Abstract
The molecular response of Pseudomonas fluorescens cells exposed to a mixture of heavy metals remains largely unknown. Here, we studied the temporal changes in the early gene expression of P. fluorescens cells exposed to three doses of a polymetallic solution over two exposure times, through the application of a customized cDNA microarray. At the lowest metal dose (MD/4), we observed a repression of the Hsp70 chaperone system, MATE and MFS transporters, TonB membrane transporter and histidine kinases, together with an overexpression of metal transport (ChaC, CopC), chemotaxis and glutamine synthetase genes. At the intermediate metal dose (MD), several amino acid transporters, a response regulator (CheY), a TonB-dependent receptor and the mutT DNA repair gene were repressed; by contrast, an overexpression of genes associated with the antioxidative stress system and the transport of chelates and sulfur was observed. Finally, at the highest metal dose (4MD), a repression of genes encoding metal ion transporters, drug resistance and alginate biosynthesis was found, together with an overexpression of genes encoding antioxidative proteins, membrane transporters, ribosomal proteins, chaperones and proteases. It was concluded that P. fluorescens cells showed, over exposure time, a highly complex molecular response when exposed to a polymetallic solution, involving mechanisms related with chemotaxis, signal transmission, membrane transport, cellular redox state, and the regulation of transcription and ribosomal activity.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Heavy metal pollution is currently one of the most serious problems for the functionality of ecosystems. Heavy metals cannot be degraded and, therefore, accumulate in the environment; on the other hand, they present a high affinity for biomolecules with concomitant adverse effects for metal-exposed organisms. Nonetheless, along evolutionary history, bacteria have acquired tolerance to heavy metal exposure through a variety of strategies: regulation of metal uptake, active efflux, intracellular sequestration, etc. (Gadd 2010).
Pseudomonas fluorescens has been proposed as a model organism for the study of bacterial adaptation to harsh environmental conditions (Lemire et al. 2010). Relevantly, many authors have found P. fluorescens as a perfect model species for the study of metal-induced bacterial responses (Sharma et al. 2006; Wasi et al. 2008, 2013; Sarma et al. 2010).
However, the mechanisms of metal toxicity and tolerance in bacteria are still not fully understood. Specifically, the molecular response of P. fluorescens cells exposed to a mixture of heavy metals remains largely unknown. Fortunately, recent advances in toxicogenomics and microarray technology are speeding up the discovery of gene regulation mechanisms in bacterial cells subjected to abiotic stresses. In particular, the development of microarray technology has facilitated the study of stress-induced metabolic responses in Pseudomonas (Reva et al. 2006; Teitzel et al. 2006; Thaden et al. 2010; Lee et al. 2014).
There are not many studies on the impact of mixtures of heavy metals on exposed organisms. Actually, most metal toxicology studies to date have dealt with the effects of one single heavy metal or, alternatively, a mixture of a few heavy metals. But polluted sites are frequently characterized by the simultaneous presence of many heavy metals, thus increasing the complexity of toxic responses in exposed organisms. Furthermore, the majority of studies have focused on “dose-effect” responses rather than on the effect of “exposure time”. Accordingly, we published a study on the temporal changes in the early gene expression profiles of Escherichia coli cells subjected to three polymetallic treatments over different exposure times (Gómez-Sagasti et al. 2014). The aim of the current study was to complement such work by means of investigating the temporal changes in the early gene expression of another Gram-negative bacterium, i.e. P. fluorescens, exposed to three doses of a polymetallic solution over two exposure times (5 and 15 min), through the application of a customized complementary DNA (cDNA) microarray. To our knowledge, this is the first study on early transcriptional responses of P. fluorescens cells simultaneously exposed to a mixture of many heavy metals using microarray technology.
Materials and methods
P. fluorescens growth conditions
P. fluorescens ATCC 13525 strain, kindly provided by Dr. Iñigo Azúa (University of the Basque Country), was maintained in Luria broth (LB) liquid medium at 30 °C. All inoculations were made at 1 % volume from an overnight LB culture. Cycloheximide (100 mg L−1) was added to the medium to prevent fungal growth.
Effect of heavy metal dose on P. fluorescens growth
The effect of heavy metal dose on the growth of P. fluorescens cells subjected to the polymetallic treatments (see below) was quantified following Moore et al. (2005) as described in Gómez-Sagasti et al. (2014).
P. fluorescens cells were exposed to three doses of a polymetallic solution containing Ag(I), Pb(II), Cd(II), Cu(II), Ni(II) and Zn(II), as nitrate salts. Specifically, P. fluorescens cells were exposed to the following metal dose (MD): 10 μM Ag(I), 10 μM Pb(II), 10 μM Cd(II), 10 μM Cu(II), 500 μM Ni(II) and 300 μM Zn(II), following Moore et al. (2005) (although As was substituted by Pb since, in our region, Pb is a much more common soil pollutant than As). The effect of a fourfold higher (4MD) and a fourfold lower (MD/4) heavy metal dose on P. fluorescens cell growth was also quantified. Control cells were grown in the absence of heavy metals. Heavy metal-induced inhibition of cell growth (%) was determined according to Gómez-Sagasti et al. (2014).
cDNA microarray studies
Under sterile conditions, P. fluorescens cells were grown at 30 °C in 250-mL Erlenmeyer flasks containing 45 mL of LB. Flasks were shaken at 180 rpm until mid-exponential phase (OD595 = 0.45) when cell cultures (in duplicate) were supplemented with 5 mL of LB containing the required metal salt concentration, in order to obtain the metal doses described above (MD/4, MD, 4MD, control). For each treatment, a 10-mL sample was taken from the flasks after 5 and 15 min, respectively (i.e. two exposure times). Samples were stabilized by using RNAProtect Bacteria (Qiagen), and then, total RNA was isolated with the RNeasy Mini Kit (Qiagen), as described in Gómez-Sagasti et al. (2014). The quantity and quality of the extracted RNA was assessed using NanoDrop-1000 (NanoDrop Technologies) and Bioanalyzer 2100 (Agilent), respectively. RNA samples were stored at −80 °C until use.
The temporal changes in the early gene expression of P. fluorescens exposed to the polymetallic treatments were investigated through the application of our own customized cDNA 8 × 15 K microarray (Design ID: 036764 Agilent, https://earray.chem.agilent.com/earray/). To this purpose, all known coding sequences from the transcriptome of P. fluorescens Pf0-1 found in the JCVI-CMR (Taxon ID 205922), DDBJ-GTPS (Pflu_PFO1: GIB00282CH01 CP000094) and JGI-IMG (Taxon ID 637000221) databases were used. For each target sequence, a probe was designed using Agilent’s eArray. Probe sequences (60-mer) were selected according to eArray’s Base Composition Methodology and synthesized on a microarray 8 × 15 K platform using Sure-Print Technology (Agilent).
The impact of multiple-metal exposure on P. fluorescens gene expression was investigated by hybridization of fluorescently labelled (with Cy3) cDNA samples to our customized microarray. cDNA synthesis, labelling and hybridization were performed following Agilent’s One-Color Microarray-Based Prokaryote Analysis—Fairplay III Microarray Labeling v. 1.3 protocol, as indicated in Gómez-Sagasti et al. (2014).
After hybridization, data were extracted using the Agilent Feature Extraction Software v. 10.7.3.1 (Agilent) following the GE1-107-Sep09 protocol, and then processed by GeneSpring GX 11.5.1 software (Agilent). Data were normalized using the quantile method (Bolstad et al. 2003) and centred by median. In order to simplify data handling, the average value of the signal intensity for each probe was transformed to log2. Using log2 signal values, the absolute fold change for each probe was calculated according to the following criteria (Leonhardt et al. 2004): if log2 signal value > 0, \( \mathrm{fold}\ \mathrm{change} = {2}^{\left({ \log}_2\ \mathrm{signal}\ \mathrm{value}\right)} \); if log2 signal value < 0, \( \mathrm{fold}\ \mathrm{change} = \left(-1\right)\times {2}^{-\left({ \log}_2\ \mathrm{signal}\ \mathrm{value}\right)} \). From here onwards, the term “gene” will be used to refer to “probes”. Microarray data were deposited in the EMBL-EBI ArrayExpress, accession E-MTAB-3094.
Statistical analysis of genes differentially expressed over exposure time
Within each heavy metal dose, statistically significant differences between exposure times (i.e. 5 vs. 15 min) were analysed using the Bayesian estimation of temporal regulation (BETR) (p < 0.01) (Aryee et al. 2009), available in MultiExperiment Viewer (MeV) Open Source Software v. 4.7.1. Gene expression patterns were visualized by the K-means clustering method (Soukas et al. 2000) integrated in the MultiExperiment Viewer (MeV) Open Source Software v. 4.7.1. The measure of figure of merit (Yeung et al. 2001) was used to estimate an appropriate value for K (i.e. number of clusters). The clustering procedure was based on Pearson distance and used the average linkage method. Out of all the clusters, we selected only those clusters showing a progressive increase (overexpression trend, 5 min < 15 min) or decrease (repression trend, 5 min > 15 min) over exposure time in terms of gene expression.
In order to establish that our temporal gene expression patterns were caused by heavy metal exposure, and not due to normal physiological changes associated with cell growth, gene expression data of metal-treated P. fluorescens cells within each treatment were compared to gene expression data of control cells (i.e. cells grown in the absence of heavy metals). Furthermore, in order to be on the safe side, we selected only those genes whose temporal gene expression response was opposite (overexpression vs. repression) to that found in control cells. On the other hand, for each cluster within each heavy metal treatment, only those genes showing a strong metal-induced molecular response over exposure time were considered: to this aim, we selected those genes whose response at 15 min was ≥2-fold higher (overexpression) or lower (repression) compared to that observed at 5 min.
Gene products were identified with the following databases: Pseudomonas Genome Database (Winsor et al. 2011), PseudoCyc (Romero and Karp 2003) and EcoGene 3.0 (Zhou and Rudd 2013). Within each cluster, we selected only those genes with a well-established gene product (i.e. with a known function), not considering in our analysis those genes with a hypothetical or unknown function. These genes were assigned to higher levels of COG (Clusters of Orthologous Groups) functional categories using the http://www-archbac.u-psud.fr/genomics/tree cogs.html website and the Pseudomonas Genome Database.
Genes differentially expressed over exposure time under more than one polymetallic treatment and that were not differentially expressed under control treatment were identified as potential biomarkers of the effect of exposure time on the early gene expression of P. fluorescens cells exposed to a polymetallic solution.
Validation of microarray results by RT-qPCR
Reverse transcription quantitative PCR (RT-qPCR) was used to validate our microarray results. Two genes were randomly selected from each of the abovementioned clusters (repression and overexpression) for all four treatments: then, a total of 16 genes were used for validation. For each gene, the two exposure times were analysed. RT-qPCRs were performed with the same RNA samples used for microarray analysis.
Specific primer pair sets for the 16 selected genes were designed using Primer3 (v.0.4.0) design software (Rozen and Skaletsky 2000). Criteria for primer design were established as in Gómez-Sagasti et al. (2014) (in this case, the size of the amplification product was established between 50 and 200 bp). Primer pair sequences are shown in Table 1.
RNA samples were treated with DNAse I (Invitrogen). cDNA synthesis was done using the High-Capacity cDNA Reverse Transcription Kit (Invitrogen) (Gómez-Sagasti et al. 2014).
Gene expression was measured by qPCR using the qPCR-SYBR Premix ExTaq Perfect Real Time (Takara Bio Inc.) in a 7500 Fast Real-Time PCR System (Applied Biosystems). Briefly, qPCRs were performed in a 25 μL reaction containing 1.5 μL cDNA and 1.5 μL of each primer (final primer concentration = 10 μM). Templates were pre-incubated at 50 °C for 2 min, denatured at 95 °C for 10 min and subjected to 40 cycles of the following thermal conditions: 95 °C (15 s) and 55 °C (60 s). For the melt curve, the conditions were 95 °C for 15 s, 60 °C for 1 min and 95 °C for 30 s. Product cycle threshold (Ct) was determined from ROX-normalized fluorescence emission and used to calculate the initial input of the template. RT-qPCR was performed with the two biological replicates of each treatment; in addition, three technical replicates were used for each biological replicate.
RT-qPCRs were analysed using GenEx qPCR analysis software v. 5.4.3 (MultiD Analyses AB). The stability of reference genes was determined using geNorm (Vandesompele et al. 2002) and NormFinder (Andersen et al. 2004) algorithms integrated in GenEx. Changes in quantification cycle data (ΔCq) of the 16 selected genes were normalized to the more stable reference gene, and then, Cq values were converted to fold expression change values according to the comparative method of Cq (Schmittgen and Livak 2008) with corrected efficiencies (Pfaffl 2006). All statistical analyses were done on log2-scaled data, except for the correlation analysis between microarray vs. RT-qPCR. Differences in fold expression change values over exposure time for individually analysed genes within each treatment were compared using one-way analysis of variance—ANOVA (LSD post hoc test, p < 0.05) (SPSS 18.0). Fold-change data for the correlation analysis between microarray vs. RT-qPCR were tested for normality using Shapiro-Wilk test; due to normality, Pearson’s correlation coefficient was calculated to determine the level of association between variables (SPSS 18.0).
Results and discussion
Effect of polymetallic treatments on P. fluorescens growth
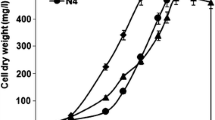
At mid-exponential phase, P. fluorescens growth was reduced by 4, 20 and 100 % at MD/4, MD and 4MD treatments, respectively, compared to control cells (Fig. 1). When E. coli cells were exposed to the same polymetallic treatments (Gómez-Sagasti et al. 2014), they showed a higher sensitivity to heavy metal exposure than our P. fluorescens strain. The genus Pseudomonas is well-known for its metabolic versatility and tolerance to the presence of organic and inorganic pollutants (Aguilar-Barajas et al. 2010).
Gene expression in the absence of metals
In the absence of metals (control treatment), 35 genes with a well-defined function were strongly repressed over exposure time (i.e. 35 genes whose response at 15 min was ≥2-fold lower compared to that observed at 5 min) (cluster I, Table 2). Similarly, 28 genes with a well-defined function were strongly overexpressed over exposure time (i.e. 28 genes whose response at 15 min was ≥2-fold higher compared to that observed at 5 min) (cluster II, Table 2).
We observed a repression of genes encoding transport proteins, such as ExbB (iron transport) and GCN5 (N-acetyltransferases) (Vetting et al. 2005). On the contrary, genes encoding a glutathione S-transferase and a spermidine/putrescine ABC transporter (Igarashi and Kashiwagi 2010) were overexpressed. Genes involved in the regulation of transcription (e.g. AraC, LacI, MerR and Fis families) were also overexpressed. By contrast, other genes involved in transcription regulation (LysR family) were repressed over exposure time.
Gene expression at the lowest metal dose (MD/4 treatment)
Under MD/4 treatment, 51 and 55 genes were strongly repressed (cluster I) and overexpressed (cluster II), respectively (Table 3). Owing to the large number of genes included in Table 3 (also in Tables 4 and 5), here and elsewhere in this paper, only those genes involved in (i) heavy metal-related processes, (ii) oxidative stress responses or (iii) relevant cellular functions under stressing conditions are discussed.
Concerning stimuli signalling, we observed the repression (cluster I) of a gene encoding a TonB-dependent copper receptor (Table 3). According to Hu et al. (2005), this type of receptor can be used as sensor of external stimuli (e.g. presence of heavy metals) and, unlike here in this study, is usually overexpressed in the presence of the stimulus. Two genes encoding sensor histidine kinase proteins located in the periplasm (Mascher et al. 2006; Krell et al. 2010) were also repressed under MD/4 treatment.
We observed the repression of genes encoding ABC membrane transporters involved in multidrug resistance (MDR) pumps, such as MatE and MdtK which belong to the MATE-family transporters for multidrug and toxic compound extrusion (Omote et al. 2006), and also the repression of genes encoding transporters from the major facilitator superfamily (MFS) (Lubelski et al. 2007; Kumar et al. 2013). MDR pumps are capable of extruding heavy metals (Silver and Phung 2005; Martínez et al. 2009). Reva et al. (2006) and Pagès et al. (2007) found a metal-induced overexpression of MFS genes in Pseudomonas putida and Pseudomonas brassicacearum, respectively.
MD/4 treatment led to a repression of hsp70 (dnaK), dnaJ and grpE genes (Table 3). In E. coli cells, these three genes are implicated in heat shock responses (Dubern et al. 2005). Sharma et al. (2008) also observed the inhibitory effect of Cd2+, Hg2+ and Pb2+ on Hsp70-assisted (DnaK/DnaJ/GrpE) protein folding. In addition, the clpA gene encoding an ATP-dependent Clp protease was also repressed under MD/4 treatment. Li et al. (2012) found that clp gene rupture results in an activation of chaperone expression and, in turn, an inhibition of enzymes related to tRNA modification.
Pertaining to cluster II (Table 3), we observed the overexpression of genes involved in (i) transport (ChaC: Ca2+/H+ antiporter putatively associated with cobalt transport; CopC: copper resistance; ABC transporter), (ii) entry of drugs and toxins (outer membrane porin proteins) and (iii) chemotaxis (EnvZ). Our analysis revealed the overexpression of two genes encoding glutamine synthetase (GS), an enzyme involved in nitrogen metabolism (Forchhammer 2007), but the role of this GS overexpression in response to the presence of metals is not clear. Pagès et al. (2007) reported a GS overexpression in Cd-exposed P. brasicacearum cells. In E. coli, we also observed the overexpression of GS under MD/4 and MD treatments (Gómez-Sagasti et al. 2014).
Gene expression at the intermediate metal dose (MD treatment)
Under MD treatment, 55 and 72 genes were strongly repressed (cluster I) and overexpressed (cluster II), respectively (Table 4). Genes involved in amino acid metabolism (e.g. arginine/ornithine antiporter, glutamine amidotransferase class I, glutamate dehydrogenase) were repressed. Like in the MD/4 treatment, a gene encoding a TonB-dependent receptor and two genes encoding a PAS/PAC sensor signal transduction histidine kinase located in the periplasm were also repressed. We also observed the repression of genes encoding MscS proteins (mechanosensitive channels), CheY protein involved in signalling and reception of stimuli, DNA topoisomerase and helicase and NUDIX hydrolase (MutT: repair of oxidative damage) (Braz and Marques 2005). In a previous work (Gómez-Sagasti et al. 2014), in E. coli cells, we observed the repression of mutY gene which functions synergistically with mutT to protect the cell from deleterious effects on DNA; then, its repression might be understood as an early symptom of metal toxicity. On the contrary, the mutT gene was overexpressed in metal-exposed Caulobacter crescentus cells (Hu et al. 2005). Finally, several families of transcription regulators (HxlR, AraC, LysR,) were repressed under this intermediate metal treatment.
Regarding cluster II (overexpression), we observed the overexpression of genes encoding the Sigma-24 factor (FecI protein) and FecR protein for the uptake and transport of ferric chelates (Potvin et al. 2007; Llamas and Bitter 2010; Saha et al. 2013). Likewise, genes encoding signalling proteins such as a heavy metal sensor histidine kinase and methyl-accepting chemotaxis proteins (MCP), which are usually methylated by protein methyltransferase CheR (Bi and Lai 2014), were overexpressed. Interestingly, sulfur metabolism was stimulated by MD treatment, as reflected by the overexpression of a sulfate permease, a sulfatase, a thiol:disulfide interchange protein and oxidorreductase (DsbE/CcmG), a sulfate/thiosulfate-binding protein and three genes encoding taurine ABC transporters (taurine is a sulfur-containing amino acid). We observed a similar response in E. coli cells exposed to MD/4 and MD treatments for some genes involved in the biosynthesis of cysteine (another sulfur-rich amino acid) (Gómez-Sagasti et al. 2014). In E. coli, it has been postulated that the Dsb system (e.g. DsbE) can repair non-native disulfide bonds in the periplasm (Collet and Bardwell 2002). Teitzel et al. (2006) also observed the induction of a thiol:disulfide interchange protein and taurine ABC permeases in Cu-exposed Pseudomonas aeruginosa. These genes could be important for cell growth in the presence of an excess of heavy metals, since some cations, particularly “sulfur lovers”, can be segregated into complex compounds by thiol-containing molecules (Nies 2003).
Similarly, genes involved in oxidative stress (e.g. 1-Cys peroxiredoxin and alkyl hydroperoxide reductase subunit), alginate biosynthesis (e.g. AlgJ), cell wall peptidase (NlpC/P60 family) (Anantharamn and Aravind 2003), and active transport systems (ExbD/TolR and MotA/TolQ/ExbB for siderophore-chelated iron) (Ma et al. 2009; Schalk et al. 2011) were overexpressed. E. coli cells under MD/4 and MD treatments exhibited the overexpression of exbB and alkyl hydroperoxide reductase, respectively (Gómez-Sagasti et al. 2014), suggesting their involvement in a possible protection mechanism against metal stress.
Finally, we found an overexpression of transcription regulators (e.g. DeoR, GntR and IclR), a RpoH heat shock transcription factor (Sigma-32) (Potvin et al. 2007) and a gene encoding the LexA transcriptional repressor (a key component of the SOS response) (Butala et al. 2009).
Gene expression at the highest metal dose (4MD treatment)
Under 4MD treatment, 356 and 375 genes were strongly repressed (cluster I) and overexpressed (cluster II), respectively (Table 5). Interestingly, some genes related to heavy metal and metalloid transport were repressed: an arsenical pump, the cobalt transporter subunit CbtA, a heavy metal-(Cd/Co/Hg/Pb/Zn)-P-type ATPase and FecR and FecI (sigma-24) genes. Moreover, we observed the repression of genes involved in signal transduction: genes encoding chemotaxis transducers and histidine kinases. Unlike MD treatment, 4MD treatment resulted in the repression of sulfatases, sulfate transporters, thiosulfate-binding proteins and taurine catabolism proteins. Similarly, some transmembrane transporters were repressed: ABC transporters (14 genes), general substrate transporters (5 genes), binding-protein-dependent transporters (13 genes), MFS proteins (6 genes), genes encoding the OmpA/MotB system (3 genes), TonB-dependent outer membrane siderophore receptors and drug resistance transporters (2 genes).
Alginate biosynthesis was strongly inhibited, as reflected in the repression of several genes: algK, algJ, alg44 and alg8 belonging to the algD-alg8-alg44-algKEGXLIJFA operon (Kiliç et al. 2010). Alginates are exopolysaccharides known to chelate metals and, hence, can increase bacterial metal tolerance (Poirier et al. 2014). By contrast, Pagès et al. (2007) found an overexpression of alginate biosynthesis in Cd-exposed P. brassicacearum.
Pertaining to transcription regulators, we detected a repression of genes belonging to the LysR (11 genes), AsnC (2 genes), GntR (2 genes), TetR (2 genes), IclR, LacI, DeoR and MerR families, as well as of the CusR heavy metal response transcriptional regulator.
Concerning gene overexpression (cluster II), 4MD treatment led to a clear activation of the antioxidative system: overexpression of genes encoding glutathione S-transferase, glutathionylspermidine synthetase, spermidine/putrescine ABC transporter, thiol peroxidase (2-Cys peroxiredoxin) and two alkyl hydroperoxide reductases. Nevertheless, genes related to glutathione S-transferase and a spermidine/putrescine ABC transporter were also overexpressed in control cells (in the absence of metals), suggesting their constitutive expression under the specific conditions of this experiment. Furthermore, our data revealed an overexpression of TonB-dependent receptors as well as membrane transporters, such as MotA/TolQ/ExbB, MscS, OmpA/MotB and OmpF. Likewise, genes related to chemotaxis were overexpressed: CheZ phosphatase, CheA signal-transducing histidine kinase and the chemotaxis response regulator CheY. On the other hand, genes involved in protein folding (chaperonin 10, chaperonin 60, Hsp70, Hsp90 and PpiC-type peptidyl-prolyl cis-trans isomerase) and genes belonging to the Clp protein family (clpX, clpA, clpP, clpS, clpA-B) were also overexpressed under 4MD treatment. Some members of the Clp protein family are implicated in proteolysis regulation; however, besides being regulators of energy-dependent proteolysis, Clp proteins may also function as molecular chaperones (Li et al. 2012). Certain Clp proteins play a decisive role in determining the destiny of proteins, not only during normal growth but also under conditions of extreme stress (Jain and Bhatt 2013). In E. coli, we found an overexpression of genes encoding heat shock proteins (e.g. clpP, hslV-clpQ, dnaK, ibpA and ibpB) when cells were exposed to the intermediate metal dose (MD) (Gómez-Sagasti et al. 2014). This similar gene expression pattern in both species (P. fluorescens and E. coli) suggests the importance of chaperone systems in response to the presence of metals. Similarly, succinate dehydrogenase A/B/C/D genes were overexpressed under 4MD treatment. Miller et al. (2009) interpreted the overexpression of succinate dehydrogenase in metal-exposed P. putida cells as a response to metal-induced oxidative stress.
In the same way, 18 LSU ribosomal protein genes, 12 SSU ribosomal protein genes and 3 Rpl/Rpm/Rps ribosomal protein genes were overexpressed. Finally, transcription regulators were either overexpressed (i.e. AraC, MarR and LuxR) or repressed (i.e., GntR, LysR, LacI and TetR), suggesting that, under 4MD treatment, P. fluorescens cells developed a complex transcriptional modulation over exposure time.
Genes differentially expressed under more than one polymetallic treatment
Genes differentially expressed under more than one polymetallic treatment (and not differentially expressed under control treatment) were identified, in an attempt to look for potential biomarkers of the effect of exposure time on the early gene expression of Gram-negative bacterial cells exposed to a polymetallic solution. Only one probe was differentially expressed under all three polymetallic treatments (MD/4, MD, 4MD): a gene encoding a GCN5-related N-acetyltransferase (Table 6). On the other hand, six genes were differentially expressed under both MD/4 and MD treatments: arginine/ornithine antiporter, IndB protein, H+ transporter ATPase, malate synthase G, P-loop ATPase and YceI protein. In turn, 24 genes were differentially expressed under both MD and 4MD treatments (Table 6), such as, for instance, genes encoding AlgJ protein, alkyl hydroperoxide reductase, arginine/ornithine antiporter, succinate semialdehyde dehydrogenase, sulfatase, taurine transporter, thiosulfate-binding protein, TonB-dependent receptor, transcription regulators (AraC, DeoR), etc. Unexpectedly, 17 genes were differentially expressed under both MD/4 and 4MD treatments; MD/4 and MD treatments shared only 6 differentially expressed genes, despite being closer in terms of heavy metal concentration (Fig. 2).
Venn diagram showing the overlap of genes differentially expressed over exposure time under the different polymetallic treatments (MD/4, MD, 4MD). MD (metal dose) = 10 μM Ag(I), 10 μM Pb(II), 10 μM Cd(II), 10 μM Cu(II), 500 μM Ni(II) and 300 μM Zn(II). MD/4 fourfold lower metal dose, 4MD fourfold higher metal dose. Venn diagrams use overlapping circles to visually represent the commonalities amongst sets of information. The number of differentially expressed genes shared by different polymetallic treatments is displayed in the overlapping circles
Comparison of gene expression patterns between P. fluorescens and E. coli
Despite being subjected to the same experimental conditions, transcriptional patterns in P. fluorescens were different to those observed in E. coli in a previous study (Gómez-Sagasti et al. 2014). In fact, over exposure time, P. fluorescens regulated a greater number of genes than E. coli at each metal dose (Fig. 3). At MD/4, both P. fluorescens and E. coli overexpressed metal-specific transporters, in order to cope with metal stress (Fig. 3). On the other hand, relevant cellular response mechanisms observed over exposure time in P. fluorescens and E. coli cells appear to occur in different cellular locations: the expression of genes involved in chemotaxis and membrane permeability (i.e. stimuli sensors and efflux pumps) was fine-tuned in P. fluorescens (i.e. some genes were upregulated while others were downregulated) whereas in E. coli, metal exposure led to the regulation of genes related to reactive oxygen species (ROS) and sulfur homeostasis in the cell interior (i.e. superoxide dismutase SodA, Fe-S cluster assembly scaffold and synthesis of cysteine).
Comparison of P. fluorescens and E. coli gene expression patterns under MD/4, MD and 4MD treatments. − number of repressed genes, + number of overexpressed genes. ROS reactive oxygen species, SOD superoxide dismutase, GSH glutathione, Glu glutamine, Cys cysteine, Tau taurine, TFs transcription factors, ↑ overexpression, ↓ repression
Under MD treatment, heavy metals increased the generation of ROS, overwhelming the antioxidant defences of P. fluorescens and E. coli cells and then resulting in oxidative stress. As a consequence, genes encoding ROS scavengers were overexpressed in both species (Fig. 3). On the other hand, P. fluorescens cells showed an overexpression of genes related to sulfur transport and iron acquisition over exposure time. This might be understood as a tolerance mechanism probably designed to promote the chelation of free metals and mitigate metal-induced iron deficiencies (Teitzel et al. 2006). Nonetheless, we observed toxicity symptoms in P. fluorescens cells due to the disruption of chemotaxis sensors, amino acid metabolism and DNA repairment. Conversely, E. coli cells overexpressed genes encoding heat shock proteins, in order to protect and recycle peptides/macromolecules in the face of metal stress. At this MD dose, in the Gram-positive bacterium Bacillus subtilis, CDF (Cation Diffusion Facilitators) family members were responsible for cell protection against sudden exposure to Zn(II), Cu, Co(II) and Ni(II) (Moore et al. 2005). In addition, pathways related to cysteine were strongly induced by metal stress in B. subtilis (Moore et al. 2005).
Finally, 4MD treatment appeared to cause a somewhat greater disturbance to E. coli cells over exposure time, as compared to P. fluorescens which activated important protection mechanisms in response to metal exposure: thus, P. fluorescens cells maintained the transcription of ROS scavengers and overexpressed chaperone-coding genes, as well as genes related with ribosome formation (Fig. 3); instead, E. coli cells did not show an early and coordinated transcriptional response in response to metal exposure. In fact, according to the growth curves obtained in the presence of metals, E. coli growth was more sensitive to the metal treatments used here than P. fluorescens.
The abovementioned transcriptional differences between both Gram-negative bacteria might be, at least in part, due to their adaptation to different environmental conditions: P. fluorescens is a common inhabitant of the soil and rhizosphere environment (Varivarn et al. 2013) and harbours a large panel of metal resistance and tolerance mechanisms (Aguilar-Barajas et al. 2010); on the contrary, E. coli is most commonly found as a commensal of the intestinal tract of warm-blooded animals (Tenaillon et al. 2010).
Validation of microarray results by RT-qPCR
Amplification efficiencies of primer pairs were between 93 and 105 % (that is to say, acceptable for quantification by RT-qPCR). Primer specificity was confirmed by melting curves analysis, which showed the amplification of a single PCR product. For most of the selected genes, Ct values were close to 30. If we consider Ct values around 35 as negative, our selected genes could be classified as low-expression genes.
Regarding the reference genes (fabD, rsd/algQ, pyr, 16S), the geNorm analysis indicated that rsd/algQ (regulator of sigma factor RpoD) and 16S (16S rRNA) genes showed a high expression stability, as reflected by the stability measure M (Vandesompele et al. 2002; Gómez-Sagasti et al. 2014). Similarly, NormFinder analysis showed rsd/algQ gene (SD = 0.0251) as an optimal reference gene for the quantification of transcriptional responses by RT-qPCR. Moreover, unlike 16S gene, Ct values for rsd/algQ were within the range of the values observed for the selected genes. In consequence, rsd/algQ was used here as a stable and accurate reference gene.
Out of the 16 genes selected for validation, 4 genes (lysR, chem, ATPase, PFKB) were not detected by the designed primers (Table 1). Out of the remaining 12 genes, 9 genes showed the same response trend (overexpression or repression) in microarray vs. RT-qPCR analysis (Fig. 4). But, according to the RT-qPCR analysis, only 2 genes (hsp70 and Zn-Alcohol) were differentially expressed over exposure time (5 vs. 15 min; ANOVA p < 0.05) (Fig. 4).
Validation of microarray results by RT-qPCR. Rectangles represent expression fold changes of selected genes (DEAD, Zn-Alcohol and mo-FAD for control treatment; chaC, GS, F3 and hps70 for MD/4 treatment; GNC5, tonB and cytC for MD treatment; met and oxo for 4MD treatment) as measured by microarray and RT-qPCR. Bars, standard deviations (n = 2)
Under both techniques, the obtained fold changes for each gene were moderately correlated (Pearson’s, r = 0.822) (Fig. 5). Correlation values between microarray and RT-qPCR results can vary between 0.48 and 0.94 (Morey et al. 2006). Morey et al. (2006) indicated that criteria for the determination of an acceptable validation of microarray results by RT-qPCR are seldom defined. Then, since nine of the selected genes showed the same response trend under RT-qPCR and microarray analysis, we considered our microarray results validated by RT-qPCR.
Correlation plot between fold changes of genes differentially expressed in the RT-qPCR analysis (Zn-Alcohol, mo-FAD, GS, F3, hsp70, tonB, cytC, met, oxo) and their corresponding fold-change values in the microarray analysis. r, Pearson’s r. Horizontal bars, standard deviations (n = 2) for microarray data; vertical bars, standard deviations (n = 2) for RT-qPCR data
Conclusions
We studied the temporal changes in the early gene expression of P. fluorescens exposed to three doses of a polymetallic solution over two exposure times (5 and 15 min). In the absence of metals, a lower number of genes, compared to metal treatments, were differentially expressed over exposure time: genes encoding chaperones, secretion proteins, membrane structural proteins, redox balance proteins and transcription regulators were differentially expressed. At the lowest heavy metal dose (MD/4), we observed the repression of TonB-dependent copper receptor, the Hsp70 protein folding system, MATE and MFS drug resistance transporters, and histidine kinases, together with the overexpression of metal transport (ChaC and CopC), chemotaxis and glutamine synthetase genes. At the intermediate dose (MD), several amino acid transporters, a stimuli receptor (CheY), a TonB-dependent receptor and the mutT DNA repair gene were repressed; by contrast, an overexpression of genes associated with the antioxidative stress system and the transport of chelates and sulfur was observed. Finally, at the highest dose, a repression of genes encoding metal ion transporters, drug resistance, and alginate biosynthesis was found, together with an overexpression of genes encoding antioxidative proteins, membrane transporters, ribosomal proteins, chaperones and proteases. Over exposure time, P. fluorescens cells showed a complex cellular response when exposed to a polymetallic solution, involving mechanisms related with chemotaxis, signal transmission, membrane transport, cellular redox state, and regulation of transcription and ribosomal activity. In contrast to gene expression patterns shown by E. coli cells exposed to the same polymetallic treatments under the same experimental conditions, P. fluorescens cells regulated the expression of genes associated with signalling and chemotaxis, alginate biosynthesis, transcription regulation and ribosomal subunit formation. As reflected by their growth curves, P. fluorescens appears to be more tolerant to early metal exposure than E. coli, possibly due to the regulation of membrane sensing and permeability, as well as the maintenance of antioxidant and chaperone systems at increasing metal doses. Finally, a gene encoding a GCN5-related N-acetyltransferase was differentially expressed under all three polymetallic treatments and, thus, could be a potential biomarker of the effect of exposure time on the early gene expression of Gram-negative bacteria exposed to a polymetallic solution; in any case, much further research is needed to ascertain the potential of this gene as biomarker of metal exposure.
References
Aguilar-Barajas E, Ramírez-Díaz MI, Riveros-Rosas H, Cervantes C. Heavy metal resistance in pseudomonads. In: Ramos JL, Filloux A, editors. Pseudomonas, vol. 6: molecular microbiology, infection and biodiversity. Dordrecht: Springer; 2010. p. 255–82.
Anantharamn V, Aravind L. Evolutionary history, structural features and biochemical diversity of the NlpC/P60 superfamily of enzymes. Genome Biol. 2003;4:R11.
Andersen CL, Jensen JL, Ørntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–50.
Aryee M, Gutierrez-Pabello J, Kramnik I, Maiti T, Quackenbush J. An improved empirical bayes approach to estimating differential gene expression in microarray time-course data: BETR (Bayesian Estimation of Temporal Regulation). BMC Bioinform. 2009;10:409. doi:10.1186/1471-2105-10-409.
Bi S, Lai L. Bacterial chemoreceptors and chemoeffectors. Cell Mol Life Sci. 2014;1-18. doi:10.1007/s00018-014-1770-5.
Bolstad BM, Irizarry RA, Åstrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–93.
Braz VS, Marques MV. Genes involved in cadmium resistance in Caulobacter crescentus. FEMS Microbiol Lett. 2005;251:289–95.
Butala M, Žgur-Bertok D, Busby SJW. The bacterial LexA transcriptional repressor. Cell Mol Life Sci. 2009;1:82–93.
Collet J-F, Bardwell JCA. Oxidative protein folding in bacteria. Mol Microbiol. 2002;44:1–8.
Dubern J-F, Lagendijk EL, Lugtenberg BJJ, Bloemberg GV. The heat shock genes dnaK, dnaJ and grpE are involved in regulation of putisolvin biosynthesis in Pseudomonas putida PCL1445. J Bacteriol. 2005;187:5967–76.
Forchhammer K. Glutamine signalling in bacteria. Front Biosci. 2007;12:358–70.
Gadd GM. Metals, minerals and microbes: geomicrobiology and bioremediation. Microbiology. 2010;156:609–43.
Gómez-Sagasti MT, Becerril JM, Martín I, Epelde L, Garbisu C. cDNA microarray assessment of early gene expression profiles in Escherichia coli cells exposed to a mixture of heavy metals. Cell Biol Toxicol. 2014;30:207–32.
Hu P, Brodie EL, Suzuki Y, McAdams HH, Andersen GL. Whole-genome transcriptional analysis of heavy metal stresses in Caulobacter crescentus. J Bacteriol. 2005;187:8437–49.
Igarashi K, Kashiwagi K. Characteristics of cellular polyamine transport in prokaryotes and eukaryotes. Plant Physiol Biochem. 2010;48:506–12.
Jain S, Bhatt A. Proteomic analysis of diversified extremophilic strains of Pseudomonas in the presence of cadmium. Agric Res. 2013;2:354–9.
Kiliç NK, Stensballe A, Otzen DE, Dönmez G. Proteomic changes in response to chromium (VI) toxicity in Pseudomonas aeruginosa. Bioresour Technol. 2010;7:2134–40.
Krell T, Lacal J, Busch A, Silva-Jiménez H, Guazzaroni M-E, Ramos JL. Bacterial sensor kinases: diversity in the recognition of environmental signals. Annu Rev Microbiol. 2010;64:539–59.
Kumar S, Floyd JT, He G, Varela MF. Bacterial antimicrobial efflux pumps of the MFS and MATE transporter families: a review. Recent Res Devel Antimicrob Agents Chemother. 2013;7:1–21.
Lee JH, Kim YG, Cho MH, Lee J. ZnO nanoparticles inhibit Pseudomonas aeruginosa biofilm formation and virulence factor production. Microbiol Res. 2014;169:888–96.
Lemire J, Mailloux R, Auger C, Whalen D, Appanna VD. Pseudomonas fluorescens orchestrates a fine metabolic-balancing act to counter aluminium toxicity. Environ Microbiol. 2010;12:1384–90.
Leonhardt N, Kwak JM, Robert N, Waner D, Leonhardt G, Schroeder JI. Microarray expression analyses of Arabidopsis guard cells and isolation of a recessive abscisic acid hypersensitive protein phosphatase 2C mutant. Plant Cell. 2004;16:596–615.
Li K, Pidatala RR, Ramakrishna W. Mutational, proteomic and metabolomic analysis of a plant growth promoting copper-resistant Pseudomonas spp. FEMS Microbiol Lett. 2012;335:140–8.
Llamas MA, Bitter W. Cell-surface signalling in Pseudomonas. In: Ramos J-L, Filloux A, editors. Pseudomonas. volume 6: molecular microbiology, infection and biodiversity. Netherlands: Springer; 2010. p. 59–95.
Lubelski J, Konings WN, Driessen AJM. Distribution and physiology of ABC-type transporters contributing to multidrug resistance in bacteria. Microbiol Mol Biol Rev. 2007;71:463–76.
Ma Z, Jacobsen FE, Giedroc DP. Metal transporters and metal sensors: how coordination chemistry controls bacterial metal homeostasis. Chem Rev. 2009;109:4644–81.
Martínez JL, Sánchez MB, Martínez-Solano L, Hernández A, Garmendia L, Fajardo A, et al. Functional role of bacterial multidrug efflux pumps in microbial natural ecosystems. FEMS Microbiol Rev. 2009;33:430–49.
Mascher T, Helmann JD, Under G. Stimulus perception in bacterial signal-transducting histidine kinases. Microbiol Mol Biol Rev. 2006;70:910–38.
Miller CD, Pettee B, Zhang C, Pabst M, McLean JE, Anderson AJ. Copper and cadmium: responses in Pseudomonas putida KT2440. Lett Appl Microbiol. 2009;49:775–83.
Moore CM, Gaballa A, Hui M, Ye RW, Helmann JD. Genetic and physiological responses of Bacillus subtilis to metal ion stress. Mol Microbiol. 2005;57:27–40.
Morey JS, Ryan JC, Van Dolah FM. Microarray validation: factors influencing correlation between oligonucleotide microarrays and real-time PCR. Biol Proced Online. 2006;8:175–93.
Nies DH. Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol Rev. 2003;27:313–39.
Omote H, Hiasa M, Matsumoto T, Otsuka M, Moriyama Y. The MATE proteins as fundamental transporters of metabolic and xenobiotic organic cations. Trends Pharmacol Sci. 2006;27:587–93.
Pagès D, Sánchez L, Conrad S, Gidrol X, Fekete A, Schmitt-Kopplin P, et al. Exploration of intraclonal adaptation mechanisms of Pseudomonas brassicacearum facing cadmium toxicity. Environ Microbiol. 2007;9:2820–35.
Pfaffl MW. Relative quantification. real time qPCR. New York: Taylor & Francis Group; 2006. p. 63–82.
Poirier I, Kuhn L, Caplat C, Hammann P, Bertrand M. The effect of cold stress on the proteome of the marine bacterium Pseudomonas fluorescens BA3SM1 and its ability to cope with metal excess. Aquat Toxicol. 2014;157:120–33.
Potvin E, Sanschagrin F, Levesque RC. Sigma factors in Pseudomonas aeruginosa. FEMS Microbiol Rev. 2007;32:38–55.
Reva ON, Weinel C, Weinel M, Böhm K, Stjepandic D, Hoheisel JD, et al. Functional genomics of stress response in Pseudomonas putida KT2440. J Bacteriol. 2006;188:4079–92.
Romero P, Karp P. PseudoCyc, a pathway-genome database for Pseudomonas aeruginosa. J Mol Microbiol Biotechnol. 2003;5:230–9.
Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–86.
Saha R, Saha N, Donofrio RS, Bestervelt LL. Microbial siderophores: a mini review. J Basic Microbiol. 2013;53:303–17.
Sarma B, Acharya C, Joshi SR. Pseudomonads: a versatile bacterial group exhibiting dual resistance to metals and antibiotics. Afr J Microbiol Res. 2010;4:2828–35.
Schalk IJ, Hannauer M, Braud A. New roles for bacterial siderophores in metal transport and tolerance. Environ Microbiol. 2011;13:2844–54.
Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat Protoc. 2008;3:1101–8.
Sharma S, Sundaram CS, Luthra PM, Singh Y, Sirdeshmukh R, Gade WN. Role of proteins in resistance mechanism of Pseudomonas fluorescens against heavy metal induced stress with proteomics approach. J Biotechnol. 2006;126:374–82.
Sharma SK, Goloubinoff P, Christen P. Heavy metal ions are potent inhibitors of protein folding. Biochem Biophys Res Commun. 2008;372:341–5.
Silver S, Phung LT. A bacterial view of the periodic table: genes and proteins for toxic inorganic ions. J Ind Microbiol Biot. 2005;32:587–605.
Soukas A, Cohen P, Socci ND, Friedman JM. Leptin-specific patterns of gene expression in white adipose tissue. Genes Dev. 2000;14:963–80.
Teitzel GM, Geddie A, De Long SK, Kiristis MJ, Whiteley M, Parsek MR. Survival and growth in the presence of elevated copper: transcriptional profiling of copper-stressed Pseudomonas aeruginosa†. J Bacteriol. 2006;188:7242–56.
Tenaillon O, Skurnik D, Picard B, Denamur E. The population genetics of commensal Escherichia coli. Nat Rev Microbiol. 2010;8:207–17.
Thaden JT, Lory S, Gardner TS. Quorum-sensing regulation of a copper toxicity system in Pseudomonas aeruginosa. J Bacteriol. 2010;192:2557–68.
Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paene A, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:research0034.0031-11.
Varivarn K, Champa LA, Silby MW, Robleto EA. Colonization strategies of Pseudomonas fluorescens Pf0-1: activation of soil-specific genes important for diverse and specific environments. BMC Microbiol. 2013;13:92.
Vetting MW, de Carvalho LPS, Yu M, Hegde SS, Magnet S, Roderick SL, et al. Structure and functions of the GNAT superfamily of acetyltransferases. Arch Biochem Biophys. 2005;1:212–26.
Wasi S, Jeelani G, Ahmad M. Biochemical characterization of a multiple heavy metal, pesticides and phenol resistant Pseudomonas fluorescens strain. Chemosphere. 2008;71:1348–55.
Wasi S, Tabrez S, Ahmad M. Use of Pseudomonas spp. for the bioremediation of environmental pollutants: a review. Environ Monit Assess. 2013;185:8147–55.
Winsor GL, Lam DK, Fleming L, Lo R, Whiteside MD, Yu NY, et al. Pseudomonas genome database: improved comparative analysis and population genomics capability for Pseudomonas genomes. Nucleic Acids Res. 2011;39(Database issue):D596–600.
Yeung KY, Haynor DR, Ruzzo WL. Validating clustering for gene expression data. Bioinformatics. 2001;17:309–18.
Zhou J, Rudd KE. EcoGene 3.0. Nucleic Acids Res. 2013;41:D613–24.
Acknowledgments
This work has been financially supported by 7/12/TK/2009/3 LURCHIP (Biscay County Council) and MINECO AGL2012-39715-CO3-01/02 projects. M.T. Gómez-Sagasti is the recipient of a Fellowship for Recent Doctors, University of the Basque Country. Technical support by Javier Etxebarria and Amaia García from GAIKER is gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Gómez-Sagasti, M.T., Becerril, J.M., Epelde, L. et al. Early gene expression in Pseudomonas fluorescens exposed to a polymetallic solution. Cell Biol Toxicol 31, 39–81 (2015). https://doi.org/10.1007/s10565-015-9294-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10565-015-9294-9