Abstract
Metastasis-associated gene or metastasis tumor antigen 1 (MTA1) is a new member of cancer progression-related gene family. It was first identified in rat mammary adenocarcinoma and later recognized as an important constituent of nucleosomal remodeling complex (NuRD), displaying dual regulatory functions as a co-repressor and co-activator for a large number of genes. Chromatin remodelers are ATP-dependent multi-protein chromatin modifying machines. These complexes alter the nucleosome positioning regulating the accessibility of genomic DNA to various transcription factors and thus modulate eukaryotic gene transcription. Since its identification two decades ago, MTA1 has been reported to be overexpressed in many cancers. Moreover, its overexpression has also been correlated with transformation and tumor progression. Furthermore, MTA1 has been shown to modulate the response of several tumor suppressor genes like p53 and oncogenes like c-myc. Taken together, current literature suggests that MTA proteins, especially MTA1, act as a master co-regulatory molecule involved in the carcinogenesis and progression of various malignant tumors. The primary focus of this review is to provide an overview of the MTA proteins with special emphasis on its role in cancer and use as a marker for cancer progression and potential target for therapy.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Cancer is a multistep and multifactorial disease driven by alterations at the level of genome and epigenome that lead to biological capabilities of increased proliferation, enhanced survival, and heightened angiogenesis and metastasis [1]. Recent advances have led to the identification of several driver genomic alterations. Starting from the discovery of oncogenic KRAS activation and inactivation of tumor suppressor genes APC and p53 driving colon transformation to the identification of oncogenic signatures across different human cancer types, we have improved our understanding of molecular tumor maps [2, 3].
Apart from genetic alterations, epigenetic changes have also been implicated in neoplasms. These heritable but reversible changes encompass covalent modifications of histones, methylation of CpG dinucleotides in DNA, and genomic imprinting [4]. The first epigenetic alteration in cancer was demonstrated by Vogelstein et al in colon cancer, in which a global reduction of 5-methylcytosine was seen in the cancerous tissues [5]. Subsequently, it has become clear that epigenetic abnormalities regulate gene expression by controlling accessibility to chromatin. One of the key regulators of the chromatin includes ATP-dependent chromatin-remodeling complexes. These include the switching defective/sucrose non-fermenting (SWI/SNF) family, the imitation SWI (ISWI) family, the nucleosome remodeling and deacetylation (NuRD)/Mi2/CHD (chromodomain, helicase, DNA binding) family, and the INO80 (inositol requiring 80) family of remodelers [4].
With the recent advances in technology, several cancer-related chromatin-remodeling complexes have been discovered. Among these is metastasis-associated gene, mta1, that was found to be highly expressed in metastatic rat mammary adenocarcinoma cell lines compared to non-metastatic cell lines using differential complementary DNA (cDNA) library screening technique [6]. This gene encodes for a protein that belongs to a family of ubiquitously expressed co-regulators that control the transcriptional milieu of the cell by directly binding to the transcription factors and interacting with histone tails to modulate chromatin accessibility.
A human homologue of rat mta1 was later found to be located on the chromosome 14q32.3 [7]. The human homologue, metastasis tumor antigen 1 (MTA1), has been demonstrated to be overexpressed in many cancers including those of breast, esophageal, gastric, colorectal, and pancreatic origins and the corresponding cell lines [8, 9]. The expression of this gene was found to correlate with progression and invasiveness of these cancers [9]. However, the functional role of this gene remained to be characterized.
Subsequently, human MTA1 cDNA was cloned by Toh et al. to characterize and understand the role of MTA1 in the tumorigenesis [10]. A large number of research studies have been able to demonstrate a significant contribution of MTA1 in malignant properties of various cancers. Recent studies have led to identification of other functions of MTA1, most notably in DNA repair and inflammation. The present review highlights MTA1 and its related proteins as a central nexus in cancer and examines it as a candidate therapeutic target.
2 MTA1 and its family members: structure and localization
In addition to MTA1, other related genes MTA2 and MTA3 have also been identified. The three genes together form the MTA family and encode for six reported isoforms (MTA1, MTA1s, MTA1-ZG29p, MTA2, MTA3, and MTA3L) with MTA1 being the founder member of the family. MTA1 and MTA2 are large polypeptides with the molecular masses of approximately 80 and 70 kDa, respectively. MTA2 shares 68.2 % protein alignment homology with MTA1, whereas the 65-kDa MTA3 is 73.2 % homologous with MTA1 [11].
Except for MTA1-ZG29p isoform that lacks N-terminal region, the MTA family members are highly homologous at the N-terminus and share four highly conserved domains. These consist of one bromo-adjacent homology (BAH) domain thought to be involved in protein-protein interactions; one SWI, ADA2, N-CoR, and TFIIIB-B (SANT) domain [11], a novel motif mostly found in eukaryotic transcriptional regulatory proteins similar to DNA-binding domain of c-myb [12] and functionally known to interact with histone N-terminal tails [13], and one egl-27 and MTA1 homology (ELM) domain with an undefined function and a GATA-type zinc finger motif and leucine zipper motif responsible for interactions with various transcription factors. Additionally, two proline rich src-homology (SH)-binding motifs responsible for interactions with signaling molecules have also been identified in MTA1, 2, and 3 protein sequences [6, 10, 11].
In addition to the different domains, MTA proteins contain basic amino acid rich nuclear localization signals (NLS) and therefore localize to nucleus in the normal adult tissues [6, 11]. However, cytoplasmic expression of MTA1 has also been recently shown in mouse embryonic tissues [14], human B cell lymphomas, endometrial carcinoma, and human hepatic carcinomas [15, 16]. MTA3 which lacks NLS localizes to both cytoplasm and nucleus, whereas spliced form MTA1 localizes only to cytoplasmic compartment [17, 18]. A very recent report by Liu et al. also shows localization of MTA1 to the nuclear envelope in a translocated promoter region (TPR)-dependent manner [14]. It has also been demonstrated that the nuclear expression of MTA1 in HCT116 promotes a less differentiation state and proliferation, whereas MTA1 knockdown induces expression of genes involved in development and differentiation [14].
3 Functions of MTA proteins
With the characterization of MTA1 as a metastasis related gene, understanding the biological role of MTA and its related proteins has become important. The role of MTA family of proteins is reviewed in the following sections.
3.1 Association with NuRD complex
Despite the availability of structural information about MTA proteins and their anticipated role as a transcriptional regulator, their exact biological functions remain elusive. In 1998, Zhang et al. isolated a protein complex called nucleosome remodeling and histone deacetylation (NuRD) containing MTA1-related polypeptide Mi2β, with histone deacetylation and ATP-dependent nucleosome remodeling activities [19]. In the same year, MTA1 was also reported as a subunit of ATP-dependent chromatin-remodeling complex NuRD providing a link between chromatin remodeling and histone deacetylase activities [20]. Later biochemical studies and immunoaffinity purification of multi-subunit chromatin remodeler NuRD revealed MTA2 to be a component of the complex that modulates enzymatic activity of the histone deacetylase core complex [21]. Similarly, Fujita et al. demonstrated MTA3 to be an estrogen-dependent component of Mi2/NuRD complex. This complex functions as a transcriptional repressor in breast epithelial cells inhibiting their growth and differentiation [17]. Subsequently, physical interaction between MTA1 and HDAC1 has also been reported in pancreatic cancer [22]. Notably, it is now speculated that the functions of MTA proteins are not restricted only to NuRD complexes.
3.2 Interaction with non-histone proteins
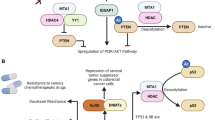
Apart from targeting chromatin histones, MTA1 is also associated with non-histone proteins such as p53 tumor suppressor gene [23]. MTA1 containing NuRD complex-mediated deacetylation of p53 attenuates its trans-activation thereby inhibiting p53-induced apoptosis in human non-small cell carcinoma and hepatoma cells [8, 24]. MTA1 expression in breast cancer has been shown to be associated with deacetylation of HIF-1α via recruitment of HDAC1 in hypoxic conditions, leading to stabilization of HIF-1α protein [25]. Hepatitis B-virus X protein (Hbx) also induced the expression of MTA1 and HDAC1 protein in hepatocellular carcinoma thereby influencing hypoxia signaling [25, 26]. MTA1 has now been shown to be a bonafide target of NF-κB maintaining the inflammatory response seen in various cancers [27].
4 MTA and cancer
MTA proteins are found in most normal tissues including brain, liver, breast, Sertoli cells, and ovary [28–30]. However, its expression increases in transformed cells. Since the identification of MTA proteins in highly metastatic breast cancer, their occurrence in several other cancers has also been reported. The spectrum of cancers with overexpressed MTA proteins in immunohistochemical evaluation has been listed in Table 1, mentioning various antibodies used for detection.
4.1 Involvement of MTA proteins in breast cancer
Role in cancer progression
MTA1 was initially discovered in rat breast cancer where its expression correlated with enhanced metastasis [6, 10]. Subsequently, the expression of MTA1 was also seen in human breast cancer cell lines and tissue specimens, correlating with tumor aggressiveness [46–48]. Consistent with these reports, inhibition of MTA1 using antisense phosphorothioate oligonucleotides in MDA-MB-231 breast cancer cell line that expresses high levels of MTA1 resulted in inhibition of its proliferative capacity [49]. Direct evidence of MTA1 contributing to malignant properties came with demonstration of anchorage-independent growth and heightened invasiveness in otherwise non-invasive MCF 7 cells after overexpression of MTA1 [29]. Further studies in animal models also demonstrated that overexpression of mammary tumor-like virus (MMTV)-MTA1 in mouse mammary gland resulted in increased ductal extension and branching, eventually developing into hyperplastic nodules and mammary tumors which then metastasized to distant organs [50].
Mazumdar et al. first identified ER-α to be a direct target of MTA1 showing its interaction with the ligand-binding domain of ER-α [29]. Stimulation with the growth factor ligand heregulin-β1 (HRG), increases the expression of MTA 1 in breast cancer cell lines. The highly expressed MTA1 is associated with HDAC2 binding to estrogen response element (ERE), repressing the trans-activation activity of ER-α. Also, on inhibiting protein synthesis using cycloheximide, high transcript levels of MTA1 was seen, suggesting the regulation of MTA1 expression by HRG [29].
Another strong evidence for the role of MTA1 in tumorigenesis came from the study of Ohshiro et al. which revealed that the induction of MTA1 in Rat1 fibroblasts was sufficient for their malignant transformation [51]. This oncogenic activity relied on the acetylation of MTA1 at Lys626 by histone acetyltransferase p300 to activate Ras-Raf pathway. This was mediated by the repression of Galphai2 regulatory element via co-repressive activity of MTA1-HDAC [51]. On stimulation with the growth factor HRG, MTA1-HDAC2 binds to estrogen response element (ERE), repressing the trans-activation activity of ER-α, thus conferring a more aggressive phenotype in ER-positive breast cancer. Similarly, MTA2 was also shown to deacetylate ER-α through its histone deacetylation activity, repressing colony formation capacity, and rendering breast tumors resistant to estradiol and antiestrogen drug tamoxifen [52]. In contrast, MTA1s, the shorter spliced cytoplasmic form of MTA1 sequesters ER-α, prevents its translocation to nucleus after ER-ligand binding and blocks malignant transformation of breast cancer cells [16].
Using a PyV-mT breast cancer model in transgenic mice, Zhang et al. monitored the expression patterns of MTA1, 2, and 3 during cancer progression. They showed that each MTA protein had a unique expression pattern which co-ordinated with others to bring about tumor progression and metastasis of the breast cancer [45].
Role in epithelial- mesenchymal transition
MTA3, reported to be a part of Mi2/NuRD complex, plays a major role in regulating epithelial to mesenchymal transition (EMT) in breast cell by controlling expression of transcriptional repressor Snail. In the absence of MTA3 or ER, aberrantly expressed Snail causes a reduction in the levels of E-cadherin resulting in invasive growth [17]. The study showed that co-repressor activity of MTA3 could not be substituted with that by MTA1. Similarly, MTA1, not MTA2, was observed to be present on the Snail promoter ascertaining its specialized function [17].
Additionally, MTA3 has also been shown to be a transcriptional target of ER-α which binds to SP1 site on the promoter region of MTA3 regulating its expression [17, 53]. In contrast to MTA1, forced expression of MTA3 in virgin and pregnant transgenic mice inhibited ductal extension and branching suggesting a protective role against transformation [54]. MTA3 also suppresses the Wnt4 pathway in an HDAC-dependent process by physically interacting with Wnt4 chromatin, inhibiting its transcription and secretion in the mammary epithelial cells [54]. In addition to co-repressor activity of MTA proteins via histone deacetylation, their role as transcriptional activators became evident from studies carried out by Gururaj et al. They demonstrated that the transcript levels of a highly amplified gene, breast cancer-amplified sequence 3 (BCAS3), was induced by MTA1 protein in breast tumors [55, 56]. Recently, it was revealed that MTA1 upregulates the expression of VEGF and its receptor Flt-1 genes and that the cross talk between VEGF and MTA1 regulates angiogenesis and metastasis associated with breast cancer [57]. Taken together, these studies suggest that the MTA proteins act as a dual regulator in breast tumors having both the transcription repressing as well as activating capabilities.
Role in chemo-resistance
Recently, Kang et al. showed in in vitro experiments that transcription factor AP-2γ (TFAP2C) and the IFN-γ-inducible protein 16 (IFI16) are components of the MTA1 complex, and these protein complexes may contribute to the epigenetic regulation of estrogen receptor 1 expression in breast cancer and may determine the chemo-sensitivity of tumors to tamoxifen therapy [58]. In addition, to the abovementioned methods, it was shown that MTA1 induces its oncogenic functions via regulation of endogenous levels of c-myc which is responsible for cellular transformation, binding, and recruitment of transcriptional co-activators on to MTA1 locus [59]. Loss of MTA1 was found to affect the oncogenic activity of c-myc [59].
4.2 Esophageal cancer and MTA1
Esophageal squamous cell carcinoma (ESCC) is one of the most common malignant tumors [60]. Upregulation of metastasis-associated protein 1 (MTA1) has been reported to contribute to the development of esophageal squamous cell carcinoma [61, 62]. While examining the surgically resected node-negative esophageal cancer, Li et al. observed that the expression of MTA1 significantly correlated with shorter disease-free survival in these patients [40]. Recently, using cell-based assays, Weng et al demonstrated that MTA1 overexpression promotes invasiveness of the human esophageal carcinoma cell line EC-9706 by inhibiting the Snail, Slug, and HDAC1-dependent promoter activity of E-cadherin, an epithelial cell marker. They also showed the upregulation of Vimentin and MM9 by MTA1. These results reveal a novel role for MTA1 in the regulation of esophageal squamous cell carcinoma invasion and provide insights into mechanisms involved in this process [63]. In another recent report, Miyashita et al. used a rat model of esophageal cancer to show that HDAC1 and MTA1 expression may be involved in neoplastic transformation of the esophageal mucosa into cancer cells with both squamous and adeno differentiation [64].
4.3 Expression of MTA in other cancers
As mentioned earlier, the overexpression of MTA1 in mammary glands of virgin transgenic mice led to the metastasis to other organs most predominantly in lymph nodes [65]. The transgenic mice developed diffuse large B cell lymphomas, inducing MTA1 mediated Pax5 transcription factor expression. An independent study on human large B cell lymphoma patients confirmed the regulation of Pax5 expression by MTA1 [66]. Additionally, a number of studies have reported overexpression of MTA proteins in other cancers like melanomas, prostate, thymomas, tonsil cancers, early-stage non-small cell lung cancer (NSCLC), and advanced lung cancer highlighting its relevance in carcinogenesis [33, 67–71]. Overexpression of MTA1 in 29/60 (48.3 %) nasopharyngeal carcinoma patients was seen which significantly correlated with tumor metastasis via the Wnt1 pathway and β-catenin activation [72]. Li et al. found miR-125b to regulate MTA1 expression and have an antagonistic effect on the migration and invasion of NSCLC cells [73]. Adding to the list of cancers showing upregulation of MTA1, recently, another report by Zhang et al. showed the importance of MTA1 in the migration and invasion of laryngeal squamous cell carcinoma (LSCC) [74]. Due to its role in stabilizing HIF-1α, MTA1 promotes angiogenesis in hepatocellular carcinoma (HCC). Moon et al., using 45 tumors, demonstrated that overexpression of MTA1 is associated with HCC growth and vascular invasion [15]. Later on, Lee et al. identified polymorphisms IVS4-81G/A in MTA1 and suggested them to be important risk factors for the recurrence of HCC [74]. Similar role of MTA1 and its related proteins have also been observed in ovarian, cervical, endometrial, and tonsillar cancers [16, 33, 34, 37, 42].
5 Clinical implications
Metastasis-associated protein 1 (MTA1) has now been shown to be a molecular marker in various solid tumors. The initial suggestion of clinical implications of MTA1 in human cancers came from the gastric and colorectal cancers in which the transcript levels MTA1 was found to be highly expressed compared to paired normal counterparts in cancer tissue specimens [9]. The expression significantly correlated with stage of the cancer defined by local invasiveness and lymph node metastases. Similarly, overexpression of MTA1 mRNA was also shown in human colorectal and highly invasive, lymph node-positive esophageal squamous cell cancers [61, 75, 76]. Subsequently, MTA1 was shown to be a predicative marker for poor prognosis after surgery of esophageal squamous cell carcinomas [76]. Further, there was a significant association between MTA1 protein overexpression and shorter disease-free interval after complete resection of histologically node-negative esophageal cancers suggesting MTA1 to be a predictor of relapse [40]. Consistent with gastric cancer, examination of small intestinal carcinomas also revealed a role of MTA1 in cancer progression, proposing it as a genetic marker for distinguishing gastric carcinoids and other gastric neoplasms and also distinguishing malignant from benign tumors [77, 78].
Similar to the gastrointestinal cancer, nuclear expression of MTA1 was found to be associated with advanced, recurrent, and metastatic disease in pancreatic cancer [31, 32, 79]. High MTA1 expression levels as determined by immunohistochemistry correlated with high-grade pancreatic endocrine tumors, suggesting that levels of MTA1 can be used to predict the clinical behavior of these tumors and can be used as biomarker for the malignant progression [79]. An association was also found between MTA1 expression and malignant behavior of hepatocellular carcinoma (HCC) and advanced stage ovarian cancer suggestive of a new prognostic marker [80].
Apart from its role in metastasis, overexpression of MTA1 was also found to be closely associated with tumor grade, angiogenesis promotion, and high risk of recurrence in breast cancers [81, 82]. Similarly, aberrant expression of MTA2 in ER-negative patients resulted in enhanced metastasis associated with poor clinical outcome [83].
Meta-analysis of various cancers provided evidence of MTA1 as a new indicator of poor cancer prognosis. Moreover, siRNA-mediated inhibition of MTA1 has been shown to lead to inhibition of invasive and migrating capacity of prostate and esophageal cancer cells [84, 85] highlighting its role in metastasis. It was also shown that silencing of MTA1 by RNAi led to an altered expression of p53, E-cadherin, and β-catenin inhibiting migration and invasion [86].
6 Future perspectives
With the advent of new technologies and relevant recent research, several biological functions of the MTA family of proteins have been discovered. MTA1 has been implicated in the DNA damage-repair response in a p53-dependent and p53-independent manner [87–90], in inflammatory response mediated by NF-kappaB pathway [91, 92] and viral- and parasite-mediated carcinogenesis [27, 93–95]. Most importantly, given its important role in cancer progression, the possibility of MTA family of proteins being used as therapeutic targets has to be considered. In fact, several studies using natural compounds like resveratrol and pterostilbene (PTER) have evaluated MTA as a potential target [96, 97]. However, many questions remain to be addressed before its utility as a therapeutic target can be achieved. MTA1 has been thought to have physiological roles in spermatogenesis, rhodopsin expression, dopamine synthesis, and hepatic proliferation [30,98–100]. An in-depth understanding of physiological functions of MTA proteins is thus required especially because although these proteins are part of an enzymatic complex, MTA1 itself does not possess any enzymatic activity. Moreover, since upregulated levels of MTA proteins increase metastatic potential in tumors and MTA1 expression is regulated by microRNA-661, it would be interesting to see if and how this control is lost in the highly aggressive tumors [101]. Additionally, identification of critical regulators maintaining steady-state levels of MTA proteins will facilitate identification of the casual factors that disrupt this equilibrium. Furthermore, it seems that MTA family members have distinct functions and unraveling their non-redundant functions will enable achieving specificity of drug treatments.
References
Hanahan, D., & Weinberg, R. A. (2011). Hallmarks of cancer: the next generation. Cell, 144(5), 646–674.
Weinberg, R. A. (1983). Oncogenes and the molecular biology of cancer. Journal of Cell Biology, 97(6), 1661–1662.
Ciriello, G., et al. (2013). Emerging landscape of oncogenic signatures across human cancers. Nature Genetics, 45(10), 1127–1133.
Esteller, M. (2011). Epigenetic changes in cancer. F1000 Biology Reports, 3, 9.
Feinberg, A. P., & Vogelstein, B. (1983). Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature, 301(5895), 89–92.
Toh, Y., Pencil, S. D., & Nicolson, G. L. (1994). A novel candidate metastasis-associated gene, mta1, differentially expressed in highly metastatic mammary adenocarcinoma cell lines. cDNA cloning, expression, and protein analyses. Journal of Biological Chemistry, 269(37), 22958–22963.
Cui, Q., et al. (2001). Assignment of the human metastasis-associated gene 1 (MTA1) to human chromosome band 14q32.3 by fluorescence in situ hybridization. Cytogenetics and Cell Genetics, 93(1–2), 139–140.
Nicolson, G. L., et al. (2003). Tumor metastasis-associated human MTA1 gene and its MTA1 protein product: role in epithelial cancer cell invasion, proliferation and nuclear regulation. Clinical and Experimental Metastasis, 20(1), 19–24.
Toh, Y., et al. (1997). Overexpression of the MTA1 gene in gastrointestinal carcinomas: correlation with invasion and metastasis. International Journal of Cancer, 74(4), 459–463.
Toh, Y., Pencil S.D., and Nicolson G.L. (1995). Analysis of the complete sequence of the novel metastasis-associated candidate gene, mta1, differentially expressed in mammary adenocarcinoma and breast cancer cell lines. Gene,159(1), 97–104.
Manavathi, B., Singh, K., & Kumar, R. (2007). MTA family of coregulators in nuclear receptor biology and pathology. Nuclear Receptor Signaling, 5, e010.
Aasland, R., Stewart, A. F., & Gibson, T. (1996). The SANT domain: a putative DNA-binding domain in the SWI-SNF and ADA complexes, the transcriptional co-repressor N-CoR and TFIIIB. Trends in Biochemical Sciences, 21(3), 87–88.
Boyer, L. A., et al. (2002). Essential role for the SANT domain in the functioning of multiple chromatin remodeling enzymes. Molecular Cell, 10(4), 935–942.
Liu, J., et al. (2014). The subcellular distribution and function of MTA1 in cancer differentiation. Oncotarget, 5(13), 5153–5164.
Moon, W. S., Chang, K., & Tarnawski, A. S. (2004). Overexpression of metastatic tumor antigen 1 in hepatocellular carcinoma: relationship to vascular invasion and estrogen receptor-alpha. Human Pathology, 35(4), 424–429.
Balasenthil, S., Broaddus, R. R., & Kumar, R. (2006). Expression of metastasis-associated protein 1 (MTA1) in benign endometrium and endometrial adenocarcinomas. Human Pathology, 37(6), 656–661.
Fujita, N., et al. (2003). MTA3, a Mi-2/NuRD complex subunit, regulates an invasive growth pathway in breast cancer. Cell, 113(2), 207–219.
Kumar, R., et al. (2002). A naturally occurring MTA1 variant sequesters oestrogen receptor-alpha in the cytoplasm. Nature, 418(6898), 654–657.
Zhang, Y., et al. (1998). The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell, 95(2), 279–289.
Xue, Y., et al. (1998). NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities.Molecular Cell, 2(6), 851–861.
Zhang, Y., et al. (1999). Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes and Development, 13(15), 1924–1935.
Iguchi, H., et al. (2000). Expression of MTA1, a metastasis-associated gene with histone deacetylase activity in pancreatic cancer.International Journal of Oncology, 16(6), 1211–1214.
Toh, Y., & Nicolson, G. L. (2009). The role of the MTA family and their encoded proteins in human cancers: molecular functions and clinical implications. Clinical and Experimental Metastasis, 26(3), 215–227.
Moon, H. E., Cheon, H., & Lee, M. S. (2007). Metastasis-associated protein 1 inhibits p53-induced apoptosis. Oncology Reports, 18(5), 1311–1314.
Yoo, Y. G., Kong, G., & Lee, M. O. (2006). Metastasis-associated protein 1 enhances stability of hypoxia-inducible factor-1alpha protein by recruiting histone deacetylase 1. EMBO Journal, 25(6), 1231–1241.
Yoo, Y. G., et al. (2008). Hepatitis B virus X protein induces the expression of MTA1 and HDAC1, which enhances hypoxia signaling in hepatocellular carcinoma cells. Oncogene, 27(24), 3405–3413.
Bui-Nguyen, T. M., et al. (2010). NF-kappaB signaling mediates the induction of MTA1 by hepatitis B virus transactivator protein HBx.Oncogene, 29(8), 1179–1189.
Li, W., et al. (2009). Expression profile of MTA1 in adult mouse tissues. Tissue and Cell, 41(6), 390–399.
Mazumdar, A., et al. (2001). Transcriptional repression of oestrogen receptor by metastasis-associated protein 1 corepressor. Nature Cell Biology, 3(1), 30–37.
Zhang, S., et al. (2012). Sertoli cell-specific expression of metastasis-associated protein 2 (MTA2) is required for transcriptional regulation of the follicle-stimulating hormone receptor (FSHR) gene during spermatogenesis. Journal of Biological Chemistry, 287(48), 40471–40483.
Dias, S. J., et al. (2013). Nuclear MTA1 overexpression is associated with aggressive prostate cancer, recurrence and metastasis in African Americans. Science Reports, 3, 2331.
Hofer, M. D., et al. (2009). Immunohistochemical and clinicopathological correlation of the metastasis-associated gene 1 (MTA1) expression in benign and malignant pancreatic endocrine tumors. Modern Pathology, 22(7), 933–939.
Park, J. O., et al. (2011). Relationships between metastasis-associated protein (MTA) 1 and lymphatic metastasis in tonsil cancer.European Archives of Oto-Rhino-Laryngology, 268(9), 1329–1334.
Prisco, M. G., et al. (2012). Prognostic role of metastasis tumor antigen 1 in patients with ovarian cancer: a clinical study. Human Pathology, 43(2), 282–288.
Tuncay Cagatay, S., et al. (2013). MTA-1 expression is associated with metastasis and epithelial to mesenchymal transition in colorectal cancer cells. Tumour Biology, 34(2), 1189–1204.
Higashijima, J., et al. (2011). Expression of histone deacetylase 1 and metastasis-associated protein 1 as prognostic factors in colon cancer. Oncology Reports, 26(2), 343–348.
Liu, T., et al. (2013). Metastasis-associated protein 1 is a novel marker predicting survival and lymph nodes metastasis in cervical cancer. Human Pathology, 44(10), 2275–2281.
Du, B., et al. (2011). Metastasis-associated protein 1 induces VEGF-C and facilitates lymphangiogenesis in colorectal cancer. World Journal of Gastroenterology, 17(9), 1219–1226.
Li, S. H., et al. (2011). Overexpression of metastasis-associated protein 1 is significantly correlated with tumor angiogenesis and poor survival in patients with early-stage non-small cell lung cancer. Annals of Surgical Oncology, 18(7), 2048–2056.
Li, S. H., Wang, Z., & Liu, X. Y. (2009). Metastasis-associated protein 1 (MTA1) overexpression is closely associated with shorter disease-free interval after complete resection of histologically node-negative esophageal cancer. World Journal of Surgery, 33(9), 1876–1881.
Sharma, G., et al. (2011). Clinical significance of Maspin promoter methylation and loss of its protein expression in invasive ductal breast carcinoma: correlation with VEGF-A and MTA1 expression. Tumour Biology, 32(1), 23–32.
Dannenmann, C., et al. (2008). The metastasis-associated gene MTA1 is upregulated in advanced ovarian cancer, represses ERbeta, and enhances expression of oncogenic cytokine GRO. Cancer Biology and Therapy, 7(9), 1460–1467.
Agboola, A.O., et al. (2014). Clinicopathological and molecular significance of Sumolyation marker (ubiquitin conjugating enzyme 9 (UBC9)) expression in breast cancer of black women.Pathology - Research and Practice, 210(1),10–7.
Liu, S. L., et al. (2012). Expression of metastasis-associated protein 2 (MTA2) might predict proliferation in non-small cell lung cancer. Targeted Oncology, 7(2), 135–143.
Zhang, H., Stephens, L. C., & Kumar, R. (2006). Metastasis tumor antigen family proteins during breast cancer progression and metastasis in a reliable mouse model for human breast cancer. Clinical Cancer Research, 12(5), 1479–1486.
Kumar, R., Wang, R. A., & Bagheri-Yarmand, R. (2003). Emerging roles of MTA family members in human cancers. Seminar in Oncology, 30(5 Suppl 16), 30–37.
Li, D. Q., et al. (2012). Metastasis-associated protein 1/nucleosome remodeling and histone deacetylase complex in cancer. Cancer Research, 72(2), 387–394.
Kumar, R. (2003). Another tie that binds the MTA family to breast cancer. Cell, 113(2), 142–143.
Nawa, A., et al. (2000). Tumor metastasis-associated human MTA1 gene: its deduced protein sequence, localization, and association with breast cancer cell proliferation using antisense phosphorothioate oligonucleotides. Journal of Cellular Biochemistry, 79(2), 202–212.
Bagheri-Yarmand, R., et al. (2004). Metastasis-associated protein 1 deregulation causes inappropriate mammary gland development and tumorigenesis. Development, 131(14), 3469–3479.
Ohshiro, K., et al. (2010). Acetylation-dependent oncogenic activity of metastasis-associated protein 1 co-regulator. EMBO Reports, 11(9), 691–697.
Cui, Y., et al. (2006). Metastasis-associated protein 2 is a repressor of estrogen receptor alpha whose overexpression leads to estrogen-independent growth of human breast cancer cells. Molecular Endocrinology, 20(9), 2020–2035.
Mishra, S. K., et al. (2004). Upstream determinants of estrogen receptor-alpha regulation of metastatic tumor antigen 3 pathway.Journal of Biological Chemistry, 279(31), 32709–32715.
Zhang, H., et al. (2006). Metastatic tumor antigen 3 is a direct corepressor of the Wnt4 pathway. Genes and Development, 20(21), 2943–2948.
Gururaj, A. E., et al. (2006). Breast cancer-amplified sequence 3, a target of metastasis-associated protein 1, contributes to tamoxifen resistance in premenopausal patients with breast cancer. Cell Cycle, 5(13), 1407–1410
Gururaj, A. E., et al. (2006). MTA1, a transcriptional activator of breast cancer amplified sequence 3. Proceedings of the National Academy of Sciences of the United States of America, 103(17), 6670–6675
Nagaraj, S.R., et al. (2013). Crosstalk between VEGF and MTA1 signaling pathways contribute to aggressiveness of breast carcinoma. Molecular Carcinogenesis. doi:10.1002/mc.22104.
Kang, H. J., et al. (2014). Differential regulation of estrogen receptor alpha expression in breast cancer cells by metastasis-associated protein 1. Cancer Research, 74(5), 1484–1494.
Zhang, X. Y., et al. (2005). Metastasis-associated protein 1 (MTA1) is an essential downstream effector of the c-MYC oncoprotein.Proceedings of the National Academy of Sciences of the United States of America, 102(39), 13968–13973
Tang, Z. Y. (2001). Hepatocellular carcinoma—cause, treatment and metastasis. World Journal of Gastroenterology, 7(4), 445–454.
Toh, Y., et al. (1999). Overexpression of metastasis-associated MTA1 mRNA in invasive oesophageal carcinomas. British Journal of Cancer, 79(11–12), 1723–1726.
LLi, S. H., et al. (2012). Metastasis-associated protein 1 nuclear expression is closely associated with tumor progression and angiogenesis in patients with esophageal squamous cell cancer. World Journal of Surgery, 36(3), 623–631.
Weng, W., et al. (2014). Metastasis-associated protein 1 promotes tumor invasion by downregulation of E-cadherin. International Journal of Oncology, 44(3), 812–818.
Miyashita, T., et al. (2014). Impact of histone deacetylase 1 and metastasis-associated gene 1 expression in esophageal carcinogenesis. Oncology Letters, 8(2), 758–764.
Bagheri-Yarmand, R., et al. (2007). Metastasis-associated protein 1 transgenic mice: a new model of spontaneous B-cell lymphomas. Cancer Research, 67(15), 7062–7067.
Balasenthil, S., et al. (2007). Identification of Pax5 as a target of MTA1 in B-cell lymphomas. Cancer Research, 67(15), 7132–7138.
Qian, H., et al. (2007). RNA interference of metastasis-associated gene 1 inhibits metastasis of B16F10 melanoma cells in a C57BL/6 mouse model. Biology of the Cell, 99(10), 573–581.
Sasaki, H., et al. (2001). Expression of the MTA1 mRNA in thymoma patients. Cancer Letters, 174(2), 159–163.
Sasaki, H., et al. (2002). Expression of the MTA1 mRNA in advanced lung cancer. Lung Cancer, 35(2), 149–154.
Zhu, X., et al. (2010). Metastasis-associated protein 1 nuclear expression is associated with tumor progression and clinical outcome in patients with non-small cell lung cancer. Journal of Thoracic Oncology, 5(8), 1159–1166.
Lin, Z., et al. (2014). EBV-encoded LMP2A Promotes EMT in nasopharyngeal carcinoma via MTA1 and mTOR signaling induction. Journal of Virology, 88(20), 11872–85.
Li, Y., et al. (2013). MTA1 promotes the invasion and migration of non-small cell lung cancer cells by downregulating miR-125b.Journal of Experimental & Clinical Cancer Research, 32, 33.
Zhang, H., et al. (2014). Metastasis-associated gene 1 promotes invasion and migration potential of laryngeal squamous cell carcinoma cells. Oncology Letters, 7(2), 399–404
Lee, S. H., et al. (2012). Single nucleotide polymorphisms associated with metastatic tumour antigen 1 overexpression in patients with hepatocellular carcinoma. Liver International, 32(3), 457–466.
Giannini, R., & Cavallini, A. (2005). Expression analysis of a subset of coregulators and three nuclear receptors in human colorectal carcinoma. Anticancer Research, 25(6B), 4287–4292.
Toh, Y., et al. (2004). Expression of the metastasis-associated MTA1 protein and its relationship to deacetylation of the histone H4 in esophageal squamous cell carcinomas. International Journal of Cancer, 110(3), 362–367
Kidd, M., et al. (2006). The role of genetic markers—NAP1L1, MAGE-D2, and MTA1—in defining small-intestinal carcinoid neoplasia. Annals of Surgical Oncology, 13(2), 253–262.
Kidd, M., et al. (2006). Utility of molecular genetic signatures in the delineation of gastric neoplasia. Cancer, 106(7), 1480–1488.
Hofer, M. D., et al. (2004). The role of metastasis-associated protein 1 in prostate cancer progression. Cancer Research, 64(3), 825–829.
Hamatsu, T., et al. (2003). The role of MTA1 gene expression in human hepatocellular carcinoma. Oncology Reports, 10(3), 599–604.
Martin, M. D., et al. (2006). Breast tumors that overexpress nuclear metastasis-associated 1 (MTA1) protein have high recurrence risks but enhanced responses to systemic therapies. Breast Cancer Research and Treatment, 95(1), 7–12.
Jang, K. S., et al. (2006). MTA1 overexpression correlates significantly with tumor grade and angiogenesis in human breast cancers.Cancer Science, 97(5), 374–379.
Covington, K.R., et al.(2013). Metastasis tumor-associated protein 2 enhances metastatic behavior and is associated with poor outcomes in estrogen receptor-negative breast cancer. Breast Cancer Research and Treatment,141( 3),375–384.
Kai, L., et al. (2011). Targeting prostate cancer angiogenesis through metastasis-associated protein 1 (MTA1). Prostate, 71(3), 268–280.
Qian, H., et al. (2005). Reduced MTA1 expression by RNAi inhibits in vitro invasion and migration of esophageal squamous cell carcinoma cell line. Clinical and Experimental Metastasis, 22(8), 653–662.
Rao, Y., et al. (2011). Silencing MTA1 by RNAi reverses adhesion, migration and invasiveness of cervical cancer cells (SiHa) viaaltered expression of p53, and E-cadherin/beta-catenin complex. Journal of Huazhong University of Science and Technology. Medical Sciences, 31(1), 1–9.
Li, D. Q., et al. (2010). Revelation of p53-independent function of MTA1 in DNA damage response via modulation of the p21 WAF1-proliferating cell nuclear antigen pathway. Journal of Biological Chemistry, 285(13), 10044–10052.
Li, D. Q., et al. (2010). Requirement of MTA1 in ATR-mediated DNA damage checkpoint function. Journal of Biological Chemistry, 285(26), 19802–19812.
Li, D. Q., et al. (2009). MTA1 coregulator regulates p53 stability and function. Journal of Biological Chemistry, 284(50), 34545–34552.
Li, D. Q., & Kumar, R. (2010). Mi-2/NuRD complex making inroads into DNA-damage response pathway. Cell Cycle, 9(11), 2071–2079.
Pakala, S. B., et al. (2010). Regulation of NF-kappaB circuitry by a component of the nucleosome remodeling and deacetylase complex controls inflammatory response homeostasis. Journal of Biological Chemistry, 285(31), 23590–23597.
Pakala, S. B., et al. (2010). MTA1 coregulator regulates LPS response via MyD88-dependent signaling. Journal of Biological Chemistry, 285(43), 32787–32792.
Bui-Nguyen, T. M., et al. (2010). Stimulation of inducible nitric oxide by hepatitis B virus transactivator protein HBx requires MTA1 coregulator. Journal of Biological Chemistry, 285(10), 6980–6986.
Nair, S. S., et al. (2011). The metastasis-associated protein-1 gene encodes a host permissive factor for schistosomiasis, a leading global cause of inflammation and cancer. Hepatology, 54(1), 285–295.
Nair, S. S., et al. (2011). Inflammatory response to liver fluke Opisthorchis viverrini in mice depends on host master coregulator MTA1, a marker for parasite-induced cholangiocarcinoma in humans. Hepatology, 54(4), 1388–1397.
Kai, L., Samuel, S. K., & Levenson, A. S. (2010). Resveratrol enhances p53 acetylation and apoptosis in prostate cancer by inhibiting MTA1/NuRD complex. International Journal of Cancer, 126(7), 1538–1548.
Li, K., et al. (2013). Pterostilbene acts through metastasis-associated protein 1 to inhibit tumor growth, progression and metastasis in prostate cancer. PLoS One, 8(3), e57542.
Reddy, S. D., et al. (2011). Multiple coregulatory control of tyrosine hydroxylase gene transcription. Proceedings of the National Academy of Sciences of the United States of America, 108(10), 4200–4205.
Manavathi, B., et al. (2007). Repression of Six3 by a corepressor regulates rhodopsin expression. Proceedings of the National Academy of Sciences of the United States of America, 104(32), 13128–13133.
Li, W., et al. (2008). Involvement of metastasis tumor antigen 1 in hepatic regeneration and proliferation. Cellular Physiology and Biochemistry, 22(1–4), 315–326.
Reddy, S. D., et al. (2009). MicroRNA-661, a c/EBPalpha target, inhibits metastatic tumor antigen 1 and regulates its functions. Cancer Research, 69(14), 5639–5642.
Acknowledgments
SD is supported by the Department of Biotechnology, Ministry of Science and Technology grant (BT/PR4020/MED/30/792/2012). EK is a Council of Scientific and Industrial Research (CSIR) fellow.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Kaur, E., Gupta, S. & Dutt, S. Clinical implications of MTA proteins in human cancer. Cancer Metastasis Rev 33, 1017–1024 (2014). https://doi.org/10.1007/s10555-014-9527-z
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10555-014-9527-z