in colorectal carcinoma. Mol Med Rep. 2015;12(5):6501–8. https://doi.org/10.3892/mmr.2015.4310.
Smolle M, Uranitsch S, Gerger A, Pichler M, Haybaeck J. Current status of long non-coding RNAs in human cancer with specific focus on colorectal cancer. Int J Mol Sci. 2014;15(8):13993–4013. https://doi.org/10.3390/ijms150813993.
Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23(13):1494–504. https://doi.org/10.1101/gad.1800909.
Yamada A, Yu P, Lin W, Okugawa Y, Boland CR, Goel A. A RNA-sequencing approach for the identification of novel long non-coding RNA biomarkers in colorectal cancer. Sci Rep. 2018;8(1):575. https://doi.org/10.1038/s41598-017-18407-6.
Yang Y, Junjie P, Sanjun C, Ma Y. Long non-coding RNAs in colorectal cancer: progression and future directions. J Cancer. 2017;8(16):3212–25. https://doi.org/10.7150/jca.19794.
Ye DZ, Field J. PAK signaling in cancer. Cell Logist. 2012;2(2):105–16. https://doi.org/10.4161/cl.21882.
Yu J, Han Z, Sun Z, Wang Y, Zheng M, Song C. LncRNA SLCO4A1-AS1 facilitates growth and metastasis of colorectal cancer through beta-catenin-dependent Wnt pathway. J Exp Clin Cancer Res. 2018;37(1):222. https://doi.org/10.1186/s13046-018-0896-y.
Zhu H, Yu J, Zhu H, Guo Y, Feng S. Identification of key lncRNAs in colorectal cancer progression based on associated protein-protein interaction analysis. World J Surg Oncol. 2017a;15(1):153. https://doi.org/10.1186/s12957-017-1211-7.
Zhu SP, Wang JY, Wang XG, Zhao JP. Long intergenic non-protein coding RNA 00858 functions as a competing endogenous RNA for miR-422a to facilitate the cell growth in non-small cell lung cancer. Aging. 2017b;9(2):475–86. https://doi.org/10.18632/aging.101171.
Funding
This work was supported by Special Project of Academic New Seedling Cultivation and Innovation Exploration of Guizhou Medical University (Grant No. [2018]5779-30), Science and Technology Fund Project of Guizhou Health and Family Planning Commission (Grant No. gzwjkj-2018-1-035), Science and Technology Fund Project of Guizhou Health and Family Planning Commission (Grant No. gzwjkj-2018-1-075), State science and technology plan project of Qiandongnan Prefecture in 2019(145) and State science and technology plan project of Qiandongnan Prefecture in 2019(146).
Author information
Authors and Affiliations
Contributions
Wei Zhan and Xin Liao and Rui Li conceived and designed the experiments, Tian Tian analyzed and interpreted the results of the experiments, Cheng Zhongsheng and LiangheLi and Lei Yu performed the experiments.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee on Drug Clinical Trials in Affiliated Hospital of Guizhou Medical University (Approval No. 2018-123-01). Written informed consent was obtained from each of the participants. All methods were performed in accordance with the relevant guidelines and regulations as per the instructions of the Ethical Research Board.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Zhan, W., Liao, X., Chen, Z. et al. LINC00858 promotes colorectal cancer by sponging miR-4766-5p to regulate PAK2. Cell Biol Toxicol 36, 333–347 (2020). https://doi.org/10.1007/s10565-019-09506-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10565-019-09506-3