Abstract
Backgrounds
Triple negative breast cancer (TNBC) is a heterogeneous disease with more aggressive clinical courses than other subtypes of breast cancer. In this study, we performed high-resolution mass spectrometry-based quantitative proteomics with TNBC clinical tissue specimens to explore the early and sensitive diagnostic signatures and potential therapeutic targets for TNBC patients.
Methods
We performed an iTRAQ labeling coupled LC–MS/MS approach to explore the global proteome in tumor tissues and corresponding para-tumor tissues from 24 patients with grade I-II and grade III primary TNBC. Relative peptide quantification and protein identification were performed by Proteome Discoverer™ software with Mascot search engine. Differentially expressed proteins were analyzed by bioinformatic analyses, including GO function classification annotation and KEGG enrichment analysis. Pathway analyses for protein–protein interactions and upstream regulations of differentially expressed candidates were performed by Ingenuity Pathway Analysis (IPA) software.
Results
Totally, 5401 unique proteins were identified and quantified in different stage of TNBCs. 845 proteins were changed in patients with grade I or II TNBC, among which 304 were up-regulated and 541 were down-regulated. Meanwhile, for patients with grade III TNBC, 358 proteins were increased and 651 proteins were decreased. Comparing to para-cancerous tissues, various signaling pathways and metabolic processes, including PPAR pathways, PI3K-Akt pathway, one-carbon metabolism, amino acid synthesis, and lipid metabolism were activated in TNBC cancer tissues. Death receptor signaling was significantly activated in grade I-II TNBCs, however, remarkably inhibited in grade III TNBCs. Western blot experiments were conducted to validate expression levels of CYCS, HMGA1 and XIAP with samples from individual patients.
Conclusions
Overall, our proteomic data presented precise quantification of potential signatures, signaling pathways, regulatory networks, and characteristic differences in each clinicopathological subgroup. The proteome provides complementary information for TNBC accurate subtype classification and therapeutic targets research.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Background
Triple negative breast cancer (TNBC) is defined as a subtype of breast cancer, which does not express estrogen receptor (ER) or progesterone receptor (PR) and lacks human epidermal growth factor receptor 2 (HER2) overexpression or amplification [1]. TNBC accounts for 15–23.8% of all breast cancer and occurs more frequently in younger women [2]. The pathological type of TNBC is usually invasive ductal carcinoma, characterized by higher degree of malignancy as well as higher rate of early distant recurrence than other subtypes of breast cancer. Surgery combined with radiation and chemotherapy are the current strategies for TNBC treatment, and targeted therapeutic treatments are still challenging due to the absence of well-defined molecular targets [3]. Hence, there is an urgent need for in-depth molecular characterization to stratify patients with early diagnosis and personalized therapy.
In the preceding decades, numerous efforts have been focused on characterizing breast cancer heterogeneity in a clinicopathological relevant manner [4,5,6]. Recently, Jiang et al. [7] created the genomic and transcriptomic landscape of a Chinese cohort of 465 primary TNBCs and subtyped them into four transcriptome-based subtypes, which highlighted the substantial genomic heterogeneity of TNBC and paved the potential approaches for anticancer therapeutics. However, none of the newly developed molecular classifications, to date, has demonstrated clinical utility [8].
Proteins, as the ultimate performer of life activities, the quantitative changes can more directly reflect the occurrence and development of disease. Protein-based researches lag far behind relative to genomics, but recent breakthroughs of mass spectrometry have offered a glimpse of the proteomic achievements on precision medicine. Liu et al. [9] reported an 11-protein prognostic signatures via comparative proteome analysis to predict TNBC patients who develop a distant metastasis in order to reduce unnecessary adjuvant systemic therapy. Lawrence et al. [10] showed the proteomic landscape of TNBC by performing label-free deep proteome analysis of 24 human breast specimens which could help to identify breast cancer subtypes at the protein level. In brief, the unbiased analysis of thousands of protein expression signatures has identified a subset of tumors, as being associated with a high degree of tumor differentiation and improved patient outcome.
Quantitative proteomics has distinct advantages, such as low abundance proteins can be identified and quantified, and multiple samples can be quantitatively analyzed simultaneously under the same experimental conditions without complicating MS data analysis [11, 12]. Herein, we performed high-resolution mass spectrometry-based quantitative proteomics, using early and locally advanced TNBC clinical tissue specimens, to explore the early and sensitive diagnostic signatures and potential therapeutic targets for TNBC patients.
Materials and methods
Ethics statement
This study and consent procedure were approved by the Ethics Committee of Fujian Medical University Union Hospital (Fujian, China). All the participants provided their written informed consent to be included in the study.
Patients and clinical specimens
The paired TNBC samples used in this study were obtained from patients who underwent breast surgery (total mastectomy or breast conserving surgery) from the department of breast surgery, Fujian Medical University Union Hospital. Patients were randomly selected from May 2015 to April 2017 upon their first hospital visit and did not undergo any neoadjuvant chemotherapy or radiotherapy prior to surgery.
Surgically resected primary tumor tissues and paired non-cancerous adjacent tissues (> 3 cm apart from tumor edge) were collected from 12 patients with grade I-II as well as 12 patients with grade III. All patients were histopathologically diagnosed with TNBC (ER, PR, and HER2 negative, HER2 status was strictly defined according to the ASCO guidelines). The overall tumor grade was determined by tubule formation, pleomorphism and mitoses via postoperative hematoxylin and eosin (H&E) staining. To ensure the accuracy, all grade scoring was performed by two experienced pathologists independently. Clinical information was collected, including gender, age, histological grade, Ki-67 labeling index, tumor number, diameter of tumor, status of axillary lymph node metastasis, status of cancer recurrence, and status of survival. After surgical resection, the tissue samples for the proteomic analysis were frozen in liquid nitrogen for storage before use.
Immunohistochemistry staining
Formalin-fixed, paraffin-embedded tissues were cut into 4-μm sections. Following deparaffinization, sections were rehydrated and subjected to antigen retrieval. Sections were incubated at 4 °C overnight with anti-CD68, anti-CD4 and anti-CD8 antibodies. Immunostaining was performed using EnVision™ Detection Kit, Peroxidase/DAB, Rabbit/Mouse (DakoCytomation) according to the manufacturer’s instructions. Protein staining was evaluated under a light microscope at 100 × and 200 × magnification respectively (Leica).
Western blot assay
To extract the proteins from the tissues, 11 paired TNBC tumor tissues and paired non-cancerous adjacent tissues stored in a liquid nitrogen tank were thawed, and then cut into pieces, grinded in liquid nitrogen with a mortar and pestle, lysed on ice using 2% SDS-lysis buffer (pH 8.0) supplemented with protease and phosphatase inhibitors (Thermo Fisher Scientific, USA) and then sonicated for 3 min (5 s on and 5 s off, amplitude 25%). Protein concentrations were determined using the BCA protein assay (Thermo Fisher Scientific). Equal amounts of the protein lysates (25 μg) were separated on 10% SDS-PAGE gels and transferred to polyvinylidene difluoride (PVDF) membranes (Merck Millipore, Germany). Membranes were then blocked in 5% fat-free milk and immunoblotted at 4 °C overnight with the following primary antibodies: Cytochrome c (CST, USA, 1:1000), HMGA1 (CST, 1:1000), ERO1A (Abcam, USA, 1:1000), XIAP(CST, 1:1000), and β-actin (Proteintech, China, 1:5000). The membranes were then incubated with the appropriate secondary antibody for 1 h at room temperature. The specific bands were detected using an ECL detection kit (GE Healthcare, USA). Relative expression was determined by normalizing with β-actin using the ImageJ software (NIH, USA).
Protein extraction and digestion
Minced breast tissues were grinded in liquid nitrogen with a mortar and pestle, lysed in 2% SDS-lysis buffer (pH 8.0) containing protease and phosphatase inhibitors (Thermo Fisher Scientific, USA), then sonicated for 3 min (5 s on and 5 s off, amplitude 25%). The lysate was centrifuged at 16,000 g for 30 min, and the supernatant was collected as whole-tissue extract. Protein concentration was determined using the BCA protein assay (Thermo Fisher Scientific).
Each aliquot of 100 μg of breast proteins was diluted to 100 μl with 100 mM TEAB solution (Sigma-Aldrich, USA). Each sample was reduced with 10 mM TCEP for 30 min at 56 °C, followed by alkylation with 20 mM iodoacetamide for 30 min in the dark at room temperature. Then, proteins were precipitated overnight at − 20 °C using analytical grade acetone (Sigma-Aldrich). The recovered proteins after pelleting by centrifugation at 8000 g for 5 min were resuspended with 100 μl 100 mM TEAB solution. Sequencing-grade rLys-C (Promega, USA) was added to a final protease:protein ratio of 1:100 (w/w) and incubated for 3 h at 37 °C. Samples were then digested by sequencing-grade modified trypsin (Promega) at ratio of 1:50 (w/w) for 16 h at 37 °C. Peptides were collected by centrifugation at 16,000 g for 20 min, and the supernatants were collected for further iTRAQ labeling.
iTRAQ labeling
Four groups of peptides were labeled with iTRAQ4-plex reagent respectively according to manufacturer’s protocol. The labeled peptides were combined and vacuum-dried. The dried peptides were desalted using a Sep-Pak C18 cartridge (Waters) and the eluted peptides were also dried in a vacuum concentrator.
Peptide fractionation
Peptides were separated on an ACQUITY UPLC BEH C18 column (130 Å, 1.7 µm, 2.1 × 150 mm) (Waters, USA). Briefly, buffer A (315 mg ammonium formate and 10 mL 25% NH3·H2O in 1 L deionized water, pH 10) and buffer B (315 mg ammonium formate and 10 mL 25% NH3·H2O in 90% ACN) were prepared. Lyophilized peptides were resuspended with 30 μl buffer A. Gradient was developed over 60 min ranging from 0 to 90% buffer B at a flow rate of 0.25 ml/min. Twelve fractions were collected then dried for further analysis by LC–MS/MS.
LC–MS/MS analysis
The dried iTRAQ-labeled peptides were dissolved in 0.1% formic acid and fractionated on an Eksigent UPLC system by low pH reversed-phase chromatography at a flow rate of 250 nl/min for a 90 min gradient elution. Peptides were injected into the Q Exactive Orbitrap mass spectrometry system and the ion source parameters were operated with the following parameters: ISVF = 2300 V and Temp = 275℃. The data acquisition mode in the information-dependent acquisition (IDA) experiments was set to obtain a MS scan range 350–1600 m/z, followed by 100 to 1250 m/z for MS/MS scans of top 40 ion candidates per cycle. The instrument was operated in high sensitivity mode with MS1 AGC 3e6 and MS2 AGC 5e5. The total LC–MS running time for each IDA injection was 120 min. Each fractionation was performed in triplicate LC–MS/MS runs.
Data analysis
Relative peptide quantification and protein identification were performed using Proteome Discoverer™ software with Mascot search engine. Search parameters were set as follows: (i) Species, Homo Sapiens; (ii) Protein database, UniProtKB/Swiss-Prot (including 20,264 sequences); (iii) Quantification Method, iTRAQ 4-plex (Peptide Labeled); (iv) Digestion, Trypsin, allowing up to two missed cleavage; (v) Static Modifications, Carbamidomethyl, iTRAQ4plex; (vi) Dynamic Modifications, Methionine Oxidation; (vii) Precursor mass tolerance = 10 ppm, fragment mass tolerance = 0.05 Da. Quantification is based on the relative intensities of reporter ions which appear in the low mass range (114—117) of MS/MS spectra. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE [13] partner repository with the dataset identifier PXD020867 (https://www.ebi.ac.uk/pride/archive/projects/PXD020867).
Proteins with FDR < 0.01 on both protein and peptide level and matching ≥ 1 unique peptide were considered as positively identified proteins. Proteins with quantification p-value < 0.05 (115:114, 117:116, 117:115) and with fold change ≥ 1.5 or ≤ 0.67 (the average fold change of three repeat experiments) were considered as differentially expressed proteins.
The gene ontologies (biological processes and molecular functions) of all IDs were searched against the Gene Ontology database using Blast2GO—Functional Annotation and Genomics (www.blast2go.com/) as well as KEGG Mapping – GenomeNet (www.genome.jp/kegg/). Pathway analyses for protein–protein interactions and upstream regulations of differentially expressed candidates were performed using Ingenuity Pathway Analysis (IPA) software (QIAGEN, USA).
Results
Identification and quantification of proteins from primary TNBC tissue samples
Twenty-four TNBC patients, ranging in age from 35 to 65 years, fulfilled the clinical criteria for inclusion in this study. The clinical characteristics of these patients are summarized in Supplementary Table S1. Histologically normal tissues adjacent to the tumor tissues were used as a control here. Representative images of H&E staining presented tumor tissues and adjacent non-tumor tissues from different stage of TNBC patients (Supplementary figure S1a). Microscopic tumor foci from TNBC patients with stage III were indicated by yellow arrows, lymphocytic infiltration was detected surrounding tumor foci, and tumor angiogenesis was found to support the rapid growth of tumor foci (Supplementary figure S1b). A representative case of TNBC tissue from stage II showed the absence of estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2) expression via IHC staining (Supplementary figure S2).
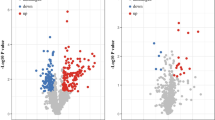
To obtain the proteomic profile, the paired tumor and corresponding para-tumor tissues (> 3 cm apart from tumor edge) from 24 TNBC patients were divided into four groups and trypsin into peptides respectively (Fig. 1a), 12 clinical specimens from grade I-II tumor tissues were pooled and labeled with iTRAQ115, while specimens from the corresponding non-cancerous adjacent tissues (NATs) were pooled and labeled with iTRAQ114. As for grade III patients, 12 specimens from tumor tissues were pooled and labeled with iTRAQ117, while specimens from the corresponding NATs were pooled and labeled with iTRAQ116. Four group iTRAQ-labeled peptides were combined and separated by high pH reversed-phase chromatography, then analyzed via LC–MS/MS approach. Scatter plots exhibited good reproducibility of the repeat experiments with the same samples (Supplementary figure S3a). A total of 5401 protein groups were identified and quantified through off-line high-pH fractionation followed by triplicate low pH LC–MS/MS runs (1% FDR rate on both protein and peptide level). Venn diagram depicted the number of identified proteins for the three replicates (Supplementary Fig. 3b). In detail, 845proteins changed in patients with Grade I or II TNBCs, among which 304 were up-regulated (fold change tumor/para-tumor ≥ 1.5, p < 0.05) and 541 were down-regulated (fold change tumor/para-tumor ≤ 0.67, p < 0.05). For patients with Grade III TNBCs, 358 proteins were increased and 651 proteins were decreased. Volcano plots in R were utilized to visualize differential expressions with proteomic results (Fig. 1b). Venn diagram comparing the number of differential expressed proteins (DEPs) quantified in different stage of TNBCs was presented in Supplementary Fig. 3c. Gene ontology (GO) analysis for cellular component of all the changed proteins interpreted that both up-regulated and down-regulated proteins significantly enriched in plasma membrane as well as cytoplasm (Fig. 1c). Differentially expressed proteins were listed in Supplementary Table S2 & S3.
iTRAQ-based quantitative proteomic analysis for TNBC tissues. a Diagram for preparation and iTRAQ-labeled analysis of paired tumor tissues and corresponding non-cancerous adjacent tissues (NATs) from 24 TNBC patients. b Volcano plots displayed differential protein expressions (log2 fold change) against -log10 (p-value) from the t-test. c GO-CC analysis for the differential expressed proteins
Proteomics features in TNBC tumor tissues compared with NATs
GO enrichment analyses of the DEPs were performed using clusterProfiler. The top 15 most significant GO terms (p < 0.05) in down-regulated and up-regulated proteins were shown in Fig. 2a. The color represented the p-adjusted values for these terms, and brighter red was more significant. The size of the plot displayed the significant genes by gene ratio from Supplementary Table S4 (# genes related to GO term / total number of sig genes). The protein activation cascade was the most significant enriched GO term, followed by acute inflammatory response, complement activation, and so forth in down-regulated proteins from both Grade I-II and Grade III TNBCs. As for up-regulated proteins, ER to Golgi vesicle-mediated transport and antimicrobial humoral response were significant enriched in Grade I-II TNBCs, while SRP-dependent cotranslational protein targeting to membrane and establishment of protein localization to endoplasmic reticulum were significant enriched in Grade III TNBCs (Fig. 2a). IPA functional analysis revealed the top 20 canonical pathways in which DEPs participated. Canonical pathway analysis in tumor vs. NATs from grade I-II TNBCs showed that the significant pathways (|z-score| > 2) were metabolic process (LXR/RXR activation and FXR/RXR activation) or immunity-related (acute phase response signaling, complement system and GP6 signaling pathway) (Fig. 2b). Figure 2c displayed the top 20 canonical pathways enriched from grade III TNBCs, among which coagulation system, EIF2 signaling and mTOR signaling were activated (z-score > 2).
GO enrichment and IPA analysis of DEPs. a Top 15 GO enrichment terms in down-regulated and up-regulated proteins from different stage of TNBCs. The dot plot showed the number of significant genes associated with the first 15 terms (size) and the p-adjusted values for these terms (color). The top 20 canonical pathways enriched in grade I-II b or grade III c TNBCs determined by IPA. The left y-axis displayed the-log of the Fischer’s exact test p-value. The right y-axis displayed the ratio of number of genes derived from our dataset, divided by the total number of genes in the pathway. The blue or orange bars depicted the significant pathways which were inhibited or activated based on p-value
Functional analyses of differentially expressed proteins from TNBCs
DEPs were further analyzed by KEGG enrichment analyses. DEPs from grade I-II TNBCs were significantly associated with PPAR signaling pathway (p = 3.47e-05), PI3K-Akt signaling pathway (p = 4.51e-04) and tyrosine metabolism (p = 2.32e-03) (Fig. 3a). Whereas genes related to ECM-receptor interaction (p = 1.39e-12), focal adhesion (p = 3.19e-06), PPAR signaling pathway (p = 6.04e-04) and glutathione metabolism (p = 8.14e-04) were significantly enriched from DEPs in grade III TNBCs (Fig. 4a).
Functional analyses of DEPs from TNBCs with grade I-II. a Pie chart represented the distribution of KEGG pathways which were obtained by enrichment analysis of DEPs in TNBCs with grade I-II. The enrichment of quantified DEPs against all KEGG database proteins were used to identify enriched pathways via two-tailed Fisher's exact tests. Pathways with corrected p values < 0.05 were considered significantly enriched. b Upstream analyses by IPA predicted which transcription factors were activated or inhibited, based on DEPs in tumor vs. para-tumor tissues from grade I-II TNBCs. Gene products were represented as nodes. The red nodes were up-regulated proteins and the green nodes were the reduced ones when compared tumor with para-tumor tissues from grade I-II TNBCs. The inner ring theme colors of orange and blue, with orange indicating “activated” and blue reflecting “inhibited”. The inset image showed the prediction legend
Functional analyses of DEPs from TNBCs with grade III. a KEGG enrichment of quantified DEPs against all KEGG database proteins were used to identify enriched pathways via two-tailed Fisher's exact tests. Pathways with corrected p values < 0.05 were considered significantly enriched. b Upstream analyses of DEPs in tumor vs. para-tumor tissues from grade III TNBCs were performed, and the statistically significant transcription factors were visualized in a network. The red nodes and the green nodes in the outer ring represented the up-regulated and down-regulated proteins respectively, and the orange nodes and blue nodes in the inner ring represented the activated and inhibited transcription factors respectively
Upstream analyses of DEPs with different stage were run by IPA respectively. The most statistically significant transcription factors were quickly prioritized and then visualized in networks. Transcription regulators including SMARCA4, NEUROG1, CPXM1 and SOCS1 were predicted inhibition while MAP3K7, IL27, HOXD10 and TFEB were predicted activation in tumor vs. NATs from grade I-II TNBCs (Fig. 3b). Regulators including TP53, BTK, BCL6, SMAD3 and MAPK1 were predicted inhibition while TLR9, NUPR1, JUN and IFNL1 were predicted activation in tumor vs. NATs from grade III TNBCs (Fig. 4b).
Next, we took a glance at the changed proteins when compared tumors from grade III with tumors from grade I-II TNBCs. KOG function classification of the significant changed proteins was predicted and categorized into 20 subcategories, indicating cell cycle control, amino acid transport, lipid metabolism, protein turnover and so on were involved when TNBC progression (Supplementary figure S4a). Upstream analysis of the changed proteins from tumor tissues with different stage was performed by IPA. Transcription regulators including SOCS1, SMARCA4, SAFB, NEUROG1, CPXM1 and CD3 were predicted inhibition while TFEB, SPDEF, RARB, RABL6, MAP3K7, IL27, IL15, HOXD10 and CXCL12 were predicted activation in grade III vs. grade I-II tumor tissues (Supplementary figure S4b). The upstream regulators shared partial overlap when we checked the list of regulators which were altered using DEPs in tumor vs. NATs from grade I-II or grade III TNBCs.
Comparison analyses of differentially expressed proteins from TNBCs
Further cross-dataset comparison analyses were used to delineated trends and similarities in the different datasets, as well as differences in their effects on signaling pathways. Cross-dataset comparison analyses demonstrated that death receptor signaling was activated in grade I-II TNBCs while inhibited in grade III TNBCs. For grade I-II TNBCs, the up-regulated levels of death ligands BID, TBID and CYCS mediated the activation of caspase-9, which then accelerated apoptosis by activating other caspases (Fig. 5a). Conversely, MKK4/7, CASP8/10 and XIAP were down-regulated in tumor tissues compared with NATs for grade III TNBCs, led to NF-κB signaling pathway inactivation and apoptosis signal transduction inhibition (Fig. 5b).
Comparison analyses of DEPs in different stage TNBCs. a Death receptor signaling was significantly enriched and activated in TNBCs with grade I-II (z-score > 2). b XIAP inhibited effector caspase activation, led to death receptor signaling cascade inhibition in TNBCs with grade III (z-score < -2). Gene products were represented as nodes and the red nodes were the up-regulated proteins while the green nodes were the reduced ones
Nest, these TNBC progression related DEPs were validated via western blot analysis. We selected 5 independent grade II TNBC patient samples and 12 independent grade III TNBC patient samples from 24 iTRAQ-labeled TNBCs, and conducted western blot experiment to examine the expression levels of cytochrome c (CYCS), high mobility group protein A1 (HMGA1), ERO1-like protein alpha (ERO1A) and E3 ubiquitin-protein ligase XIAP (Fig. 6a-d). Our proteomic results showed that CYCS was expressed much higher in tumors than in NATs from grade III TNBCs (T/N ratio = 2.24, p < 0.05), while a bit higher in tumors than in NATs from grade I-II TNBCs (T/N ratio = 1.55, p < 0.05). Western blot analysis confirmed proteomic features in grade II TNBCs as well as in grade III TNBCs that CYCS was extremely increased in tumor tissues compared with NATs. Proteomic results presented that HMGA1 was expressed much higher in tumors than in NATs from grade III TNBCs (T/N ratio = 2.59, p < 0.05), while remained unchanged both in tumors and in NATs from grade I-II TNBCs. Western blot analysis exhibited HMGA1 were significantly up-regulated in both grade II and grade III TNBCs when compared tumor tissues with NATs. Our proteomic results demonstrated that ERO1A was expressed higher in tumors than in NATs from grade III TNBCs (T/N ratio = 1.84, p < 0.05), while a bit higher in tumors than in NATs from grade I-II TNBCs (T/N ratio = 1.59, p < 0.05). However, western blot analysis showed ERO1A expression was not significantly up-regulated in grade II TNBCs or grade III TNBCs when compared tumor tissues with NATs. As for XIAP, proteomic results exhibited XIAP in tumors from grade III TNBCs was decreased compared with the paired NATs (T/N ratio = 0.59, p < 0.05), which was confirmed by western blot analysis. These findings highlighted the important molecular events involved in TNBC progression.
Representative DEPs in tumors and NATs from different stage of TNBCs. Western blot analysis of CYCS, HMGA1, ERO1A and XIAP in 5 independent grade II TNBC patient samples a and 12 independent grade III TNBC patient samples b-c collected with tumor (T) and their matched NATs (N). d Densitometry analysis by Quantity One shows quantitation of CYCS, HMGA1, ERO1A and XIAP levels, normalized against β-actin (*** p < 0.001). e Immunohistochemistry staining for CD68 in tumor tissues from different grade TNBCs (Left panel: magnification × 100; Right panel: magnification × 200)
Human immune system fights against tumor and sends various immune cells to the tumor tissue to induce antitumor immune response. Macrophages are the most common type of tumor-infiltrating immune cells. Here we conducted IHC experiments to examine the expression of CD68 from TNBC tissues with different stage. TNBC tissues from stage III showed more CD68-positive macrophages infiltrations surrounding tumor foci compared with TNBC tissues from stage I-II (Fig. 6e). IHC staining for CD4 and CD8 positive T cells infiltrations surrounding tumor foci from different stage of TNBCs were examined (Supplementary figure S5).
Additionally, networks developed by IPA integrated DEPs when compared tumor tissues with NATs from TNBC patients. For grade I-II TNBCs, IPA top score networks were predicted, one showed numerous immune related genes around two central hubs (NF-κB and Creb), the other with three central hubs (APOA1, CAV1 and P38 MAPK) associated with lipid metabolism and molecular transport (Supplementary figure S6a). For grade III TNBCs, one with two central hubs (CASP8 and CYCS) associated with inflammatory disease, and the other was focused on 30 DEPs also associated with lipid metabolism (Supplementary figure S6b). IPA top score networks indicated inflammatory disorder and abnormality of lipid metabolism were involved in different stage of TNBCs.
Discussion
The clinical behavior of TNBC is remarkably complex and heterogeneous. Due to the poor prognosis of patients with TNBC, great efforts have been fostered to discover specific molecular signatures and actionable therapeutic targets for TNBC early diagnosis and personalized treatment. Targeted agents, such as PI3K inhibitors, PARP inhibitors, HSP 90 inhibitors and HDAC inhibitors, are currently under clinical investigation in TNBC patients [14,15,16]. To data, functional proteomics provide a rich resource for unique information and knowledge mining toward the clinicopathological characteristic differences as well as the accurate classification for cancer subtypes [17, 18].
In the present study, we applied the unbiased analysis of protein expression signatures in tumor and NATs from TNBC patients with the early and advanced stages. A total of 5401 unique proteins were identified and quantified in different stages of TNBC. For the KEGG pathway enrichment analysis (Fig. 3a and Fig. 4a), the regulated proteins found to be enriched in various signaling pathways, including PPAR pathway, PI3K-Akt pathway, carbon metabolism, amino acid synthesis, and lipid metabolism, which indicate that these pathways were highly relevant to the pathogenesis of TNBC. Especially, IPA canonical pathway as well as regulatory network analyses depicted that lipid metabolism deregulation and inflammatory response disorder were closely involved in different stages of TNBC (Fig. 2b-c and Supplementary figure S6).
PPAR pathway has emerged as unavoidable links between lipid metabolism, inflammation and cancer [19, 20]. PPARs regulate gene expression by binding with RXR, and the PPAR-RXR heterodimer are activated by fatty acids and their derivatives. PPARs also connect lipid metabolism to inflammation under certain pathological conditions including cancer, and breast tissues are prone to inflammation when lipid homeostasis is disturbed [21]. Besides, the activity of PPAR is also regulated by phosphorylation events which we could investigated later in our system.
Here, four group iTRAQ-labeled peptides were combined and analyzed via LC–MS/MS approach. The pooling step combined 12 patients into 1 sample for each group which would eliminate the patient variations, therefore we performed western blot experiment to validate the proteomics results (Fig. 6a-d). In both grade II and grade III TNBCs, CYCS and HMGA1 were extremely increased in tumor tissues compared with NATs. XIAP expression was significantly reduced in grade III TNBCs when compared tumor tissues with NATs, but remained unchanged in grade II TNBCs. All above were consistent with proteomics results, however, western blot analysis showed ERO1A expression was not significantly up-regulated in grade II TNBCs or grade III TNBCs when compared tumor tissues with NATs. These findings highlighted the important molecular events involved in TNBC progression.
Notably, cross-dataset comparison analyses delineated that death receptor signaling was activated in grade I-II TNBCs while inhibited in grade III TNBCs (Fig. 5). The death receptor family which is part of the tumor necrosis factor (TNF) receptor superfamily can be triggered by death ligands to result in apoptotic or survival signals [22]. The ligands mediate the apoptotic effects via their respective cell membrane death receptors, and the most well-characterized death receptors are TNFR-1 and Fas. Upon the recruitment of Fas-associating protein with death domain (FADD), caspase-8 is activated through self-cleavage, leading to the initiation of the caspase cascade, which is critical for transduction of the apoptotic signal [23, 24]. In our dataset, the death ligands BID, TBID and CYCS mediated the activation of caspase cascade, which then transduced apoptotic signals for grade I-II TNBCs. Conversely, effector caspase activation was inhibited in tumor tissues compared with NATs for TNBCs with grade III, led to NF-κB signaling pathway inactivation and apoptosis signal transduction inhibition. This inhibition of the death receptor signaling transduction events provides a regulatory mechanism of the TNBC progression.
Conclusion
In summary, we performed a highly sensitive quantitative proteomic approach to explore the global proteome in tumor and corresponding non-cancerous adjacent tissues from patients with grade I-II and III TNBC and lay the foundation of precise targeted therapy for TNBC.
Data Availability
Raw proteomics data have been uploaded to the ProteomeXchange Consortium via the PRIDE partner repository with the subproject ID PXD020867 (https://www.ebi.ac.uk/pride/archive/projects/PXD020867). All data generated or analyzed during this study are included in this article (and its supplementary information files).
Abbreviations
- TNBC:
-
Triple negative breast cancer
- ER:
-
Estrogen receptor
- PR:
-
Progesterone receptor
- HER2:
-
Human epidermal growth factor receptor 2
- LC/MS:
-
Liquid chromatography-mass spectrometry
- IDA:
-
Information-dependent acquisition
- GO:
-
Gene ontology
- IPA:
-
Ingenuity Pathway Analysis
- DEP:
-
Differential expressed protein
- PPAR:
-
Peroxisome proliferator-activated receptor
- CYCS:
-
Cytochrome c
- HMGA1:
-
High mobility group protein A1
- ERO1A:
-
ERO1-like protein alpha
- TNF:
-
Tumor necrosis factor
- FADD:
-
Fas-associating protein with death domain
References
Foulkes WD, Smith IE, Reis JS (2010) Triple-negative breast cancer. N Engl J Med 363(20):1938–1948
Carey L, Winer E, Viale G, Cameron D, Gianni L (2010) Triple-negative breast cancer: disease entity or title of convenience? Nature Reviews Clinical Oncology 7(12):683–692
Bianchini G, Balko JM, Mayer IA, Sanders ME, Gianni L (2016) Triple-negative breast cancer: challenges and opportunities of a heterogeneous disease. Nature Reviews Clinical Oncology 13(11):674–690
Mertins P, Mani DR, Ruggles KV, Gillette MA, Clauser KR, Wang P et al (2016) Proteogenomics connects somatic mutations to signalling in breast cancer. Nature 534(7605):55–62
Huang KL, Li S, Mertins P, Cao S, Gunawardena HP, Ruggles KV et al (2017) Proteogenomic integration reveals therapeutic targets in breast cancer xenografts. Nat Commun 8:14864
Johansson HJ, Socciarelli F, Vacanti NM, Haugen MH, Zhu Y, Siavelis I et al (2019) Breast cancer quantitative proteome and proteogenomic landscape. Nat Commun 10(1):1600
Jiang YZ, Ma D, Suo C, Shi J, Xue M, Hu X et al (2019) Genomic and transcriptomic landscape of triple-negative breast cancers: subtypes and treatment strategies. Cancer Cell 35(3):428–440.e5
Weigelt B, Pusztai L, Ashworth A, Reis-Filho JS (2011) Challenges translating breast cancer gene signatures into the clinic. Nat Rev Clin Oncol 9(1):58–64
Liu NQ, Stingl C, Look MP, Smid M, Braakman RB, De Marchi T et al (2014) Comparative proteome analysis revealing an 11-protein signature for aggressive triple-negative breast cancer. J Natl Cancer Inst 106(2):djt376
Lawrence RT, Perez EM, Hernández D, Miller CP, Haas KM, Irie HY et al (2015) The proteomic landscape of triple-negative breast cancer. Cell Rep 11(4):630–644
Wiese S, Reidegeld KA, Meyer HE, Warscheid B (2007) Protein labeling by iTRAQ: a new tool for quantitative mass spectrometry in proteome research. Proteomics 7(3):340–350
Köcher T, Pichler P, Schutzbier M, Stingl C, Kaul A, Teucher N et al (2009) High precision quantitative proteomics using iTRAQ on an LTQ Orbitrap: a new mass spectrometric method combining the benefits of all. J Proteome Res 8(10):4743–4752
Perez-Riverol Y, Csordas A, Bai JW, Bernal-Llinares M, Hewapathirana S, Kundu DJ et al (2019) The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res 47(D1):D442–D450
Di Cosimo S, Baselga J (2010) Management of breast cancer with targeted agents: importance of heterogeneity. Nat Rev Clin Oncol 7(3):139–147
Mayer IA, Abramson VG, Lehmann BD, Pietenpol JA (2014) New strategies for triple-negative breast cancer–deciphering the heterogeneity. Clin Cancer Res 20(4):782–790
Torres-Adorno AM, Lee J, Kogawa T, Ordentlich P, Tripathy D, Lim B et al (2017) Histone deacetylase inhibitor enhances the efficacy of MEK inhibitor through NOXA-mediated MCL1 degradation in triple-negative and inflammatory breast cancer. Clin Cancer Res 23(16):4780–4792
Akbani R, Ng PK, Werner HM, Shahmoradgoli M, Zhang F, Ju Z et al (2014) A pan-cancer proteomic perspective on The Cancer Genome Atlas. Nat Commun 5:3887
Tyanova S, Albrechtsen R, Kronqvist P, Cox J, Mann M, Geiger T (2016) Proteomic maps of breast cancer subtypes. Nat Commun 7:10259
Michalik L, Wahli W (2008) PPARs mediate lipid signaling in inflammation and cancer. PPAR Res 2008:134059
Wahli W, Michalik L (2012) PPARs at the crossroads of lipid signaling and inflammation. Trends Endocrinol Metab 23(7):351–363
Doan TB, Graham JD, Clarke CL (2017) Emerging functional roles of nuclear receptors in breast cancer. J Mol Endocrinol 58(3):R169–R190
Fuchs Y, Steller H (2011) Programmed cell death in animal development and disease. Cell 147(4):742–758
Peter ME, Krammer PH (2003) The CD95(APO-1/Fas) DISC and beyond. Cell Death Differ 10(1):26–35
Hughes MA, Powley IR, Jukes-Jones R, Horn S, Feoktistova M, Fairall L et al (2016) Co-operative and hierarchical binding of c-FLIP and caspase-8: a unified model defines how c-FLIP isoforms differentially control cell fate. Mol Cell 61(6):834–849
Acknowledgements
We sincerely appreciate Guoquan Yan for his expert help in the quantitative proteomics experiment, and Dr. Yang Zhang for his technical assistance and insightful comments.
Funding
This work was supported by the Natural Science Foundation of China (81802766), the National Key Research and Development Program of China (2018YFA0507501, 2017YFA0505001), the China Postdoctoral Science Foundation (2017M621347) and the Joint Funds for the Innovation of Science and Technology of Fujian province (2017Y9033).
Author information
Authors and Affiliations
Contributions
LY, FF, WC, YP and SH conceived and designed the experiments. LL, HA, XJ, JM and GR performed the experiments. LL, WZ and YL analyzed the data. LY and LL wrote the manuscript. All authors contributed toward data analysis, drafting and critically revising the paper, gave final approval of the version to be published, and agreed to be accountable for all aspects of the work.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethics approval
This study and consent procedure were approved by the Ethics Committee of Fujian Medical University Union Hospital (Fujian, China). All the participants provided their written informed consent to be included in the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
10549_2020_5916_MOESM1_ESM.tif
Supplementary file1 Fig. S1 Representative images of HE-stained sections were presented. (a) Histological images of tumor tissue and adjacent non-tumor tissue from different stage of TNBCs (original magnification ×100, scale bars, 200 μm). (b) Representative images from TNBC patients with stage III showed more tumor foci (yellow arrows). In the middle panel, lymphocytic infiltrations were detected surrounding tumor foci which were indicated by arrows. Neovascularization was shown in right panel to support the rapid growth of tumor foci. (TIF 6524 kb)
10549_2020_5916_MOESM2_ESM.tif
Supplementary file2 Fig. S2 IHC staining for ER, PR and HER2 in tumor tissues (Left panel: magnification × 100; scale bars, 200 μm; Right panel: magnification × 200, scale bars, 100 μm). (TIF 16104 kb)
10549_2020_5916_MOESM3_ESM.tif
Supplementary file3 Fig. S3 Overview of proteomics dataset of different stage TNBCs and quality control for the mass spectrometry. a Scatter plots for replicate proteome profiling of 24 paired tumor and para-tumor samples from TNBC patients. b Venn diagram comparing the number of identified proteins for the three replicates and their relationship. c Venn diagram comparing the number of differential expressed proteins quantified in different stage of TNBC tissues. (TIF 980 kb)
10549_2020_5916_MOESM4_ESM.tif
Supplementary file4 Fig. S4. Functional analyses of changed proteins from tumor tissues with different stages. a Classification of KOG function for significant changed proteins which were aligned to the KOG database to predict and categorize possible functions. Proteins were divided into 20 subcategories and represented by different colors. b Activating (z-sore>2) and inhibiting (z-sore<-2) of upstream transcription factors were exhibited when comparing grade III tumor tissues with grade I-II tumor tissues. The red nodes and the green nodes in the outer ring represented the up-regulated and down-regulated proteins respectively. The inner ring theme colors of orange and blue, with orange indicating “activated” and blue reflecting “inhibited”. The inset image showed the prediction legend. (TIF 2068 kb)
10549_2020_5916_MOESM5_ESM.tif
Supplementary file5 Fig. S5. IHC staining were performed to examine the expression of CD4 and CD8 (Left panel: magnification × 100; Right panel: magnification × 200) (TIF 5416 kb)
10549_2020_5916_MOESM6_ESM.tif
Supplementary file6 Fig. S6. Networks developed by IPA integrated DEPs when compared tumor with para-tumor tissues from TNBC patients with grade I-II a and from TNBC patients with grade III b. Generated networks, which were ordered by score meaning significances, were associated to diseases and functions according to both up- and down-regulated identifiers. IPA top two score networks were presented in which gene products were represented as nodes and direct biological relationships between two nodes as a line. The red nodes were the up-regulated proteins while the green nodes were the reduced ones (TIF 1305 kb)
Rights and permissions
About this article
Cite this article
Lin, Y., Lin, L., Fu, F. et al. Quantitative proteomics reveals stage-specific protein regulation of triple negative breast cancer. Breast Cancer Res Treat 185, 39–52 (2021). https://doi.org/10.1007/s10549-020-05916-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-020-05916-8