Abstract
Bufalin is the primary component of the traditional Chinese medicine “Chan Su,” which has been widely used for cancer treatment at oncology clinics in certain countries. Evidence suggests that this compound possesses potent antitumor activities, although the exact molecular mechanism(s) require further elucidation. Therefore, this study aimed to further clarify the in vitro and in vivo antiglioma effects of bufalin and the molecular mechanism underlying the regulation of drug sensitivity. The anticancer effects of bufalin were determined by colony formation assays, apoptosis assays, and cellular redox state tests of glioma cells. Confocal microscopy was performed to determine the expression changes of the DNA damage biomarker γ-H2AX and the nuclear translocation of p53 in glioma cells. Western blotting and RT-PCR were used to detect the protein and gene expression levels, respectively. Here, we report that bufalin induced glioblastoma cell apoptosis and oxidative stress and triggered DNA damage. The critical roles of the sodium pump α1 subunit (ATP1A1) in mediating the XPO1-targeted anticancer effect of bufalin in human glioma were further confirmed. Mechanistic studies confirmed the important roles of Src and p53 signaling in mediating bufalin-induced apoptosis. Importantly, bufalin also inhibited the growth of glioma xenografts. In conclusion, our study indicated that therapies targeting the ATP1A1 and p53 signaling-mediated mitochondrial apoptotic pathways regulated by bufalin might be potential treatments for human glioma, and these findings will provide molecular bases for developing bufalin into a drug candidate for the treatment of malignant glioma.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Bufalin, an endogenous cardiac glycoside, is a soluble digoxin-like immunoreactive component of Chan Su (a traditional Chinese medicine), which is an extract of dried toad venom from the skin glands of Bufogargarizans or Bufomelanostictus. Bufalin was shown to have antitumor activity in various tumors, such as liver cancer, lung cancer, intestinal cancer, gastric cancer, gynecological cancer, and pancreatic cancer (Takai et al. 2012). However, the precise underlying mechanisms remain unknown, and more efforts should be directed toward clarifying its effects on human glioblastoma cells (Shen et al. 2014).
Bufalin treatment was shown to lead to simultaneous initiation of apoptosis and ER stress, and both mitochondrial and ER-mediated apoptotic pathways are involved in glioma cell death induced by bufalin (Shen et al. 2014). A recent study showed that bufalin resulted in a DNA double strand break (DSB) response, ROS induction, and apoptosis within 24 h in a rapid response in cancer cells (Fujii et al. 2018). DSBs could lead to phosphorylation of ataxia telangiectasia mutated (ATM). As a result, a histone variant H2AX is phosphorylated beginning within seconds, which is termed γ-H2AX (Rogakou et al. 2000; Houtgraff et al. 2006). Thus, γ-H2AX is frequently used as a DNA damage biomarker.
Interestingly, the early wave of apoptosis following DNA damage is completely dependent on the nuclear accumulation of p53 (Phesse et al. 2014). The tumor suppressor gene p53 has also been shown to exert its anticancer function by inducing cell apoptosis in cancer cells. Studies have shown that γ-H2AX triggers the checkpoint kinase 2 (CHK2) signal transduction pathway that activates p53 (Yuan et al. 2010). DSB-induced phosphorylated ATM can also directly phosphorylate p53 (Yuan et al. 2010). Thus, phosphorylated p53 transcriptionally activates the CDK inhibitor p21 and various downstream factors and arrests the cell cycle at G1/S (Kurz and Lees-Miller 2004). Although p53 has been demonstrated to be critical for various chemotherapeutic drugs, its precise roles in the anticancer effect of bufalin and its regulatory effects on bufalin resistance have not been elucidated.
Sodium pumps (Na+/K+-ATPase) are widespread in eukaryotic cell membranes, consisting of equimolar ratios of two main subunits in mammals, the catalytic α and regulatory β polypeptides. Specifically, four different α isoforms (ATP1A1-1A4) and three distinct β isoforms (ATP1B1-1B3) have been identified in humans (Mijatovic et al. 2007a, b). Na+/K+-ATPase could also function as a signal transducing receptor, and this function of signal transduction is mostly attributed to the α1 subunit (ATP1A1) (Li et al. 2011). Cardiac glycosides can bind and inhibit the α-catalytic subunit of the human Na+/K+-ATPase encoded by ATP1A1, ATP1A2, ATP1A3, or ATP1A4 (Mijatovic et al. 2007a, b). These molecules have also been shown to kill melanoma cells by inhibitingATP1A1 (Eskiocak et al. 2016). Furthermore, our previous research found that bufalin inhibited the expression of ATP1A1 in glioblastoma by promoting the activation of proteasomes and the subsequent protein degradation of ATP1A1, which could suppress glioma proliferation (Lan et al. 2018). In addition, there are various other steroidal inhibitors of Na+/K+-ATPase with strong anticancer effects. Ouabain was recently suggested to have potential anticancer effects (Xiao et al. 2017). Recent crystallographic studies have provided insight into how ouabain inhibits sodium pumps (Yatime et al. 2011; Ogawa et al. 2009), and several pathways, including key cell regulators, such as MAPK, PI-3K, AKT, PLC, and Src, were shown to be influenced by ouabain treatment (Xie and Cai 2003). Furthermore, oleandrin, which was the first cardiotonic steroids (CTS) to be tested in anticancer clinical trials, demonstrated a significant inhibitory effect on the proliferation of human cancer cells without significantly reducing the viability of normal cells (Pan et al. 2017). Hellebrin, which is currently commercially available with high selectivity for ATP1A1 (Moreno et al. 2013), was also found to be an anticancer agent that inhibited ATP1A1. Thus, ATP1A1 might be a potential target of the anticancer agent bufalin. However, the precise mechanisms remain to be elucidated.
Protein localization is tightly linked to its function (Mellman and Nelson 2008; Clarke and Zhang 2008). Improper localization of a nuclear protein to the cytoplasm can damage its functions, and protein localization in cells is tightly regulated by various transporters (Mellman and Nelson 2008; Clarke and Zhang 2008). Import of proteins inside the nucleus is carried by importins, while export of proteins is carried by exportins (Kau et al. 2004). Currently, among the several known exportins in mammalian cells, exportin 1 (XPO1, also called CRM1) is the most studied prototype (Nigg 1997; Fukuda et al. 1997). Increased expression of XPO1 protein has been found in various cancers, including pancreatic cancer (Huang et al. 2009), cervical cancer (van der Watt et al. 2009), ovarian cancer (Noske et al. 2008), mantle cell lymphoma (Zhang et al. 2013), and glioma (Shen et al. 2009). Cancer cells utilize XPO1 to export factors such as p53, APC, p21, p27, and various others to the cytoplasm (Turner and Sullivan 2008). Restriction of these key gatekeeper proteins to the cytoplasmic compartment prevents them from suppressing tumor growth. In addition, restoring nuclear p53 function through inhibition of XPO1 has been found to trigger cell apoptosis (Wood et al. 2007; Grasso et al. 2012). These results suggest that XPO1 might be an attractive target in glioblastoma.
Bufalin could reduce p53 expression in certain types of cancer cells (Wu et al. 2014) while enhancing p53 expression in others (Yu et al. 2008). For cardiac glycosides, the drug sensitivity to p53 reduction is cancer cell line-dependent but independent of p53status (wild-type or mutant). Mechanistically, the drug-induced p53 decrease occurred not at the mRNA level but at the protein level as a result of reduced synthesis rather than enhanced degradation (Wang et al. 2009). Significantly, the drug-induced p53 reduction can be rescued by Src inhibitors, suggesting the involvement of Src-activated protein kinase (MAPK) signaling pathways, initiated upon drug binding to Na+/K+-ATPase (Wang et al. 2009). Importantly, Na+/K+-ATPase-mediated cellular signaling is initialized in the cholesterol and sphingomyelin-rich plasma membrane subfractions of the caveolae, in which ATP1A1 interacts with various signaling proteins. Src is a primary target of ATP1A1 and is responsible for communication between Na+/K+-ATPase and other proteins. When the Na+/K+-ATPase-Src complex is activated, different signaling pathways, including the MAPK pathways, are initiated in a specific manner depending on the stimulus and cell type (Reinhard et al. 2013). A previous study also described an antitumor signaling mechanism of a potential ATP1A1-targeted drug, perillyl alcohol, mediated via inhibition of Na+/K+-ATPase and Src activation in a glioblastoma cell line (Garcia et al. 2015). All of these findings suggest that Na+/K+-ATPase could be a target for anticancer drugs and help clarify the downstream molecular mechanisms.
Thus, in the present study, we investigated how ATP1A1, XPO1, P53, and Src regulated bufalin-induced apoptosis in glioblastoma cells and, more importantly, explored the critical roles of ATP1A1 and p53 in bufalin sensitivity.
Materials and methods
Materials
Bufalin (purity > 98%) was purchased from Yuanye Biotech. (Jinan, Shandong, China), diluted with DMSO and stored at − 20 °C as the stock solution. Bufalin was diluted in culture medium to obtain the desired concentration, which was stable in DMSO. Antibodies specific to Bax, Bcl-2, cleaved caspase-3, cleaved caspase-9, cleaved PARP, ATP1A1, XPO1, p53, Src, pSrc, β-actin, and lamin B1 and all the secondary antibodies were obtained from Cell Signaling Technology (Cell Signaling Technology, Inc., USA). The primary antibodies for cytosolic cytochrome c (cyto c) and mitochondrial cyto c were obtained from Santa Cruz Biotechnology (Santa Cruz, CA, USA). The p53 inhibitor pifthrin-α (Pif) was purchased from the Beyotime Institute of Technology (Shanghai, China). Trypsin, Dulbecco’s modified Eagle’s medium (DMEM) and fetal bovine serum (FBS) were obtained from HyClone Laboratories (HyClone Laboratories, Inc.). Phosphate-buffered saline (PBS), protease inhibitor cocktail, and 5-diphenyltetrazolium bromide (MTT) were purchased from Sigma Chemical Co. (St. Louis, MO, USA). All other chemicals were purchased from Sigma Chemical Co. (St. Louis, MO, USA) unless otherwise specified.
Cell lines and cell culture
The human glioma cell lines U87, U251, LN229, A172, and U118 were utilized. Cells were cultured in DMEM supplemented with 10% fetal calf serum at 37 °C with a humidified atmosphere of 5% CO2 and 95% air.
Cell viability assay
Cell viability was determined by MTT assays. Briefly, the glioma cells were seeded in 96-well plates at a density of 6 × 103 cells per well. The cells were allowed to adhere overnight before being placed in fresh medium containing various concentrations of bufalin dissolved in DMSO (final concentration, 0.1%). After the cells had incubated for 48 h, we measured their growth. We quantified the effects of bufalin on cell viability by comparing the percentage of viable cells in the treated group with that in the untreated control group.
Colony formation assay
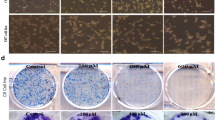
U87 and U25 cells (1 × 103 per well) were seeded in six-well plate incomplete medium and cultured for 24 h. Then, we removed the medium, and cells were exposed to various concentrations of bufalin (0, 50, and 100 nM). After 24 h, the cells were washed with PBS and supplemented with fresh medium containing 10% FBS. After incubation for 14 days, cells grew into macroscopic colonies. We removed the medium, and the colonies were counted after staining with 0.1% crystal violet.
Flow cytometry analysis
To determine the proportion of apoptotic cells, we performed flow cytometric analysis using a flow cytometer (BD FACS Accuri C6, CA, USA). After a 24-h treatment with bufalin (0, 50, and 100 nM), the cells were collected, washed with PBS, and fixed with ice-cold 70% ethanol at 4 °C for 4 h. The cells were stained with propidium iodide (PI) staining buffer (0.2% Triton X-100, 100 μg/mL DNase-free RNase A, and 50 μg/mL PI in PBS) in the dark for 30 min. For the apoptosis examination, the cells were washed with PBS, collected, and stained using an Annexin V-FITC Apoptosis Detection Kit in the dark at room temperature for 15 min. The fraction of apoptotic cells was determined using a FACS analysis system. Each experiment was performed in triplicate.
Investigation of the cellular redox state
Dihydroethidium (DHE) was used to determine the cellular ROS level. Single-cell suspensions of cells treated with different concentrations of bufalin (or DMSO as a blank control, 10 μM H2O2 for 2 h as a positive control) for 24 h were washed and incubated with 1 μM DHE at 37 °C for 30 min. Cells were then washed three times with probe-free PBS, and the fluorescence intensities of cells were measured by flow cytometry.
Measurement of extracellular oxygen consumption rate
Changes in the extracellular oxygen consumption rate (OCR) of cells were monitored using the Extracellular O2Consumption Assay Kit (cat#ab197243, Abcam). Briefly, U87 cells (6 × 104/well) were plated into 96-well (black wall) clear bottom plates and incubated in a CO2 incubator at 37 °C overnight. Then, the culture medium was removed and replaced with 150 μL of fresh culture media supplemented with the indicated doses of bufalin. An aliquot of 10 μL reconstituted Extracellular O2 Consumption reagent and two drops pre-warmed high-sensitivity mineral oil were added into each well, and the plate was immediately monitored using a multidetection microplate reader (Infinite M1000 Pro, TECAN). The extracellular O2 consumption signal was measured at 1.5 min intervals for 90–120 min at 37 °C using excitation and emission wavelengths of 380 and 650 nm, respectively.
Mitochondrial membrane potential assay
The JC-1 probe was used to test mitochondrial depolarizationin glioma cells. After the indicated treatments, cells cultured in six-well plates were incubated with an equal volume of JC-1staining solution (5 μg/mL) at 37 °C for 20 min, followed by two rinses with PBS. Mitochondrial membrane potentials were monitored by counting the relative amounts of dual emissions from mitochondrial JC-1monomers or aggregates using an Olympus fluorescence microscope under argon ion 488 nm laser excitation. Mitochondrial depolarization was shown by a trend of decrease in the red/green fluorescence intensity ratio.
Na+/K+-ATPase assay in glioblastoma cells
The Na+/K+-ATPase assay in cell culture was based on the measurement of non-radioactive Rb+ incorporation according to a previous publication (Gill et al. 2004). After the samples were digested with nitric acid, Rb+ measurements were performed in an inductively coupled plasma-optical emission spectrometer (Jobin-Yvon). Different concentrations of bufalin and 0.5 mM ouabain were added to specific wells. These drugs were obtained from Sigma. Bufalin was dissolved in DMSO and used in the cell treatments at maximum concentrations of 0.1%. After a 30 min incubation at 37 °C, the cells were washed three times with PBS, and 0.6 mL per well of the cell lysis solution (0.15% SDS) was added to each well. The Na+/K+-ATPase activity was expressed as the difference between Rb+ incorporation in the absence or presence of 0.5 mM ouabain.
Comet assay
The extent of DNA damage in glioma cells treated with bufalin was determined by using alkaline single-cell gel electrophoresis (Comet) assay (Roy et al. 2018). In brief, cells were treated with bufalin for indicated hours. Untreated cells were used as control. Cells were harvested by using trypsin (GIBCO, USA) and pelleted down by centrifugation. The cells were suspended in 1 × phosphate-buffered saline (PBS) and 40 μL of cell suspension was mixed with 60 μL of 0.5% low melting point agarose (LMPA). Then a thin smear of cell suspension was prepared in pre-coated LMPA. The agarose was allowed to solidify in the dark at 4 °C for 10 min, the slides were submerged in lysis solution (2.5 M sodium chloride, 100 mM disodium EDTA, 10 mM TRIS and 1 g sodium dodecyl sulfate, and 1% Triton X-100 at pH 10) and incubated for 4 h in dark at 4 °C. After lysis, cells were transferred to an electrophoresis unit containing freshly prepared electrophoresis buffer (10 N sodium hydroxide and 200 mM disodium EDTA, (pH > 13)) and left for unwinding of DNA for 30 min at 15 V, 300 mA of current. The slides were then gently lifted from the electrophoresis buffer and placed on a staining tray. The slides were carefully flooded with neutralizing TRIS buffer (0.4 M Tris, pH 7.5) for 5 min and finally the buffer was drained out. The process was then repeated for two more times followed by several washes with distilled water and was stained with propidium iodide. The images from the stained slides were captured by an Olympus BX61 fluorescence microscope (Olympus Inc., Japan) using the Cytovision7.2 software.
Western blot analysis
The cell lysate proteins were separated by electrophoresis on a 7.5–12% sodium dodecyl sulfate polyacrylamide minigel (SDS-PAGE) and then electrophoretically transferred to a PVDF membrane. The blots were probed with the appropriate specific antibodies, and the protein bands were detected by enhanced chemiluminescence. Similar experiments were performed at least three times. The total protein concentration was determined using a BCA Protein Assay Kit (Beyotime Biotechnology, China). The protein density values were determined by ImageJ (National Institutes of Health, Bethesda, MD). Percentages are expressed as percentages of the untreated groups (100%).
Reverse-transcriptase polymerase chain reaction (RT-PCR)
Total RNA was extracted from bufalin-treated U87 and U118 cells using the TRIzol reagent according to the kit protocol (TaKaRa Bio, Dalian, China). The cDNA was reverse-transcribed using the PrimeScript RT Reagent Kit (TaKaRa Bio, Dalian, China) according to the manufacturer’s instructions. The primer pairs were as follows: ATP1A1, forward: 5′-GGCCTTTAAGGTTGGACGTG-3′ and reverse: 5′-CACAGTAACATTGAGAACCCCC-3′; β-actin, forward: 5′-GGCACCCAGCACAATGAA-3′ and reverse: 5′-TAGAAGCATTTGCGGTGG-3′. The amplification products were analyzed using 1.5% agarose gel electrophoresis, stained with ethidium bromide, and photographed under ultraviolet light.
Confocal immunofluorescence
Briefly, bufalin-treated U87 cells were grown on chamber slides, fixed with 4% paraformaldehyde and permeabilized with 0.2% Triton X-100. The samples were probed with specific antibodies againstγ-H2AX (Cell Signaling Technology®, Danvers, MA) and fluorescein isothiocyanate- and rhodamine-conjugated secondary antibodies. Subsequently, the stained samples were mounted with 4′,6-diamidino-2-phenylindole (DAPI) to counterstain the cell nuclei. After five additional 5-min washes, the samples were examined under a Leica DM 14000B confocal microscope.
Molecular docking
Docking studies were performed to explore the potential binding mode between bufalin and ATP1A1. First, molecular docking of the existing ATP1A1 3D model and bufalin was performed. We downloaded the 3D structure of bufalin from PubChem (https://pubchem.ncbi.nlm.nih.gov/), and then, SystemsDock (http://systemsdock.unit.oist.jp/iddp/home/index) was used to assess the molecular docking process. The optimized structure of bufalin was docked to the active site of Na+/K+-ATPase (PDB Code: 4HQJ). The genetic algorithm was used to search for 100 binding conformations, and then, cluster analysis based on the docking position was performed. Second, molecular docking of our own constructed 3D model of ATP1A1 (human) and bufalin was also performed. Because there is currently no crystal structure for human ATP1A1, we used the I-Tasser (Roy et al. 2010) tool to perform homologous modeling of human ATP1A1 and bufalin for the first time. After the construction of a homologous model, we obtained the structure of the ATP1A1 protein (human); however, further determination of the possible location of its binding with bufalin was needed. Therefore, the PDB database was searched and compared, and we found that the ATP1A1 protein derived from Susscrofa had a crystal complex with an ouabain (a type of cardioside) molecule (PDBID: 4HQJ). Then, we overlapped the crystal structure of 4HQJ with our human ATP1A1 protein structure model at bufalin binding sites, and the comparison could thus be revealed. Then, we removed the ouabain from the crystal structure and used this position as the center of the protein binding pocket. The docking research was carried out by the Glide module of the commercial software Schrodinger. The protein was treated by “hydrogenation > dehydration > protein structure optimization” in the pretreatment process of Glide protein. The treatment of small molecules was carried out using the Epik mode of Ligprep. The lattice file was then selected from the center of the ouabain, and the 15 Åresidue around it (the size was 15 Å × 15 Å × 15 Å) was selected as the pocket. Then, we used Glide to obtain the only docking conformation with the highest precision docking (XP) and selected the first (− 5.561) conformation with the highest score for further analysis. The binding pattern analysis indicated that hydrogen bonding was formed between the target small molecule bufalin and the Arg979 amino acid of the human ATP1A1 protein.
Animal studies
Male nude mice (BALB/c nu/nu, 4 weeks old, 18–20 g) were obtained from the Experimental Animal Center of Dalian Medical University (Dalian, China). All animal maintenance and treatment protocols were in accordance with local institutional guidelines for the care and use of laboratory animals. Mice were randomly divided into three groups (n = 5). The control group was given 0.5% CMC-Na for 19 days and received the vehicle (DMSO) alone from the 8th day to the 12th day. The bufalin group was treated with 0.5% CMC-Na for 19 days and administered bufalin (Lan et al. 2018) intraperitoneally (1 mg/kg, dissolved in DMSO) from the 8th day to the 12th day. The Pif groups were administered Pif (Turcato et al. 2018) intraperitoneally (2 mg/kg, dissolved in 0.5% CMC-Na) for 19 days and treated with or without bufalin (1 mg/kg, dissolved in DMSO) from the 8th day to the 12th day. The tumors were measured using a caliper every 2 days, and the tumor volume was calculated according to the formula V = 1/2 (width2 × length). The body weights were also recorded. After treatments, all experimental mice were terminated with ether anesthesia on the 20th day, and the total weight of the tumors in each mouse was measured. For determination of the expression of ATP1A1, nuclear p53, Bax, Bcl-2, cleaved caspase-9, cleaved PARP, β-actin, and lamin B1, the tumor tissues were removed for Western blotting analysis.
All animals were given free access to sterilized food and water and were habituated for 7 days before the experiments. All procedures were performed in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals (National Institutes of Health, Bethesda, MD, USA). The protocol was approved by the Animal Care and Ethics Committee of Dalian Medical University.
Statistical analysis
All results are shown as the mean ± SEM. Data were analyzed using SPSS 19.0. A typical image from at least three similar experiments was presented. Statistical analysis was performed by an independent Student’s t test. Two-way analysis of variance (ANOVA), followed by Bonferroni’s test for multiple comparisons, was performed to analyze the data for Fig. 1a, d, f, g. One-way ANOVA, followed by Tukey’s post-test for multiple comparisons, was performed to analyze the data for Figs. 1b and 4a, c. For most of the in vitro and animal experiments, Student’s t tests were used to calculate the P value. P values less than 0.05 were considered statistically significant: *P < 0.05, **P < 0.01.
Bufalin inhibited glioblastoma cell growth and induced mitochondria-mediated apoptosis. a Cancer cells (U87, U251, LN229, A172, and U118) were cultured with the indicated concentrations of bufalin for the indicated time, and then, cell viability was determined by MTT assays (*P < 0.05, **P < 0.01 vs. control; #P < 0.05, ##P < 0.01 vs. other cell lines). b U87 and U251 cells were cultured with bufalin for 48 h. The induced colony formation was analyzed, and the colony formation numbers were calculated. The data are presented as the mean ± SD of at least three separate experiments (**P < 0.01, bufalin treatment vs. vehicle control groups). c After treatment with bufalin for 48 h, the levels of apoptosis were determined in U87 and U251 cells by FACS analysis, and the percentage of apoptotic cells was calculated. d Effect of bufalin on cellular ROS production. The results are expressed as a ratio of relative fluorescent intensity. Each bar represents the mean ± SD from three experiments (*P < 0.05 and **P < 0.01 versus DMSO control). e Western blot analysis of the expression of Bax, Bcl-2, cytosolic cytochrome c (cyto c), mitochondrial cyto c, cleaved caspase-9, and cleaved caspase-3. f U87 cells were exposed to the indicated doses of bufalin for 24 h, and the reduction of oxygen consumption rate (OCR) induced by bufalin was examined. g Changes in the mitochondrial membrane potential were determined by JC-1 staining, and a quantitative analysis of the shift of mitochondrial orange-red fluorescence to green fluorescence among the groups (red/green fluorescence ratio) was conducted. Data from three independent experiments are shown. *P < 0.05, **P < 0.01 vs. control
Results
Bufalin inhibited the proliferation of glioblastoma cell sand induced mitochondria-mediated apoptosis
First, the effects of bufalin on cell proliferation were examined in several cancer cell lines with various concentrations of bufalin (0 nmol/L, 20 nmol/L, and 50 nmol/L) for 24 h, 48 h, and 72 h. The results indicated that bufalin significantly inhibited glioblastoma cell proliferation (U87, U251, LN229, A172, and U118 cells) to different degrees (Fig. 1a). Because significant inhibitory effects of bufalin on the proliferation of U87 and U251 cells were observed and both cell lines had relatively high expression levels of ATP1A1 (data not shown), we selected these cell lines for further mechanistic investigations. Bufalin (20 and 50 nmol/L) significantly reduced the cell viability of both U87 and U251 cells in a time-dependent manner over 24 h, 48 h, and 72 h of culture (Fig. 1a). U87 cells were more sensitive to bufalin treatment than U251 cells, and U118 cells were significantly more resistant to bufalin treatment than other cells. Colony formation (Fig. 1b) and apoptosis assays (Fig. 1c) also revealed similar results in both cells after bufalin treatment for 48 h. To determine if bufalin exposure led to oxidative stress, we quantified ROS in U87cell cultures exposed for 24 h to up to 50 nmol/L bufalin. As shown in Fig. 1d, bufalin treatment induced a significant dose-dependent ROS increase compared to that of the control cells. The extent of the increase in ROS in U87 cells was also significantly greater than that in U251 cells. Because mitochondrial dysfunction is closely related to oxidative stress, we next detected mitochondrial apoptosis-related proteins in U87 cells treated with bufalin. The potential cell signaling pathways related to apoptosis, including Bax, Bcl-2, cleaved caspase-3, cleaved caspase-9, cytosolic cyto c, and mitochondrial cyto c, were also investigated in U187 cells. As shown in Fig. 1e, bufalin treatment significantly increased the expression levels of Bax, cleaved caspase-3, cleaved caspase-9, and cytosolic cyto c and decreased the expression levels of Bcl-2 and mitochondrial cyto c in U87 cells at a concentration of 50 nmol/L. These results indicated that the mitochondrial apoptosis pathway contributes to bufalin-induced glioblastoma cell death. Moreover, decreases in OCR indicators of mitochondrial function could also be induced by bufalin (Fig. 1f). JC-1, a fluorescent probe, was used to test the mitochondrial membrane potential. Significantly, increased dose-dependent cell death was inversely correlated with decreased mitochondrial membrane potential in bufalin-induced glioma cells based on the results of the JC-1 assay. In the current study, treatment with the indicated doses of bufalin for 24 h caused a significant decrease in the mitochondrial membrane potential, as detected by enhanced green intensity and reduced red intensity in JC-1 assays (Fig. 1g).
Bufalin triggered DNA DSBs to induce glioblastoma apoptosis
The fluorescence of phosphorylated H2AX (γ-H2AX)-positive cells was observed and measured under a laser scanning confocal microscope, and the fluorescence intensity significantly increased in a time-dependent manner (Fig. 2a). Western blotting analysis showed similar results (Fig. 2b). In addition, we found that the upward trend of γ-H2AX expression level was consistent with that of apoptosis rate, suggesting that the upregulation of γ-H2AX was positively related to the increased apoptosis induced by bufalin treatment (Fig. 2c). The above data indicated that bufalin induced apoptosis by triggering DNA damage (DSBs) through phosphorylation of H2AX in U87 cells. In addition, inhibition of ROS with the antioxidant N-acetyl-cysteine (NAC) attenuated bufalin-induced upregulation of γ-H2AX (Supplementary Fig. S1). Besides, to exert direct measurement of DNA damage and validate the consequence of accumulated intracellular oxidative stress in bufalin-treated glioma cells, we have also analyzed the extent of DNA damages by comet assay at different time points. The extent of DNA damage was determined from the tail length of the comet. Our results showed that the severity of glioma cell DNA damage was prominent after 24 h treatment of bufalin (Fig. 2d). Representative images of comet assay are provided in Fig. 2d. These results indicated that ROS stress could be involved in bufalin-induced DNA damage.
Detection of γ-H2AX expression in U87 cells by immunofluorescence assays and immunoblot analysis after treatment with bufalin for the indicated time. a Confocal laser scanning microscopic observation. Values represent the mean ± SD (n = 5). **P < 0.001 vs. blank control. b, c The upregulation of γ-H2AX was positively related to the increased apoptosis induced by bufalin treatment. b Western blot analysis of γ-H2AX in U87 and U251 cells. c The percentage of apoptotic cells after treatment with bufalin for the indicated hours. d Determination of DNA damage in bufalin-treated U87 cells at different time points of treatment. Comet assay was performed to visualize the extent of DNA damage in U87 cells after bufalin treatment. The DNA damage effects were determined from visual scores of comet assay as explained in the “Materials and methods” section. Fluorescent images of comet assay are obtained from 20 different fields and best obtained image has been represented (20 × magnification)
Bufalin increased nuclear translocation of p53
The DNA-binding tumor suppressor p53 serves as a cornerstone in the cellular response to stress (Van Maerken et al. 2014). This protein activates apoptosis-related genes to mediate programmed cell death, cell cycle control, and cell apoptosis mechanisms. If the DNA damage is limited, p53 activation induces a cellular program to allow DNA repair, whereas in cases of severe DNA damage, the p53 response will induce the cell to undergo apoptosis (Van Maerken et al. 2014). Thus, we next examined whether bufalin induced DNA damage in glioblastoma cells via activation of the p53 signaling pathway. After treatment with the indicated doses of bufalin, total p53 expression in U87 and U251 cells decreased significantly (Fig. 3a). Total p21 expression in U87 and U251 cells also significantly decreased (Fig. 3a). Intriguingly and more importantly, nuclear p53 was upregulated both in U87 and U251 cells, while cytosolic p53 was downregulated (Fig. 3b). Furthermore, bufalin (50 nmol/L) significantly increased nuclear p53 expression in both U87 and U251 cells in a time-dependent manner at 24 h,48 h, 72 h, 96 h, 120 h, and 144 h of culture (Fig. 3c). We also found that the upward trend of nuclear p53 expression was consistent with that of the apoptosis rate, suggesting that the upregulation of nuclear p53 was positively related to the increased apoptosis induced by bufalin treatment.
Positive relationship between nuclear p53 expression levels and apoptosis rates induced by bufalin treatment for the indicated hours. a Western blot analysis for the expression of total p53 and p21 in U87 and U251 cells after treatment with bufalin for 48 h. b Western blot analysis for the expression of nuclear and cytosolic p53. c Levels of p53, lamin B1, and β-actin in the cytoplasmic and nuclear fractions of U87 cells as assessed by Western blot analysis. The upregulation of nuclear p53 was positively related to the increased apoptosis induced by bufalin treatment for the indicated hours. Upper, Western blot analysis for the expression of nuclear p53 in U87 and U251 cells. Lower, the percentage of apoptosis after treatment with bufalin for the indicated hours
Changes in p53 expression and subcellular localization could impact bufalin sensitivity under the regulation of XPO1
As mentioned above, nuclear p53 expression could be related to bufalin-induced apoptosis in human glioblastoma cells. Thus, we further examined the precise effect of p53 on the treatment efficacy of bufalin. As shown in Fig. 4a, we found that the p53 mRNA level in U118(p53mt) cells was substantially lower than that in U87(p53wt) cells (upper panel). Bufalin treatment had no effect on p53 activation in either U87 or U118 cells according to the RT-PCR results. Significantly, bufalin decreased p53 expression in U87 cells with wild-type p53 (p53wt) but had nearly no effect in U118 cells with mutant p53 (p53mt) (lower panel). Thus, we concluded that p53 status or expression levels exert a critical effect on the bufalin-induced cytotoxic effect on human glioblastoma. Using the p53-specific inhibitor pifithrin-α (Pif), we found that bufalin-induced apoptosis in U87(p53wt) cells decreased; however, in U118(p53mt) cells, there was no change after treatment with Pif, suggesting that p53 has an important role in the apoptotic effect of bufalin (Fig. 4b). Then, we silenced p53 in U87(p53wt) cells to examine whether p53 inhibition could affect the cytotoxic effect of bufalin (Fig. 4c), and intriguingly, the results further confirmed the role of p53 in bufalin-induced glioblastoma apoptosis. As mentioned previously, XPO1 could export p53 from the nucleus to the cytoplasm. Thus, we further examined whether XPO1 affected the bufalin-induced p53 expression changes. Bufalin treatment decreased XPO1 expression significantly (Fig. 4d), and bufalin was found to promote the nuclear retention of p53 the protein, showing similar results as leptomycin B, which is a known potent and selective inhibitor of XPO1 (Fig. 4e). Furthermore, silencing XPO1 significantly increased nuclear p53 expression, and intriguingly, XPO1 overexpression attenuated the increased nuclear p53 after treatment with bufalin (Fig. 4f). These results indicated that bufalin could inhibit XPO1, and XPO1 inhibition was partly responsible for the increased nuclear p53 retention induced by bufalin. Thus, we concluded that bufalin affects p53 nuclear transport by blocking XPO1.
Bufalin inhibited nucleocytoplasmic transport of the p53 protein by blocking XPO1. a Bufalin had no effect on p53 activation in both U87 and U118 cells according to the RT-PCR results. We found that the p53 mRNA level in U118 cells was significantly lower than that in U87 cells. Significantly, bufalin decreased p53 expression in U87 cells with wild-type p53 (p53wt) but had nearly no effect in U118 cells with mutant p53(p53mt). **P < 0.01 compared with the control group; ##P < 0.01 compared with the U87 cells. b Suppressing p53 activation using the p53 inhibitor pifithrin-α (Pif) eliminated the increased apoptosis induced by bufalin treatment (50 nM) in U87(p53wt). In U118(p53mt) cells, the apoptosis-inducing effect of bufalin was weak, and Pif had nearly no impact on the apoptotic effect of bufalin. c Western blot analysis for the expression of XPO1 in U87 cells after treatment with the indicated doses of bufalin. d Bufalin promoted nuclear retention of p53, and leptomycin B, a known potent and selective inhibitor of XPO1, also restored nuclear p53 expression. e Silencing XPO1 significantly increased nuclear p53 expression (left panel). In addition, Western blot analysis confirmed that bufalin could lead to nuclear retention of p53, while overexpression of XPO1 could attenuate this effect (right panel). All values are denoted as the mean ± SD. n = 3. *P < 0.05, **P < 0.01 compared with the control group; ##P < 0.01 compared with the bufalin treatment group
The sodium pump α1 subunit mediated the XPO1-targeted anticancer effects of bufalin
Because ATP1A1 could be a potential target of bufalin, we further examined whether bufalin affects XPO1 via ATP1A1. U87cells were treated with bufalin for 48 h before cell harvest. Western blot analysis showed that bufalin treatment markedly suppressed ATP1A1 expression levels (Fig. 5a). Then, the effect of ATP1A1 knockdown on the anticancer effect of bufalin was determined in U87cells. Cells were treated with si-ATP1A1to knockdown ATP1A1 expression (Fig. 5b) and treated with bufalin at the indicated doses, and then, we measured the IC50 values of bufalin. As shown in Fig. 5c, ATP1A1 knockdown markedly changed the IC50 values of bufalin for glioblastoma cells. In addition, Na+/K+-ATPase activity in glioblastoma cells was tested based on the incorporation of Rb+ by cells in the absence and presence of 0.5 mM ouabain. Na+/K+-ATPase activity, which was expressed as the difference between Rb+ incorporation in the absence or presence of 0.5 mM ouabain, was inhibited by bufalin (50, 100, 150, 200, 300, and 400 nM) in a dose-dependent manner (Fig. 5d). Furthermore, bufalin-induced suppression of XPO1 and downstream p21 expression was impeded by ATP1A1 knockdown (Fig. 5e). These findings indicated that ATP1A1 knockdown could block bufalin-induced XPO1 inhibition and the anticancer effect of bufalin. Taken together, these results suggest that bufalin could inhibit human glioblastoma growth via binding to ATP1A1. Molecular docking was used to determine the potential binding mode between bufalin and Na+/K+-ATPase (Fig. 5f). Currently, the crystal structure of CTS-bound Na+/K+-ATPase has been resolved; however, there are many forms of Na+/K+-ATPase structures and bufalin/Na+/K+-ATPase binding structures with different binding scores. Using SystemsDock (http://systemsdock.unit.oist.jp/iddp/home/index), we selected the best binding pattern with the highest binding score (Supplementary Fig. S2). The docking analysis was performed based on the binding energy, conformation, and electrostatic interaction. From the docking conformational distribution, with low binding energy, bufalin was embedded in the active site and showed a good fit with the formation of two hydrogen bonds with amino acid residues His613 and Asn713 of the enzyme. This model elucidated the likely interaction of bufalin with ATP1A1 (Fig. 5f, upper panel). Because there is currently no crystal structure for human ATP1A1, for the first time, we used the I-Tasser (Roy et al. 2010) tool to perform homologous modeling of human ATP1A1 and bufalin. After the construction of the homologous model, molecular docking was carried out using the Glide module of the commercial software Schrodinger. The binding pattern analysis also indicated that hydrogen bonding was formed between bufalin and the Arg979 amino acid of the human ATP1A1 protein (Fig. 5f, lower panel). Furthermore, the comparison at the binding site between the existing ATP1A1 3D model and our own constructed model is shown in Supplementary Fig. S3.
ATP1A1 could mediate the XPO1-targeted anticancer effect of bufalin. a Bufalin inhibited ATP1A1 protein expression in human glioma cells. U87 cells were treated with bufalin for 48 h, and cells were harvested for ATP1A1 level measurement by Western blot analysis. b, c) Knocking down ATP1A1 markedly changed the IC50 values of bufalin in U87 cells. U87cells were treated with si-ATP1A1 to knockdown ATP1A1 expression (b). Then, the cells were treated with bufalin at the indicated doses, and an MTT assay was performed to determine cell viability (c) and determine whether ATP1A1 knockdown could hinder the anticancer effect of bufalin inhuman glioma cells. d The effect of bufalin on the activity of Na+/K+-ATPase in the U87 and U251 cell lines. Cells were treated with bufalin (50–450 nM) for 30 min. The Na+/K+-ATPase activity was expressed as the difference between the Rb+ uptake in the absence or presence of 0.5 mM ouabain. Each point represents the mean ± SD from at least three different experiments conducted in triplicate. e Bufalin-induced inhibition of XPO1 expression could be impeded by ATP1A1 knockdown. Western blotting was used to determine the expression levels of XPO1 in U87cells treated with bufalin under ATP1A1 knockdown. All values are denoted as the mean ± SD. n = 3. **P < 0.01 compared with the control cultures. f Docking conformations of bufalin and ATP1A1. (1) Interactions between bufalin and the key residues of existing 2D models and 3D models of Na+/K+-ATPase with low binding energy have been presented. H-bond interactions between bufalin and the active site residues (His613 and Asn713) of ATP1A1 formed. (2) Human ATP1A1structure modeling and docking. Human ATP1A1 was modeled, and the molecular docking of our own constructed 3D model of ATP1A1 (human) and bufalin was also performed. g–i Nuclear p53 retention caused by XPO1 inhibition of bufalin was mediated by Src. g Activation of Src upon bufalin exposure. Cells were serum starved for 36 h, exposed to bufalin (50 nmol/L) for the indicated periods of time, and analyzed by Western blotting. pSrc, the phosphorylated form of Src. h p53 reduction rescued by the Src inhibitor PP2. Cells were left untreated or treated with bufalin alone or in combination with increasing concentrations of PP2 for 24 h. Cell lysates were prepared for Western blotting. i Co-IP showed that ATP1A1 directly interacts with Src
Bufalin reduced total p53 expression levels by triggering and activating the Src signaling pathway
The Na+/K+-ATPase was shown to be preassembled with its partners in caveolae (Bai et al. 2016), and the binding of cardiac glycosides to the pump activates the signalosome to transduce the signals via multiple pathways, including Src, FAK, MAPK, PI-3K, and EGFR (Wang et al. 2009; Jansson et al. 2012; Newman et al. 2008; Xie and Cai 2003; Liang et al. 2006). We first determined whether bufalin could activate Src in U87 cells. As shown in Fig. 5g, treatment of cells with bufalin for 20 min caused Src activation, indicated by the increased phosphorylation at the activation sites (pSrc-Y418). We then determined the effect of Src on drug-induced p53 reduction because Src might be an upstream molecule activated upon bufalin binding. In U87 cells, the potent Src tyrosinekinase inhibitor PP2 (Hanke et al. 1996) blocked p53 reduction by bufalin in a dose-dependent manner (Fig. 5h). We next explored the interaction between XPO1 and Src. First, we used U87extracts to immunoprecipitate Src and detected XPO1 by immunoblot analysis. Then, we immunoprecipitated XPO1 and detected Src by immunoblotting (Fig. 5i). The results indicated that Src directly interacted with XPO1. Thus, we concluded that bufalin increased Src activation, suppressing the activation of XPO1, which is important for the translocation of nuclear p53 to the cytoplasm.
Inhibition of p53 eliminated bufalin-induced mitochondrial apoptosis in vivo
Bufalin has been shown to exert apoptosis-inducing effects in vitro. Therefore, we further examined its anticancer effect in animals, and the effects of p53 suppression on bufalin-induced tumor suppression, mitochondrial apoptosis, and p53 expression changes were detected in a bufalin-treated mouse model (Fig. 6a). As shown in Fig. 6b, bufalin treatment inhibited glioblastoma growth in vivo in all groups. Specifically, in models using U87(p53wt) cells, Pif had an obvious protective effect against bufalin-induced tumor inhibition; however, in models using U118(p53mt) cells, the effect of bufalin was much weaker, and Pif had nearly no obvious protective effect against bufalin. Measurements of tumor weights showed similar results as those of tumor volume (Fig. 6c). Then, the tumor tissues from three different groups were extracted and prepared for Western blotting analysis (Fig. 6d). The results indicated that bufalin treatment promoted nuclear p53 retention and increased the expression of apoptosis-related proteins such as Bax, cleaved caspase 9, and cleaved caspase PARP. However, Pif treatment significantly attenuated these protein expression changes. These results further confirmed that bufalin treatment promoted nuclear retention of p53 protein and induced mitochondrial apoptosis, while Pif attenuated this nuclear translocation and reversed the changes in apoptosis-related protein expression induced by bufalin treatment.
In vivo data supporting the critical role of p53 in mediating the effect of ATP1A1 on bufalin resistance. a The flow chart of experiments. b Tumor volumes were measured on the indicated days. c The rats were euthanized 24 h after completion of treatment, and tumor weights were measured. d Western blot analysis for the expression of ATP1A1, nuclear p53, Bax, Bcl-2, cleaved caspase 9, and cleaved PARP in the mouse models. The values are presented as the mean ± SD, n = 5. (**P < 0.01 compared with the control cultures; ##P < 0.01 compared with the bufalin treatment group)
In addition, the cytotoxic mechanisms of bufalin regarding its effect on the ATP1A1/p53 signaling pathway are represented graphically in Fig. 7. Bufalin was found to induce mitochondrial apoptosis, ROS induction, and DNA DSBs in glioblastoma cells in this study. ATP1A1 is a potential target of the bufalin-induced anticancer effects (Lan et al. 2018), and we found that ATP1A1/p53 signaling could mediate the bufalin sensitivity of human glioblastoma cells for the first time. Mechanically, we found that bufalin-induced ATP1A1-targeted nuclear transport of p53 protein was mediated by XPO1. Previously, the Na+/K+-ATPase was reported to be preassembled with its partners in caveolae (Bai et al. 2016), and the binding of cardiac glycosides to Na+/K+-ATPase could activate the signalosome to transduce the signals via multiple pathways, including Src, FAK, MAPK, PI-3K, Ras, caveolin-1, and EGFR (Wang et al. 2009; Jansson et al. 2012; Bai et al. 2016; Newman et al. 2008; Xie and Cai 2003; Liang et al. 2006). For the first time, we also found that bufalin activates Src in caveolae in glioma cells, followed by suppressed activation of XPO1. Furthermore, XPO1 inhibition was found to be important for the subcellular translocation of p53 mediated by bufalin in this study.
Discussion
Cardiac glycosides are a class of natural products that have been used for medical purposes for a long time. Three well-known cardiac glycosides, digoxin, ouabain, and digitoxin, are used for the treatment of congestive heart failure as they inhibit Na+/K+-ATPase to increase intracellular calcium concentrations (Ahmed et al. 2008; Gjesdal et al. 2008). In addition to treatment of heart failure patients, the drugs were also found to be beneficial to breast cancer patients (Stenkvist et al. 1979) and were associated with a reduced risk of leukemia, lymphoma, kidney, urinary tract cancer (Haux et al. 2001), and glioma (Lan et al. 2018). In recent years, research on bufalin in cancer treatment has attracted increasing attention from drug researchers worldwide, and this compound has shown sufficient promise to emerge as a potent, novel, and effective anticancer therapy for cancer patients (Wang et al. 2018).
In the present study, we found that bufalin effectively inhibited glioblastoma cell growth and enhanced apoptosis induction in a dose-dependent manner. Furthermore, we showed that the effects of bufalin on cell apoptosis were mediated by blocking the ATP1A1 signaling pathway and activating the cyto c/caspase-dependent apoptotic pathway. In this process, downstream p53 status and subcellular localization could obviously affect bufalin-induced apoptosis. In addition, XPO1 had a critical effect on the bufalin-induced inhibition of p53 nucleocytoplasmic transport. Furthermore, targeted inhibition of XPO1 by bufalin was mediated by Src, and these results showed that activation of the Src signaling pathway triggered by the binding of bufalin to the sodium pump was responsible for p53 reduction.
Intriguingly, for the first time, our studies indicated that p53 status and expression level might in part be responsible for bufalin sensitivity in human glioma. In U87 cells with the wild-type p53 gene, restoring nuclear p53 function through inhibition of XPO1 induced by bufalin triggered cell apoptosis, and treatment with the p53-specific inhibitor Pif obviously eliminated bufalin-induced apoptosis; however, in U118 cells with the mutant p53 gene, bufalin-induced apoptosis was limited, and p53 suppression could not attenuate bufalin-induced apoptosis-related signaling activation. These results indicated that p53 status/expression might in part determine drug sensitivity for glioma treatment with bufalin or other chemotherapeutic drugs with similar targets.
For the first time, we reported the precise molecular mechanisms regarding XPO1-targeted regulation of p53 in human glioblastoma. The role of XPO1 has only been previously reported by Liu et al. (2016), who identified XPO1 as an attractive molecular target for the treatment of human glioma and indicated that XPO1 inhibition might represent a novel treatment approach. Here, we report that XPO1-mediated regulation of p53 translocation is responsible for bufalin-induced glioblastoma cell apoptosis. Accumulating lines of evidence have suggested that the misregulation of nuclear protein export dynamics is involved in cancer cell survival, tumor progression, and drug resistance (Mor et al. 2014; Liu et al. 2015). These observations, together with previous findings, have stimulated considerable interest in drugs targeting the nuclear export of proteins.
In addition, the current study found that the U87 cell line was more sensitive to bufalin than the U251 cell line, and more intriguingly, U118 cells were significantly far more resistant to bufalin treatment than other cells; thus, the anticancer effects of bufalin could be cell type dependent, according to current research and our previous studies. The p53 gene status of various glioma cell lines has already been established (Yee et al. 2001). Specifically, U251 and U118 were reported to have p53 mutations, while U87 had wild-type p53. Previously, we reported that ATP1A1 could partly determine the sensitivity of bufalin treatment (Lan et al. 2018), while its downstream factor, p53, was also found to be an important determinant for bufalin treatment efficacy in the current study. The clear correlation between p53 status or expression levels and resistance to bufalin suggests that p53 detection in tumor biopsy specimens might allow the development of a specific biomarker that predicts the response to bufalin or various other similar candidate drugs. Our results suggested that when clinical treatment efficacy of bufalin is tested in glioma patients, this opportunity should be seized to evaluate p53 levels as a resistance marker. Various observations have indicated that p53 status can be an important factor in radiation-induced apoptosis in response to radiotherapy (Yee et al. 2001), while our data argue that p53 expression should be measured in pretreatment biopsies of these different cancer types before bufalin treatment. We predict that the approach presented in our study may continue to yield clinically relevant resistance mechanisms that identify promising candidates for other much-needed predictive biomarkers to properly stratify individual patients before therapy.
In conclusion, our findings revealed the novel signaling pathway of bufalin in cancer cells. The results partially explain the different sensitivities of cancer cells toward bufalin treatment; that is, different cancer cells exhibit different expression levels or significant variations of p53 proteins. Thus, p53 status and expression level might be the key factors responsible for the sensitivities of different cancer cells to bufalin. ATP1A1 is a potential target for the bufalin-induced antiglioma effect, and ATP1A1 knockdown could block the anticancer effect of bufalin. Mechanically, bufalin-induced ATP1A1-targeted nuclear transport of p53 protein was found to be mediated by XPO1. These findings provide new insights into the molecular mechanisms of bufalin against cancer cells and provide some vital guidelines for the rational use of bufalin to treat cancer patients based on ATP1A1 and p53 expression patterns in cancer cells.
References
Ahmed A, Pitt B, Rahimtoola SH, Waagstein F, White M, Love TE, et al. Effects of digoxin at low serum concentrations on mortality and hospitalization in heart failure: a propensity-matched study of the DIG trial. Int J Cardiol. 2008;123(2):138–46.
Bai Y, Wu J, Li D, Morgan EE, Liu J, Zhao X, et al. Differential roles of caveolin-1 in ouabain-induced Na+/K+-ATPase cardiac signaling and contractility. Physiol Genomics. 2016;48(10):739–48.
Clarke PR, Zhang C. Spatial and temporal coordination of mitosis by Ran GTPase. Nat Rev Mol Cell Biol. 2008;9(6):464–77.
Eskiocak U, Ramesh V, Gill JG, Zhao Z, Yuan SW, Wang M, et al. Synergistic effects of ion transporter and MAP kinase pathway inhibitors in melanoma. Nat Commun. 2016;7:12336.
Fujii E, Inada Y, Kakoki M, Nishimura N, Endo S, Fujiwara S, et al. Bufalin induces DNA damage response under hypoxic condition in myeloma cells. Oncol Lett. 2018;15(5):6443–9.
Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M, Yanagida M, et al. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature. 1997;390(6657):308–11.
Garcia DG, de Castro-Faria-Neto HC, da Silva CI, de Souza e Souza KF, Gonçalves-de-Albuquerque CF, Silva AR, et al. Na/K-ATPase as a target for anticancer drugs: studies with perillyl alcohol. Mol Cancer. 2015;14:105.
Gill S, Gill R, Wicks D, Despotovski S, Liang D. Development of an HTS assay for Na+, K+-ATPase using nonradioactive rubidium ion uptake. Assay Drug Dev Technol. 2004;2(5):535–42.
Gjesdal K, Feyzi J, Olsson SB. Digitalis: a dangerous drug in atrial fibrillation? An analysis of the SPORTIF III and V data. Heart. 2008;94(2):191–6.
Grasso CS, Wu YM, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, et al. The mutational landscape of lethal castration resistant prostate cancer. Nature. 2012;487(7406):239–43.
Hanke JH, Gardner JP, Dow RL, Changelian PS, Brissette WH, Weringer EJ, et al. Discovery of a novel, potent, and Src family-selective tyrosine kinase inhibitor. Study of Lck- and FynT-dependent T cell activation. J Biol Chem. 1996;271(2):695–701.
Haux J, Klepp O, Spigset O, Tretli S. Digitoxin medication and cancer; case control and internal dose-response studies. BMC Cancer. 2001;1:11.
Houtgraff JH, Versmissen J, van der Giessen WJ. A concise review of DNA damage check points and repair in mammalian cells. Cardiovasc Revasc Med. 2006;7(3):165–72.
Huang WY, Yue L, Qiu WS, Wang LW, Zhou XH, Sun YJ. Prognostic value of CRM1 in pancreas cancer. Clin Invest Med. 2009;32(6):E315.
Jansson K, Nguyen AN, Magenheimer BS, Reif GA, Aramadhaka LR, Bello-Reuss E, et al. Endogenous concentrations of ouabain act as a cofactor to stimulate fluid secretion and cyst growth of in vitro ADPKD models via cAMP and EGFR-Src-MEK pathways. Am J Physiol Renal Physiol. 2012;303(7):F982–90.
Kau TR, Way JC, Silver PA. Nuclear transport and cancer: from mechanism to intervention. Nat Rev Cancer. 2004;4(2):106–17.
Kurz EU, Lees-Miller SP. DNA damage-induced activation of ATM and ATM-dependent signaling pathways. DNA Repair (Amst). 2004;3(8–9):889–900.
Lan YL, Wang X, Lou JC, Xing JS, Yu ZL, Wang H, et al. Bufalin inhibits glioblastoma growth by promoting proteasomal degradation of the Na+/K+-ATPase α1 subunit. Biomed Pharmacother. 2018;103:204–15.
Li Z, Zhang Z, Xie JX, Li X, Tian J, Cai T, et al. Na/K-ATPase mimetic pNaKtide peptide inhibits the growth of human cancer cell. J Biol Chem. 2011;286(37):32394–403.
Liang M, Cai T, Tian J, Qu W, Xie ZJ. Functional characterization of Src-interacting Na/K-ATPase using RNA interference assay. J Biol Chem. 2006;281(28):19709–19.
Liu X, Chong Y, Liu H, Han Y, Niu M. Novel reversible selective inhibitor of CRM1 for targeted therapy in ovarian cancer. J Ovarian Res. 2015;8:35.
Liu X, Chong Y, Tu Y, Liu N, Yue C, Qi Z, et al. CRM1/XPO1 is associated with clinical outcome in glioma and represents a therapeutic target by perturbing multiple core pathways. J Hematol Oncol. 2016;9(1):108.
Mellman I, Nelson WJ. Coordinated protein sorting, targeting and distribution in polarized cells. Nat Rev Mol Cell Biol. 2008;9(11):833–45.
Mijatovic T, Roland I, Van Quaquebeke E, Nilsson B, Mathieu A, Van Vynckt F, et al. The alpha1 subunit of the sodium pump could represent a novel target to combat nonsmall cell lung cancers. J Pathol. 2007a;212(2):170–9.
Mijatovic T, Van Quaquebeke E, Delest B, Debeir O, Darro F, Kiss R. Cardiotonic steroids on the road to anti-cancer therapy. Biochim Biophys Acta. 2007b;1776(1):32–57.
Mor A, White MA, Fontoura BM. Nuclear trafficking in health and disease. Curr Opin Cell Biol. 2014;28:28–35.
Moreno Y, Banuls L, Katz A, Miklos W, Cimmino A, Tal DM, et al. Hellebrin and its aglycone form hellebrigenin display similar in vitro growth inhibitory effects in cancer cells and binding profiles to the alpha subunits of the Na+/K+-ATPase. Mol Cancer. 2013;12:33.
Newman RA, Yang P, Pawlus AD, Block KI. Cardiac glycosides as novel cancer therapeutic agents. Mol Interv. 2008;8(1):36–49.
Nigg EA. Nucleocytoplasmic transport: signals, mechanisms and regulation. Nature. 1997;386(6627):779–87.
Noske A, Weichert W, Niesporek S, Röske A, Buckendahl AC, Koch I, et al. Expression of the nuclear export protein chromosomal region maintenance/exportin 1/Xpo1 is a prognostic factor inhuman ovarian cancer. Cancer. 2008;112(8):1733–43.
Ogawa H, Shinoda T, Cornelius F, Toyoshima C. Crystal structure of the sodium-potassium pump (Na+, K+-ATPase) with bound potassium and ouabain. Proc Natl Acad Sci U S A. 2009;106(33):13742–7.
Pan L, Zhang Y, Zhao W, Zhou X, Wang C, Deng F. The cardiac glycoside oleandrin induces apoptosis in human colon cancer cells via the mitochondrial pathway. Cancer Chemother Pharmacol. 2017;80(1):91–100.
Phesse TJ, Myant KB, Cole AM, Ridgway RA, Pearson H, Muncan V, et al. Endogenous c-Myc is essential for p53-induced apoptosis in response to DNA damage in vivo. Cell Death Differ. 2014;21(6):956–66.
Reinhard L, Tidow H, Clausen MJ, Nissen P. Na(+), K (+)-ATPase as a docking station: protein-protein complexes of the Na(+), K (+)-ATPase. Cell Mol Life Sci. 2013;70:205–22.
Rogakou EP, Nieves-Neira W, Boon C, Pommier Y, Bonner WM. Initiation of DNA fragmentation during apoptosis induces phosphorylation of H2AX histone at serine 139. J Biol Chem. 2000;275(13):9390–5.
Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5(4):725–38.
Roy S, Kar M, Roy S, Saha A, Padhi S, Banerjee B. Role of β-catenin in cisplatin resistance, relapse and prognosis of head and neck squamous cell carcinoma. Cell Oncol. 2018;41(2):185–200.
Shen A, Wang Y, Zhao Y, Zou L, Sun L, Cheng C. Expression of XPO 1 in human gliomas and its significance in p27 expression and clinical prognosis. Neurosurgery. 2009;65(1):153–9 discussion 159-60.
Shen S, Zhang Y, Wang Z, Liu R, Gong X. Bufalin induces the interplay between apoptosis and autophagy in glioma cells through endoplasmic reticulum stress. Int J Biol Sci. 2014;10(2):212–24.
Stenkvist B, Bengtsson E, Eriksson O, Holmquist J, Nordin B, Westman-Naeser S. Cardiac glycosides and breast cancer. Lancet. 1979;1(8115):563.
Takai N, Kira N, Ishii T, Yoshida T, Nishida M, Nishida Y, et al. Bufalin, a traditional oriental medicine, induces apoptosis in human cancer cells. Asian Pac J Cancer Prev. 2012;13(1):399–402.
Turcato F, Kim P, Barnett A, Jin Y, Scerba M, Casey A, et al. Sequential combined treatment of pifithrin-α and posiphen enhances neurogenesis and functional recovery after stroke. Cell Transplant. 2018;27(4):607–21.
Turner JG, Sullivan DM. CRM1-mediated nuclear export of proteins and drug resistance in cancer. Curr Med Chem. 2008;15(26):2648–55.
van der Watt PJ, Maske CP, Hendricks DT, Parker MI, Denny L, Govender D, et al. The karyopherin proteins, XPO 1 and karyopherin beta1, are overexpressed in cervical cancer and are critical for cancer cell survival and proliferation. Int J Cancer. 2009;124(8):1829–40.
Van Maerken T, Rihani A, Van Goethem A, De Paepe A, Speleman F, Vandesompele J. Pharmacologic activation of wild-type p53 by nutlin therapy in childhood cancer. Cancer Lett. 2014;344(2):157–65.
Wang Z, Zheng M, Li Z, Li R, Jia L, Xiong X, et al. Cardiac glycosides inhibit p53 synthesis by a mechanism relieved by Src or MAPK inhibition. Cancer Res. 2009;69(16):6556–64.
Wang J, Xia Y, Zuo Q, Chen T. Molecular mechanisms underlying the antimetastatic activity of bufalin. Mol Clin Oncol. 2018;8(5):631–6.
Wood LD, Parsons DW, Jones S, Lin J, Sjöblom T, Leary RJ, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318(5853):1108–13.
Wu SH, Wu TY, Hsiao YT, Lin JH, Hsu SC, Hsia TC, et al. Bufalin induces cell death in human lung cancer cells through disruption of DNA damage response pathways. Am J Chin Med. 2014;42(3):729–42.
Xiao Y, Meng C, Lin J, Huang C, Zhang X, Long Y, et al. Ouabain targets the Na+/K+-ATPase α3 isoform to inhibit cancer cell proliferation and induce apoptosis. Oncol Lett. 2017;14(6):6678–84.
Xie Z, Cai T. Na+-K+-ATPase-mediated signal transduction: from protein interaction to cellular function. Mol Interv. 2003;3(3):157–68.
Yatime L, Laursen M, Morth JP, Esmann M, Nissen P, Fedosova NU. Structural insights into the high affinity binding of cardiotonic steroids to the Na+, K+-ATPase. J Struct Biol. 2011;174(2):296–306.
Yee D, Hao C, Cheung HC, Chen HT, Dabbagh L, Hanson J, et al. Effect of radiation on cytokine and cytokine receptor messenger-RNA profiles in p53 wild and mutated human glioblastoma cell lines. Clin Invest Med. 2001;24(2):76–82.
Yu CH, Kan SF, Pu HF, JeaChien E, Wang PS. Apoptotic signaling in bufalin- and cinobufagin-treated androgen-dependent and -independent human prostate cancer cells. Cancer Sci. 2008;99(12):2467–76.
Yuan J, Adamski R, Chen J. Focus on histone variant H2AX: to be or not to be. FEBS Lett. 2010;584(17):3717–24.
Zhang K, Wang M, Tamayo AT, Shacham S, Kauffman M, Lee J, et al. Novel selective inhibitors of nuclear export CRM1 antagonists for therapy in mantle cell lymphoma. Exp Hematol. 2013;41(1):67–78.e4.
Funding
This work is supported by grants from the National Natural Science Foundation of China (Nos. 81372714, 81672480), Liaoning Provincial Natural Science Foundation of China (No. 201602244), Distinguished Professor Project of Liaoning Province, Special Grant for Translational Medicine, Dalian Medical University (No. 2015002), and Basic Research Projects in Colleges and Universities of Liaoning Province (No. LQ2017033).
Author information
Authors and Affiliations
Contributions
BZ and YD conceived and supervised the study and revised the manuscript; YLL, YJZ, and JCL designed and performed the experiments, analyzed the data, and drafted the manuscript; and JSX, XW, SZ, and BBM collected data and revised the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(PDF 329 kb)
Rights and permissions
About this article
Cite this article
Lan, YL., Zou, YJ., Lou, JC. et al. The sodium pump α1 subunit regulates bufalin sensitivity of human glioblastoma cells through the p53 signaling pathway. Cell Biol Toxicol 35, 521–539 (2019). https://doi.org/10.1007/s10565-019-09462-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10565-019-09462-y