Abstract
Purpose
Estrogen receptor positive (ER+) breast cancer constitutes almost 85% of all breast cancer patients and are a genetically highly heterogenic group. Data on the association of somatic alterations to outcome and prognosis are however sparse. In this neoadjuvant endocrine phase II trial including postmenopausal breast cancer patients with ER+, HER2 normal breast cancer, we investigated the rate of pathogenic mutations before and after treatment as well as the association with treatment response and survival.
Methods
Pretreatment and posttreatment tumour samples from 109 patients treated with neoadjuvant letrozole were collected and analysed with Next Generation Sequencing utilizing a panel of 12 genes (ALK, BRAF, EGFR, ERBB2, ERBB3, ESR1, KIT, KRAS, NRAS, PDGFRA, PIK3CA, and RAF1). Residual disease was assessed by a modified Miller Payne scale and the Residual Cancer Burden index. Survival data were collected prospectively.
Results
Among the 109 patients, 52 had at least one pathogenic mutation in the pretreatment sample and 60 in the posttreatment sample. The most frequently mutated gene was PIK3CA, followed by EGFR and KRAS. Twelve different pathogenic PIK3CA mutations were identified, primarily in exon 20 and exon 9. An altered PIK3CA mutation profile from the pre- to the posttreatment specimen was significantly associated to improved pathological outcome. Overall and Disease-Free Survival benefits in PIK3CA mutated patients was observed.
Conclusion
Considerable heterogeneity was identified both among patients and between pre- and posttreatment samples. PIK3CA has the potential to be a predictive biomarker. To further assess the implications of a treatment related altered PIK3CA mutation profile, more data are needed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Estrogen Receptor positive (ER+) breast cancer patients are the numeric largest and the most heterogenous group in terms of gene expression, genetic alterations and copy number changes [1]. ER+ breast tumours contain a significant higher number of somatic mutations compared to HER2 positive and triple negative subtypes [1,2,3]. Strikingly however, the amount of genomic data generated on ER+ cancer is sparse when compared to triple negative and HER2 positive breast cancers [2,3,4,5,6], and the clinical implications of pathogenic driver mutations is yet to be determined, including the impact of the most frequently mutated gene PIK3CA.
Pathogen mutations in PIK3CA have been reported in 25–40% of cases in ER+, HER2 normal breast cancer [1,2,3, 7]. PIK3CA mutations are associated with a favourable prognosis for ER+, HER2 normal breast cancer patients [3, 7,8,9] and it is a possible future predictive biomarker for endocrine treatment [2]. PIK3CA is commonly mutated in exon 20 and exon 9, being the kinase and helical domain, respectively. Both domains harbour well-known gain-of-function mutations and while the survival benefit of PIK3CA mutations is established, it’s still unclear if the effect is domain specific [9,10,11,12].
Although the positive prognostic association is well known, the underlying mechanism is unclear. One possible explanation is the association of favourable clinicopathological variables with PIK3CA mutations [2]. There are little data on treatment related dynamics in mutations and the potential implications hereof.
Postmenopausal women with operable ER+, HER2 normal breast cancer are routinely offered adjuvant endocrine treatment alone or in combination with chemotherapy. However, not all patients benefit from endocrine therapy and identifying these patients are of critical importance. Neoadjuvant endocrine treatment (NET) trials constitute a unique opportunity to evaluate the mechanisms of response and to correlate the variation in expression of biomarkers including Ki67 and Tumour Infiltrating Lymphocytes (TILs) with treatment response and survival [13].
In this study, we included postmenopausal women with operable ER+, HER2 normal breast cancer in a neoadjuvant phase II trial and collected core needle biopsies before, and surgically resected tissue after four months of letrozole. Tumour tissues were investigated for a panel of pathogenic somatic mutations in order to investigate the prevalence and dynamics of pathogenic mutations and the association to clinicopathological variables during NET.
Material and methods
Study population and samples
Patients were treated with neoadjuvant letrozole for four months prior to curative intended surgery as part of a clinical phase II study conducted by the Danish Breast Cancer Group (DBCG) [14]. The trial was conducted between 2009 and 2012 at nine institutions in Denmark. The study design and clinical results have been published previously [15, 16]. In the phase II study, the endpoints were clinical and pathological outcome. A total of 119 patients were registered to receive neoadjuvant letrozole. Eligible patients had histological confirmed invasive ER+, HER2 normal operable breast cancer. They met the following criteria: tumour size ≥ 1 cm, ≥ 60 years at entry, Eastern cooperative Oncology Group Score 0 – 2 and Charlson comorbidity index 0 – 2. Patients with prior cytotoxic treatment including aromatase inhibitors and prior malignant disease were not eligible. Four patients were excluded prior to study initiation, two cases due to HER2 positivity at central testing and two patients withdrew consent. An additional three were excluded from the intention to treat population, Fig. 1a.
Pretreatment formalin fixed paraffin embedded (FFPE) core biopsies and posttreatment FFPE surgical tumour samples were prospectively collected from 112 patients. In 109 (97%) cases it was possible to collect paired samples for NGS analysis. After quality assessment of the procedures, paired data were successfully retrieved from 83 patients, Fig. 1b.
Pathological response assessment
Pathological complete response (pCR) is the standard endpoint in neoadjuvant trials. pCR is however infrequent after NET [17]. For pathological response assessment we therefore used a modified Miller Payne grading system for residual disease used by the DBCG [18]. On the modified scale grade 1 equals no invasive cells present in the tumour bed,pathological complete response (pCR). Grade 2 more than 90% loss of tumour cells and grade 3 between 30 and 90% reduction in tumour cells. Grade 4 is defined as less than 30% loss of tumour cells and was considered no response. As DBCG guidelines for assessment of residual disease changes from 2020 we also assessed residual disease after The Residual Cancer Burden Index using the guidelines presented by the BIG-NABCG collaboration [19, 20]. In brief the RCB index combines the bidimensional diameter of the primary tumour with the percentage of invasive cells in the tumour, corrected for the percentages of in situ carcinoma, and number of positive lymph nodes including the diameter of the largest lymph node metastasis in a generalized linear model. The higher RCB index, and corresponding RCB-Class, the more extensive residual disease load. For the RCB index calculations in this study we used the online Residual Cancer Burden Calculator provided by the MD Andersson Cancer Center [21].
Pathological and clinical data
Patients were registered in the DBCG database upon study entry and updated prospectively. Assessment of ER, PGR, HER2 and Ki67 were performed centrally using current international standards [22,23,24]. TILs were assessed by use of the guidelines of the international Immuno-Oncology Biomarker Working Group on Breast Cancer [25]. All patients are followed for 10 years after accrual, biannually after surgery for five years, and annually for the following five years or until event or death. Event is defined as either relapse (local, regional or distant), contralateral breast cancer, other malignancy or death as first event. Data were extracted from the DBCG database January 31th 2020.
Next Generation Sequencing
DNA extraction
Haematoxylin and eosin stained slides from FFPE sample blocks were reviewed by a senior pathologist and marked for DNA extraction. DNA was extracted from unstained FFPE tissue sections with the Gene Read DNA FFPE kit. A total of 40 ng DNA was used for target enrichments for the GeneRead QIAact Actionable Insights Tumour Panel (Qiagen, Hilden, Germany). The panel includes 330 amplicons covering 16.7 kb, containing 773 unique variant positions in 12 genes (ALK, BRAF, EGFR, ERBB2, ERBB3, ESR1, KIT, KRAS, NRAS, PDGFRA, PIK3CA, and RAF1). Following target enrichment, library preparation and clonal amplification was done using the Gene Read DNA Library Q Kit and the Gene Read Clonal Amp Q Kit. Sequencing was performed on the Gene Reader instrument with the Gene Read UMI Advanced Sequencing Q Kit. Qiagen QCI Analyze Software was used for variant calling. The average sequencing coverage was 500x. Output data were manually filtered to remove polymorphisms (variants present in > 1% of the general population) before variant annotation.
Variant annotation
All variants were manually reviewed. Only rare non-synonymous variants were considered and annotated as pathogenic mutations, variant of unknown significance (VUS) or benign using literature search and the publicly available databases JAX, Clinvar and OMIM [26,27,28].
Statistical analysis
The prevalence of pathogenic and non-pathogenic mutations was determined in the complete cohort as described above. Pre- and posttreatment analysis were made on all patients with pre- and posttreatment data. Sensitivity tests were performed only on patients who had paired analysis finding parallel results. For baseline characterization standard clinicopathologic variables were categorized. Associations between PIK3CA mutations and baseline characteristics were investigated with χ2-test. Estimated potential follow-up was calculated with the reverse Kaplan Meier method [29]. Correlation between genetic alterations and the continuous variables TILs and Ki67 were assessed with the point biserial correlation coefficient (rpb). Association between PIK3CA mutations, domains and change in mutational profile with pathological outcome (Miller Payne and RCB as categorical variables) were investigated with χ2-test. As RCB as continuous variable were not normally distributed its association with pretreatment mutation profile was assessed with the Wilcoxon signed rank test. Disease free survival (DFS) was determined as the interval between initial diagnosis and detection of the first relapse regardless of its site (local, regional, or distant), contralateral breast cancer, other malign disease and death from any cause. Overall survival, defined as time from diagnosis to death from any cause, was estimated by the Kaplan–Meier method and log rank p-values are presented. All P values are 2-sided, with a P value ≤ 5% considered to be statistically significant. No correction for multiple testing was applied. Statistical analysis was performed using SAS enterprise guide version 7.15 (Cary, NC, USA).
Results
Patient characteristics and mutation frequencies
Patient characteristics are shown in Table 1. The median age was 67 years at study entry (range 60 – 87). Of the 109 patients, 58 had pathological response, including one patient with pCR, when assessed by the modified Miller Payne system. When assessed after the RCB index; One patient had RCB-class 0, six had RCB-Class 1, 88 had RCB-Class 2 and 12 RCB-Class 3, two patients did not have RCB assessment due to lack of surgical specimens.
After an estimated potential follow-up of 8.3 years, 27 deaths and 30 events were recorded (14 patients were diagnosed with relapse, 2 with contralateral breast cancer, 6 with other malignancies and 8 patients died as first event).
Fifty patients had at least one pathogenic mutation in their pretreatment sample and 60 in the posttreatment sample, with considerable heterogeneity within the group as well as within the pre- and posttreatment samples, Fig. 2. The most frequently mutated genes were PIK3CA in both pre- and posttreatment samples (n = 42 (46%) and n = 48 (48%), respectively), followed by EGFR (n = 7 and n = 10) and KRAS (n = 4 and n = 8). When visually inspecting the data presented in Fig. 2 patients with mutations in KRAS or EGFR didn’t present with a clear pattern of either positive or negative predictive or prognostic inclinations. Noteworthy is that in the patients harbouring mutations in ALK, KIT, ERBB2, ESR1 and NRAS almost all had relapsed or died at data extraction.
Somatic mutations in 83 paired pre- and posttreatment samples from menopausal ER+, HER2 normal breast cancer patients treated with neoadjuvant letrozole between 2009 and 2012. Samples are arranged by pathological response and survival data, and subsequently by mutation frequencies. Each column denotes a patients pre- and posttreatment sample. Each row represents one gene. Survival data and mutation status are shown by colour as indicated
PIK3CA characteristics
Association of PIK3CA status and baseline characteristics are presented in Table 1. A ductal subtype was significantly associated with having a PIK3CA mutation compared to lobular and other invasive subtypes (p = 0.01). No other variables were associated with harbouring a PIK3CA mutation.
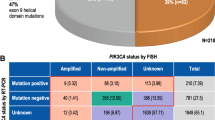
We identified 12 different pathogenic PIK3CA mutations, as presented in Table 2. Mutations were predominantly located in hotspots in exons 20 (kinase domain) and exon 9 (helical domain) followed the C2 domain (exon 5 and 8). The most common mutations were c.3140A > G (p.H1047R) in the kinase domain found in 34 samples and c.1633G > A (p. E545K) in the helical domain, found in 23 samples. Mutational domain was not correlated to any baseline variables (Table 3).
Fourteen of the 83 patients with paired samples had an altered PIK3CA mutation profile after treatment i.e. either lost or gained a pathogenic PIK3CA mutation during letrozole treatment. In patients with an unaltered mutational profile the domain predominantly remained the same. A weak but non-significant correlation was identified between changes in TILs and KI67 during treatment and association to PIK3CA mutation status (rpb 0.10; p = 0.35 and rpb 0.01; p = 0.93, respectively).
PIK3CA mutations and relation to pathological response
Pathological assessment by the modified Miller Payne system
PIK3CA mutation status was not significantly associated with achieving pathological response; however, numerically patients with a PIK3CA mutation did achieve a pathological response more frequently with a response rate of 64% of mutated vs. 47% in mutation negative tumours (p = 0.09) (Table 4).
It appeared beneficial for treatment response to have a mutation in the kinase domain, as compared to mutations in the C2 or the helical domain, p = 0.07 (Table 4). Seventy-nine percent of patients with an altered PIK3CA mutational profile achieved pathological response compared to 48% in patients with an unaltered PIK3CA profile, p = 0.04 (Table 4). Whether patients gained a pathogen mutation or became mutation negative after treatment had no impact on the association to treatment response (data not shown).
Pathological assessment by the Residual Cancer Burden index
There was no difference in the RCB index between patients with pathogenic PIK3CA mutation or mutation negative patients before treatment p = 0.54 (Table 4). PIK3CA mutational status, domain of mutation or if patients lost or gained a pathogen variant during treatment was not associated to RCB-Class, as presented in Table 4.
PIK3CA mutations and association to DFS and OS
Comparing patients with and without a pathogen PIK3CA mutation they had very similar 8-year DFS rates (79 and 75%, respectively). Patients with a pathogen PIK3CA mutation had an 8-year OS of 82% compared to 70% in mutation negative patients, p = 0.41, (Table 5).
Discussion
In this study, we applied targeted NGS on FFPE tumour tissue from the diagnostic core biopsies and posttreatment surgical specimens from patients treated with neoadjuvant letrozole. By focusing on hotspot mutations and in order to achieve high quality data with deep sequencing results, we used a panel including 12 genes targetable with FDA approved or investigational drugs.
A considerable heterogeneity of pathogenic mutations was observed. PIK3CA was, as expected, the most frequently mutated gene. PIK3CA mutations were associated to a ductal subtype which is a known positive predictive factor [15, 30, 31]. Low rate of pCR in this population makes dichotomic assessment of response difficult and similarly a low rate of events leaves us without enough power to estimate OS and DFS. However, our data support the consensus that PIK3CA mutations are positively associated with prognosis in the ER+, HER2 normal breast cancer population [7,8,9] and that PIK3CA might be a potential predictive biomarker [2]. In our subset analysis of mutated domain, it seemed to be favourable with mutations in the kinase domain for response to treatment. Our analyses showed that patients with an altered PIK3CA mutation profile after treatment had significantly improved response to endocrine treatment. There was no difference in whether patients gained a pathogenic mutation or became mutation negative suggesting it could be the instability itself that provides a favourable outcome. We speculated that this might be due to enhanced recognition by the immune system with changing neoantigens. However, this was not the case, as we found no association when assessing changes in TILs as a proxy for immunogenicity and Ki67 for proliferation.
Strengths of our study include a large group of matched patients’ samples and prospectively collected clinical data, centralized performed pathological procedures and long follow-up. The study also has some weaknesses, we rely on limited and selected amount of tumour tissue and because of that, we have an intrinsic risk of underestimating tumour heterogeneity as neoplasms consist of multiple subclonal populations [32] and both domain and alteration analyses are based on small numbers and must be viewed as hypothesis generating.
The phosphatidylinositol 3-kinase (PI3K) pathway mediates key cellular functions, including growth, proliferation, survival, angiogenesis, and motility. When PIK3CA is mutated the PIK3/AKT pathway is hyperactivated causing oncogenic transformation [7, 9, 11]. Although a higher mutation rate is seen in ER+ breast cancer, typical markers of pathway activation (e.g. phosphorylation of AKT and pS6) are not as elevated, as in basal-like and HER2 enriched breast cancers, so despite PI3K/AKT pathway activation, downstream mTORC1 signalling are not elevated (transcriptional nor biochemical). This apparent disconnect has been documented by several groups, but are yet not fully understood [1, 8, 11].
Additional efforts to identify the mechanism of protection by PIK3CA mutations are important as PIK3/AKT pathway inhibitors proceed through clinical development for targeted therapy, with alpelisib, an α-specific PI3K inhibitor that selectively inhibits p110α, FDA approved late 2019 for advanced disease in combination with fulvestrant [33, 34]. With more data coming in, it is very likely that genome analysis in the future will push treatment decisions towards a more personalized therapy regime, also in the adjuvant setting for ER+, HER2 normal breast cancer patients.
In conclusion, we have successfully utilized a targeted NGS panel in analysing paired samples from prospectively followed patients who participated in a phase 2 trial. We found that genomic instability of PIK3CA correlated with response to neoadjuvant endocrine therapy, a finding that needs to be further examined. In our population DFS and OS hazard ratios agreed with the consensus that pathogenic mutations in PIK3CA is beneficial for ER+, HER2 normal breast cancer patients, a mechanism not yet fully understood, but increasingly interesting to pursue due to the potential targeted treatment options for this population of postmenopausal breast cancer patients.
Data availability
The data supporting all the figures, tables and supplementary tables in the published article, are not publicly available due to institutional restrictions. The dataset can be made available to qualified researchers through application to the Danish Breast Cancer Group. Please contact dbcg.rigshospitalet@regionh.dk.
References
Koboldt DC, Fulton RS, McLellan MD, Schmidt H, Kalicki-Veizer J, McMichael JF et al (2012) Comprehensive molecular portraits of human breast tumours. Nature 490(7418):61–70
Luen SJ, Asher R, Lee CK, Savas P, Kammler R, Dell’Orto P et al (2018) Association of somatic driver alterations with prognosis in postmenopausal, hormone receptor-positive, HER2-negative early breast cancer: a secondary analysis of the BIG 1–98 randomized clinical trial. JAMA Oncol 4(10):1335–1343
Griffith OL, Spies NC, Anurag M, Griffith M, Luo J, Tu D et al (2018) The prognostic effects of somatic mutations in ER-positive breast cancer. Nature Commun 9(1):1–16
Selli C, Turnbull AK, Pearce DA, Li A, Fernando A, Wills J, et al (2019) Molecular changes during extended neoadjuvant letrozole treatment of breast cancer: distinguishing acquired resistance from dormant tumours. Breast Cancer Res 21(1):2.
Dunbier AK, Ghazoui Z, Anderson H, Salter J, Nerurkar A, Osin P et al (2013) Molecular profiling of aromatase inhibitor-treated postmenopausal breast tumors identifies immune-related correlates of resistance. Clin Cancer Res 19(10):2775–2786
Miller WR, Larionov A, Anderson TJ, Evans DB, Dixon JM (2012 ) Sequential changes in gene expression profiles in breast cancers during treatment with the aromatase inhibitor, letrozole. Pharmacogenom J 12(1):10–21
Cizkova M, Susini A, Vacher S, Cizeron-Clairac G, Andrieu C, Driouch K et al (2012) PIK3CA mutation impact on survival in breast cancer patients and in ERα, PR and ERBB2-based subgroups. Breast Cancer Res 14(1):1–9
Loi S, Haibe-Kains B, Majjaj S, Lallemand F, Durbecq V, Larsimont D et al (2010) PIK3CA mutations associated with gene signature of low mTORC1 signaling and better outcomes in estrogen receptor–positive breast cancer. Proc Natl Acad Sci USA 107(22):10208–10213
Kalinsky K, Jacks LM, Heguy A, Patil S, Drobnjak M, Bhanot UK et al (2009) PIK3CA mutation associates with improved outcome in breast cancer. Clin Cancer Res 15(16):5049–5059
Ellis MJ, Lin L, Crowder R, Tao Y, Hoog J, Snider J et al (2009) Phosphatidyl-inositol-3-kinase alpha catalytic subunit mutation and response to neoadjuvant endocrine therapy for estrogen receptor positive breast cancer. Breast Cancer Res Treat 119(2):379
Stemke-Hale K, Gonzalez-Angulo AM, Lluch A, Neve RM, Kuo W-L, Davies M et al (2008) An integrative genomic and proteomic analysis of PIK3CA, PTEN, and AKT mutations in breast cancer. Cancer Res 68(15):6084–6091
Papaxoinis G, Kotoula V, Alexopoulou Z, Kalogeras KT, Zagouri F, Timotheadou E et al (2015) Significance of PIK3CA mutations in patients with early breast cancer treated with adjuvant chemotherapy: a Hellenic Cooperative Oncology Group (HeCOG) study. PLoS ONE 10(10):e0140293
Chia YH, Ellis MJ, Ma CX (2010 Sep 7) Neoadjuvant endocrine therapy in primary breast cancer: indications and use as a research tool. Br J Cancer 103(6):759–764
Christiansen P, Ejlertsen B, Jensen M-B, Mouridsen H (2016) Danish breast cancer cooperative group. Clin Epidemiol 8:445
Skriver SK, Laenkholm A-V, Rasmussen BB, Handler J, Grundtmann B, Tvedskov TF et al (2018) Neoadjuvant letrozole for postmenopausal estrogen receptor-positive, HER2-negative breast cancer patients, a study from the Danish Breast Cancer Cooperative Group (DBCG). Acta Oncol 57(1):31–37
Skriver SK, Jensen M-B, Knoop AS, Ejlertsen B, Laenkholm A-V (2020) Tumour-infiltrating lymphocytes and response to neoadjuvant letrozole in patients with early oestrogen receptor-positive breast cancer: analysis from a nationwide phase II DBCG trial. Breast Cancer Res 22(1):46
Kaufmann M, von Minckwitz G, Mamounas EP, Cameron D, Carey LA, Cristofanilli M et al (2012) Recommendations from an international consensus conference on the current status and future of neoadjuvant systemic therapy in primary breast cancer. Ann Surg Oncol 19(5):1508–1516
Ogston KN, Miller ID, Payne S, Hutcheon AW, Sarkar TK, Smith I et al (2003) A new histological grading system to assess response of breast cancers to primary chemotherapy: prognostic significance and survival. Breast 12(5):320–327
Symmans WF, Peintinger F, Hatzis C, Rajan R, Kuerer H, Valero V et al (2007) Measurement of residual breast cancer burden to predict survival after neoadjuvant chemotherapy. J Clin Oncol 25(28):4414–4422
Bossuyt V, Provenzano E, Symmans WF, Boughey JC, Coles C, Curigliano G et al (2015) Recommendations for standardized pathological characterization of residual disease for neoadjuvant clinical trials of breast cancer by the BIG-NABCG collaboration. Ann Oncol 26(7):1280–1291
Residual Cancer Burden Calculator [Internet]. MD Anderson Cancer Center. https://www3.mdanderson.org/app/medcalc/index.cfm?pagename=jsconvert3. Accessed 7 Oct 2019
Hammond MEH, Hayes DF, Dowsett M, Allred DC, Hagerty KL, Badve S et al (2010) American Society of Clinical Oncology/College of American Pathologists Guideline Recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. JCO 28(16):2784–2795
Wolff AC, Hammond MEH, Hicks DG, Dowsett M, McShane LM, Allison KH et al (2013) Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J Clin Oncol 31(31):3997–4013
Dowsett M, Nielsen TO, A’Hern R, Bartlett J, Coombes RC, Cuzick J et al (2011) Assessment of Ki67 in breast cancer: recommendations from the International Ki67 in Breast Cancer Working Group. J Natl Cancer Inst 103(22):1656–1664
Dieci MV, Radosevic-Robin N, Fineberg S, van den Eynden G, Ternes N, Penault-Llorca F et al (2018) Update on tumor-infiltrating lymphocytes (TILs) in breast cancer, including recommendations to assess TILs in residual disease after neoadjuvant therapy and in carcinoma in situ: a report of the International Immuno-Oncology Biomarker Working Group on Breast Cancer. Semin Cancer Biol 1(52):16–25
Krupke DM, Begley DA, Sundberg JP, Richardson JE, Neuhauser SB, Bult CJ (2017) The Mouse Tumor Biology Database (MTB): a comprehensive resource for mouse models of human cancer. Cancer Res 77(21):e67–70
McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University (Baltimore, MD). Online Mendelian Inheritance in Man, OMIM® [Internet]. https://omim.org/
Landrum MJ, Lee JM, Benson M, Brown GR, Chao C, Chitipiralla S, et al (2018) ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res 46(Database issue):D1062–D1067
Schemper M, Smith TL (1996) A note on quantifying follow-up in studies of failure time. Control Clin Trials 17(4):343–346
Loibl S, Volz C, Mau C, Blohmer J-U, Costa SD, Eidtmann H et al (2014) Response and prognosis after neoadjuvant chemotherapy in 1,051 patients with infiltrating lobular breast carcinoma. Breast Cancer Res Treat 144(1):153–162
Delpech Y, Coutant C, Hsu L, Barranger E, Iwamoto T, Barcenas CH et al (2013) Clinical benefit from neoadjuvant chemotherapy in oestrogen receptor-positive invasive ductal and lobular carcinomas. Br J Cancer 108(2):285–291
Ellsworth RE, Blackburn HL, Shriver CD, Soon-Shiong P, Ellsworth DL (2017) Molecular heterogeneity in breast cancer: state of the science and implications for patient care. Semin Cell Dev Biol 1(64):65–72
André F, Ciruelos E, Rubovszky G, Campone M, Loibl S, Rugo HS et al (2019) Alpelisib for PIK3CA-mutated, hormone receptor-positive advanced breast cancer. N Engl J Med 380(20):1929–1940
Verret B, Cortes J, Bachelot T, Andre F, Arnedos M (2019) Efficacy of PI3K inhibitors in advanced breast cancer. Ann Oncol 30(Suppl 10):x12–x20
Funding
This study was funded by The Danish Cancer Society (Grant No. R146-A9562), The Harboe Foundation, University of Copenhagen’s Foundation for cancer research (Grant No. 2019-0018) and The Sejer Persson and Lis Klüver foundation. The founding sources are all non-commercial and had no role in the design or execution of the study and have not reviewed the data or manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
SKS declares she has no conflict of interest. MBJ has received institutional grants from Nanostring Technologies Inc and Oncology Venture. JOE declares he has no conflict of interest none, LBA declares she has no conflict of interest, ASK has received an institutional grant from Roche and is on advisory board for: Novartis, Astra Zeneca, MDS, Roche, Pfizer and ELI LILLY DANMARK A/S. MR is on advisory board for Astra Zeneca, BE has received institutional grants from: Nanostring Technologies Inc, Novartis and Roche. AVL has received research grants from Nanostring Technologies Inc. and Roche.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. The study is registered on clinicaltrials.gov (NCT00908531). The Biomedical Research Ethics of the Danish Capital Region approved the protocol (H-15012740). The genomic investigations presented here were subsequently approved (H-16031391).
Informed consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Skriver, S.K., Jensen, MB., Eriksen, JO. et al. Induction of PIK3CA alterations during neoadjuvant letrozole may improve outcome in postmenopausal breast cancer patients. Breast Cancer Res Treat 184, 123–133 (2020). https://doi.org/10.1007/s10549-020-05833-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-020-05833-w