Abstract
Purpose
Although breast cancer in young women accounts for <10% of diagnoses annually, tumors in young patients exhibit more aggressive characteristics and higher mortality rates. Determination of the frequency of germline mutations in cancer predisposition genes is needed to improve the understanding of breast cancer etiology in young women.
Methods
All female patients enrolled in the Clinical Breast Cancer Project between 2001 and 2015 and diagnosed with invasive breast cancer before age 40 were included in this study. Family history was classified using the NCCN Familial Risk Assessment guidelines. Targeted sequencing of 94 cancer predisposition genes was performed using peripheral blood DNA. Variants were detected using VariantStudio and classified using ClinVar.
Results
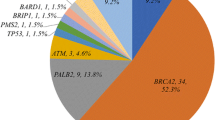
Seven percent (141/1980) of patients were young women and 44 had a significant family history. Sequencing was completed for 118 women with genomic DNA. Pathogenic mutations were present in 27 patients: BRCA1 (n = 10), BRCA2 (n = 12), TP53 (n = 1), and CHEK2 (n = 4). Mutations classified as pathogenic were also detected in APC (n = 1) and MUTYH (n = 2). Variants of uncertain significance (VUS) were detected in an additional 17 patients in ten genes.
Discussion
Pathogenic mutations in high- and moderate-risk breast cancer genes were detected in 23% of young women with an additional 3% having pathogenic mutations in colon cancer predisposition genes. VUS were observed in 14% of women in genes such as ATM, BRCA2, CDH1, CHEK2, and PALB2. Identification of those non-genetic factors is critical to reduce the burden of breast cancer in this population.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
More than 10,000 young women (<40 years of age) in the United States are diagnosed with breast cancer every year [1, 2] where the disease is associated with poor prognosis [3, 4]. Factors associated with breast cancer in young women include having children at an early age, ≥6 months oral contraceptive use, and breastfeeding [5, 6]. African ancestry has also been associated with increased risk of breast cancer, especially ER−, in young women [5]. Breast cancer in young women may have a hereditary component: for example, mutation rates for BRCA1 and BRCA2 are higher in young women (13%) compared to women aged 40–49 (2.2%) in the United Kingdom and in the United States (reviewed in [7]). The National Comprehensive Cancer Network (NCCN) guidelines include a diagnosis ≤45 years of age as a criterion for risk assessment, genetic counseling, and genetic testing of BRCA1 and BRCA2 [8].
Breast cancer genes other than BRCA1 and BRCA2 may account for breast cancer risk in young women. NCCN guidelines suggest that patients diagnosed before 31 years of age may be eligible for genetic testing of TP53, with up to 5% of young women with breast carcinomas harboring pathogenic mutations in TP53 [9]. The contribution to breast cancer risk in young women from other genes, including those associated with moderate risk for breast cancer and other genes associated with other types of cancer, is not well studied. Thus, to improve our understanding of the role of heritability in breast cancer in young women, panel testing of 94 cancer predisposition genes was performed in 118 young women diagnosed with breast cancer <40 years of age.
Methods
Between 2001 and 2015, 1980 female patients with invasive breast cancer enrolled in the Clinical Breast Care Project (CBCP). All subjects voluntarily agreed to participate in the CBCP and gave written informed consent. To be eligible to participate in this study, individuals were required to be (1) at least 18 years of age, (2) mentally competent and willing to sign the informed consent documents, and (3) a patient at Walter Reed National Military Medical Center, Bethesda, MD, Anne Arundel Medical Center, Annapolis, MD, or the Joyce Murtha Breast Care Center, Windber, PA, with evidence of current or past breast disease. Blood samples were collected with approval from the Walter Reed National Military Medical Center Human Use Committee and Institutional Review Board.
Once informed consent was granted, nurse researchers interviewed enrollees in person to collect over 500 fields of demographic data, including family cancer histories through third-degree relatives. Presence of a family history was determined using the NCCN Familial Risk Assessment criteria [10]. Age at diagnosis was calculated by subtracting the date of birth from the date of the diagnostic biopsy. Pathologic evaluation included determination of tumor stage, size, grade, and lymph node status [11,12,13]. ER, progesterone receptor (PR), and HER2 status were determined using ASCO/CAP guidelines [14, 15].
Genomic DNA was isolated from blood clots using the Gentra Clotspin and Puregene DNA purification kits (Qiagen, Valencia, CA). Samples were quantitated using the Qubit™ 3.0 Fluorometer (ThermoFisher Scientific, Waltham, MA). Libraries were created from 50 ng of DNA using the TruSight Rapid Capture kit and TruSight Cancer panel and sequenced on a MiSeq (Illumina, Inc, San Diego, CA) according to manufacturer’s protocols. Data were analyzed using VariantStudio version 3.0. (Illumina, Inc, San Diego, CA) and filtered to include only those variants with a read depth of ≥10 and a minor allele frequency of ≥0.25. Variants representing missense or frameshift mutations, stop gains or losses, initiator codons, in-frame insertions or deletions, and splice site alterations were included in the analysis. The predicted effect of variants was evaluated using the ClinVar database (http://www.clinvar.com/) and classified as pathogenic, likely pathogenic, uncertain significance (VUS), likely benign, or benign.
Results
Clinicopathological characteristics
One hundred forty-one (7%) patients with invasive breast cancer enrolled in the CBCP were diagnosed <40 years of age; 119 enrolled at the time of diagnosis, while the other 22 enrolled in the CBCP as post-treatment survivors. Forty-four patients (31%) had a family history of cancer. The majority of patients (90%) had unilateral disease and surgical options were divided between unilateral mastectomy (35%), breast-conserving surgery (34%), and double mastectomy (31%). Forty-two percent of young women underwent clinical testing for BRCA1 and BRCA2 mutations. Most of the patients were self-described European American (66%) followed by African American (28%; Table 1); European American women were more likely to report a family history of cancer (31%) than African American women (15%) although this difference was not significant (P = 0.467). The majority of patients were diagnosed more than two years after the last childbirth, although three patients (2%) had children after diagnosis.
Pathologically, the majority of the tumors were size T1 or T2 at diagnosis, poorly differentiated, lymph node negative, and ER+HER2−. Twenty-nine percent of women were diagnosed with stage III or IV tumors and 25% with TNBC. Sixteen percent of young women died of disease with a mean survival of 4.77 years (range 10 months–20.1 years) and patients alive without disease had a mean follow-up of 9.36 years. No patients died of causes other than breast cancer.
Mutation status
Of the 118 samples subjected to targeted sequencing, 117 (99%) had at least 30× coverage, with the remaining sample having 28.3× coverage. The average number of reads passing filter was 974,861. The average Q30 score was 94.3%.
Pathogenic or likely pathogenic mutations or VUS were detected in 15/94 (16%) cancer predisposition genes (Supplemental Table 1). DNA variants represented (1) pathogenic mutations in known breast cancer genes, (2) pathogenic mutations in colon cancer genes, and (3) VUS in a range of cancer predisposition genes. Pathogenic or likely pathogenic genes associated with hereditary breast cancer-predisposing syndromes were detected in 27/118 (23%) patients and included mutations in BRCA1 (n = 10), BRCA2 (n = 12), CHEK2 (n = 4), and TP53 (n = 1) (Table 2). Three patients had pathogenic mutations in the hereditary colon cancer genes APC (n = 1) and MUTYH (n = 2) (Table 3). Four patients with pathogenic mutations also carried VUS in BRCA2, BRIP1, CHEK2, MSH2, and PMS2. An additional 17 patients (14%) harbored VUS in 10 genes including APC (n = 1), ATM (n = 4), BRCA2 (n = 1), BRIP1 (n = 2), CDH1 (n = 2), CHEK2 (n = 1), MLH1 (n = 2), MSH6 (n = 1), PALB2 (n = 1), and PMS2 (n = 2) (Table 4). All patients with BRCA1 mutations and ER, PR, and HER2 data available had TNBC and 80% had a family history of cancer. In patients with BRCA2 mutations and ER, PR, and HER2 available, one patient with a 5104delAA mutation had TNBC, while all others had ER+ tumors with 80% ER+/HER2− and 20% ER+/HER2+ (luminal-HER2); 50% of young women with a mutation in BRCA2 had a significant family history. Mutation carriers were more common in African American women (29%) compared to European American women (22%) and the VUS in BRCA2 (S2059N) was detected in an African American woman with a luminal-HER2 tumor but without a significant family history of cancer.
Effect of mutational status on clinicopathological features
Clinicopathological factors were compared between young women with pathogenic mutations and those with no mutations detected in any of the 94 cancer predisposition genes; given the uncertain status of women with VUS, these 17 patients were excluded from the analyses. Presence of a family history was significantly higher (P < 0.002) in mutation carriers (57%) compared to non-carriers (24%); no other clinicopathological factors differed significantly (Table 5).
Discussion
Mutation frequencies in BRCA1 and BRCA2 in young women from the United States, Canada, Europe, China, and Brazil range from 9 to 23% [16,17,18,19,20,21,22,23,24]. Panel testing increases the frequency of detection of pathogenic mutations to 18–26% [25,26,27,28]. In our study which represents, to our knowledge, the first study to utilize panel testing to identify germline mutations in cancer predisposition genes in young women unselected for family history, ethnicity, or subtype, we detected a mutation frequency of 23% in known breast cancer genes and 3% in genes associated with other familial cancers.
Clinical management will differ between young women with and without pathogenic mutations in breast cancer genes. The majority of women did not have germline mutations and should have received standard care appropriate to her tumor subtype and/or somatic mutations. In contrast, options for the 22 young women with BRCA1 and BRCA2 mutations include increased surveillance, prophylactic mastectomy of the unaffected breast, and oophorectomy. Of the 50 women in this study that had clinical testing for BRCA1 and BRCA2, 12 harbored pathogenic BRCA1 or BRCA2 mutations. One additional woman was tested for and found to harbor a TP53 mutation. Within these young women who underwent clinical testing, surgical treatments differed significantly (P = 0.024) between those who had pathogenic mutations and those with negative BRCA1 and BRCA2 results. All of the mutation carriers opted for mastectomy: seven (54%) had a unilateral mastectomy of which four developed contralateral breast cancer and one recurred; the other 46% of carriers had a double mastectomy. In those women who tested negative for BRCA1 and BRCA2 mutations, 26% opted for breast-conserving surgery, 24% for unilateral mastectomy, and 50% for double mastectomy. Rates of oophorectomy did not differ significantly between mutation carriers and non-carriers.
Utilization of gene panels, rather than single-gene tests, identified an additional four young women who carried germline pathogenic mutations in CHEK2. The CHEK2 1100delC represents a high-risk allele associated with greater than twofold increase in risk and poor prognosis; in contrast, the I157T allele is associated with moderate risk (1.4-fold) and more favorable prognosis [29]. Functional assays demonstrate that the R117G allele encodes a functionally defective protein; however, the magnitude of risk is not well characterized. Although none of the patients received clinical testing for CHEK2, both women with the I157T allele opted for double mastectomy, as did the woman with the 1100delC mutation.
In addition to identification of women with BRCA1 and BRCA2 mutations who had not had clinical testing and women with CHEK2 mutations, we also identified three young women with breast cancer who had mutations in hereditary colon cancer genes. While these mutations may be secondary to the development of breast tumors, they are associated with increased risk for colon cancer. One woman died of breast cancer within three years of her breast cancer diagnosis, but the other two may benefit from increased colon cancer screening. Other studies have demonstrated that identification of mutations in non-breast cancer genes through panel testing may result in changes in clinical management. DNA samples from 198 women referred for BRCA1 and BRCA2 testing were subjected to panel testing of 42 cancer-associated genes; 29% of the women carried BRCA1 or BRCA2 mutations, while an additional 15 women had pathogenic mutations in other genes [30]. Six of these women had actionable mutations in CDH1, MLH1, and MUTYH and were advised to undergo frequent colonoscopy or endoscopy; one of these women was found to have a tubular adenoma, an incidental tumor that may not have been discovered had panel testing not been performed.
While the percentage of young women with invasive breast cancer enrolled in the CBCP (7%) is similar to national estimates [1], this represents only 141 patients enrolled over a 14-year period. Larger sample sizes may alter frequency estimates or patterns of germline mutations; however, our finding of a mutation frequency of 23% pathogenic mutations is similar to larger studies when subpopulations of younger women were considered. Patients in the CBCP enroll as individuals, and thus medical records and DNA samples from family members are not available to determine segregation patterns for the identified risk alleles, thereby limiting the ability to improve classification of the 17 VUS identified in this cohort. Finally, the use of a targeted sequencing approach precludes the ability to detect large rearrangements, which have been shown to account for up to 6% of BRCA1 and BRCA2 mutations in high-risk individuals from the United States [31]. Given this frequency, a small number (2–3) of young women with a family history of breast cancer may have undetected large rearrangements in BRCA1 or BRCA2, thus increasing the mutation rate in known breast cancer genes from 23 to 26%.
In conclusion, etiology of breast cancer has been associated with pathogenic mutations in 23% of known breast cancer predisposition genes, with an additional 3% of young women harboring mutations in hereditary colon cancer genes and 14% having VUS in cancer predisposition genes. While 43% of young women with family histories harbored pathogenic mutations, 15% of young women without a significant family history of cancer also harbored pathogenic mutations. The majority of mutations were detected in BRCA1 and BRCA2, associated with TNBC and ER+ tumors, respectively, and no other cancer predisposition genes accounted for a significant proportion of tumors in young women. Together, these data suggest that the majority of breast tumors in young women are not linked to germline mutations but may be attributable to non-genetic factors.
References
American Cancer Society (2015) Breast cancer facts & figures 2015-2016. American Cancer Society Inc, Atlanta
Brinton LA, Sherman ME, Carreon JD, Anderson WF (2008) Recent trends in breast cancer among younger women in the United States. J Natl Cancer Inst 100(22):1643–1648. doi:10.1093/jnci/djn344
Gnerlich JL, Deshpande AD, Jeffe DB, Sweet A (2009) Elevated breast cancer mortality in women younger than age 40 years compared with older women is attributed to poorer survival in early-stage disease. J Am Coll Surg 208(3):341–347
Nixon AJ, Neuberg D, Hayes DF, Gelman R, Connolly JL, Schnitt S, Abner A, Recht A, Vicini F (1994) Relationship of patient age to pathologic features of the tumor and prognosis for patients with stage I or II breast cancer. J Clin Oncol 12(5):888–894
Althuis MD, Brogan DD, Coates RJ, Daling JR, Gammon MD, Malone KE, Schoenberg JB, Brinton LA (2003) Breast cancers among very young premenopausal women (United States). Cancer Causes Control 14(2):151–160
Chollet-Hinton L, Anders CK, Tse CK, Bell MB, Yang YC, Carey LA, Olshan AF, Troester MA (2016) Breast cancer biologic and etiologic heterogeneity by young age and menopausal status in the Carolina Breast Cancer Study: a case-control study. Breast Cancer Res 18(1):79. doi:10.1186/s13058-016-0736-y
Gewefel H, Salhia B (2014) Breast cancer in adolescent and young adult women. Clin Breast Cancer 14(6):390–395. doi:10.1016/j.clbc.2014.06.002
National Comprehensive Cancer Network (2016) Genetic/familial high-risk assessment: breast and ovarian cancer, vol 2.2017. National Comprehensive Cancer Network, Fort Washington, PA.
Lee DS, Yoon SY, Looi LM, Kang P, Kang IN, Sivanandan K, Ariffin H, Thong MK, Chin KF, Mohd Taib NA, Yip CH, Teo SH (2012) Comparable frequency of BRCA1, BRCA2 and TP53 germline mutations in a multi-ethnic Asian cohort suggests TP53 screening should be offered together with BRCA1/2 screening to early-onset breast cancer patients. Breast Cancer Res 14(2):R66. doi:10.1186/bcr3172
National Comprehensive Cancer Network (2016) Breast cancer risk reduction. 1.2017 edn. National Comprehensive Cancer Network, Fort Washington, PA.
American Joint Committee on Cancer (2010) AJCC cancer staging manual, vol 7th. Springer, New York
Bloom HJ, Richardson WW (1957) Histological grading and prognosis in breast cancer; a study of 1409 cases of which 359 have been followed for 15 years. Br J Cancer 11(3):359–377
Elston CW, Ellis IO (1991) Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology 19(5):403–410
Hammond MEH, Hayes DF, Dowsett M, Allred DC, Hagerty KL, Badve S, Fitzgibbons PL, Francis G, Goldstein NS, Hayes M, Hicks DG, Lester S, Love R, Mangu PB, McShane L, Miller K, Osborne CK, Paik S, Perlmutter J, Rhodes A, Sasano H, Schwartz JN, Sweep FCG, Taube S, Torlakovic EE, Valenstein P, Viale G, Visscher D, Wheeler T, Williams RB, Wittliff JL, Wolff AC (2010) American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J Clin Oncol 28(16):2784–2795
Wolff AC, Hammond ME, Hicks DG, Dowsett M, McShane LM, Allison KH, Allred DC, Bartlett JM, Bilous M, Fitzgibbons P, Hanna W, Jenkins RB, Mangu PB, Paik S, Perez EA, Press MF, Spears PA, Vance GH, Viale G, Hayes DF, American Society of Clinical O, College of American P (2013) Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J Clin Oncol 31(31):3997–4013. doi:10.1200/JCO.2013.50.9984
Carraro DM, Koike Folgueira MA, Garcia Lisboa BC, Olivieri EHR, Krepischi ACV, de Carvalho AF, de Carvalho Mota LD, Puga RD, do Socorro Maciel M, Michelli RA, de Lyra EC, Grosso SH, Soares FA, Achatz MI, Brentani H, Moreira-Filho CA, Brentani MM (2013) Comprehensive analysis of BRCA1, BRCA2 and TP53 germline mutation and tumor characterization: a portrait of early-onset breast cancer in Brazil. PLoS One 8(3):e57581. doi:10.1371/journal.pone.0057581
de Sanjose S, Leone M, Berez V, Izquierdo A, Font R, Brunet JM, Louat T, Vilardell L, Borras J, Viladiu P, Bosch FX, Lenoir GM, Sinilnikova OM (2003) Prevalence of BRCA1 and BRCA2 germline mutations in young breast cancer patients: a population-based study. Int J Cancer 106(4):588–593. doi:10.1002/ijc.11271
Musolino A, Bella MA, Bortesi B, Michiara M, Naldi N, Zanelli P, Capelletti M, Pezzuolo D, Camisa R, Savi M, Neri TM, Ardizzoni A (2007) BRCA mutations, molecular markers, and clinical variables in early-onset breast cancer: a population-based study. Breast 16(3):280–292. doi:10.1016/j.breast.2006.12.003
Meindl A, German Consortium for Hereditary B, Ovarian C (2002) Comprehensive analysis of 989 patients with breast or ovarian cancer provides BRCA1 and BRCA2 mutation profiles and frequencies for the German population. Int J Cancer 97(4):472–480
Tonin PN, Perret C, Lambert JA, Paradis AJ, Kantemiroff T, Benoit MH, Martin G, Foulkes WD, Ghadirian P (2001) Founder BRCA1 and BRCA2 mutations in early-onset French Canadian breast cancer cases unselected for family history. Int J Cancer 95(3):189–193
Malone KE, Daling JR, Neal C, Suter NM, O’Brien C, Cushing-Haugen K, Jonasdottir TJ, Thompson JD, Ostrander EA (2000) Frequency of BRCA1/BRCA2 mutations in a population-based sample of young breast carcinoma cases. Cancer 88(6):1393–1402
Langston AA, Malone KE, Thompson JD, Daling JR, Ostrander EA (1996) BRCA1 mutations in a population-based sample of young women with breast cancer. N Engl J Med 334(3):137–142. doi:10.1056/NEJM199601183340301
FitzGerald MG, MacDonald DJ, Krainer M, Hoover I, O’Neil E, Unsal H, Silva-Arrieto S, Finkelstein DM, Beer-Romero P, Englert C, Sgroi DC, Smith BL, Younger JW, Garber JE, Duda RB, Mayzel KA, Isselbacher KJ, Friend SH, Haber DA (1996) Germ-line BRCA1 mutations in Jewish and non-Jewish women with early-onset breast cancer. N Engl J Med 334(3):143–149. doi:10.1056/NEJM199601183340302
Loman N, Johannsson O, Kristoffersson U, Olsson H, Borg A (2001) Family history of breast and ovarian cancers and BRCA1 and BRCA2 mutations in a population-based series of early-onset breast cancer. J Natl Cancer Inst 93(16):1215–1223
Tung N, Lin NU, Kidd J, Allen BA, Singh N, Wenstrup RJ, Hartman AR, Winer EP, Garber JE (2016) Frequency of germline mutations in 25 cancer susceptibility genes in a sequential series of patients with breast cancer. J Clin Oncol 34(13):1460–1468. doi:10.1200/JCO.2015.65.0747
Couch FJ, Hart SN, Sharma P, Toland AE, Wang X, Miron P, Olson JE, Godwin AK, Pankratz VS, Olswold C, Slettedahl S, Hallberg E, Guidugli L, Davila JI, Beckmann MW, Janni W, Rack B, Ekici AB, Slamon DJ, Konstantopoulou I, Fostira F, Vratimos A, Fountzilas G, Pelttari LM, Tapper WJ, Durcan L, Cross SS, Pilarski R, Shapiro CL, Klemp J, Yao S, Garber J, Cox A, Brauch H, Ambrosone C, Nevanlinna H, Yannoukakos D, Slager SL, Vachon CM, Eccles DM, Fasching PA (2015) Inherited mutations in 17 breast cancer susceptibility genes among a large triple-negative breast cancer cohort unselected for family history of breast cancer. J Clin Oncol 33(4):304–311. doi:10.1200/JCO.2014.57.1414
Churpek JE, Walsh T, Zheng Y, Moton Z, Thornton AM, Lee MK, Casadei S, Watts A, Neistadt B, Churpek MM, Huo D, Zvosec C, Liu F, Niu Q, Marquez R, Zhang J, Fackenthal J, King MC, Olopade OI (2015) Inherited predisposition to breast cancer among African American women. Breast Cancer Res Treat 149(1):31–39. doi:10.1007/s10549-014-3195-0
Lin PH, Kuo WH, Huang AC, Lu YS, Lin CH, Kuo SH, Wang MY, Liu CY, Cheng FT, Yeh MH, Li HY, Yang YH, Hsu YH, Fan SC, Li LY, Yu SL, Chang KJ, Chen PL, Ni YH, Huang CS (2016) Multiple gene sequencing for risk assessment in patients with early-onset or familial breast cancer. Oncotarget 7(7):8310–8320. doi:10.18632/oncotarget.7027
Muranen TA, Blomqvist C, Dork T, Jakubowska A, Heikkila P, Fagerholm R, Greco D, Aittomaki K, Bojesen SE, Shah M, Dunning AM, Rhenius V, Hall P, Czene K, Brand JS, Darabi H, Chang-Claude J, Rudolph A, Nordestgaard BG, Couch FJ, Hart SN, Figueroa J, Garcia-Closas M, Fasching PA, Beckmann MW, Li J, Liu J, Andrulis IL, Winqvist R, Pylkas K, Mannermaa A, Kataja V, Lindblom A, Margolin S, Lubinski J, Dubrowinskaja N, Bolla MK, Dennis J, Michailidou K, Wang Q, Easton DF, Pharoah PD, Schmidt MK, Nevanlinna H (2016) Patient survival and tumor characteristics associated with CHEK2:p.I157T—findings from the Breast Cancer Association Consortium. Breast Cancer Res 18(1):98. doi:10.1186/s13058-016-0758-5
Kurian AW, Hare EE, Mills MA, Kingham KE, McPherson L, Whittemore AS, McGuire V, Ladabaum U, Kobayashi Y, Lincoln SE, Cargill M, Ford JM (2014) Clinical evaluation of a multiple-gene sequencing panel for hereditary cancer risk assessment. J Clin Oncol 32(19):2001–2009. doi:10.1200/JCO.2013.53.6607
Palma MD, Domchek SM, Stopfer J, Erlichman J, Siegfried JD, Tigges-Cardwell J, Mason BA, Rebbeck TR, Nathanson KL (2008) The relative contribution of point mutations and genomic rearrangements in BRCA1 and BRCA2 in high-risk breast cancer families. Cancer Res 68(17):7006–7014. doi:10.1158/0008-5472.CAN-08-0599
Acknowledgements
We thank Darrell L. Ellsworth, PhD for his kind review of this manuscript. The views expressed in this article are those of the authors and do not reflect the official policy of the Department of Defense or U.S. Government. This research was supported by a grant from the Office of the Congressionally Directed Medical Research Programs (Department of Defense Breast Cancer Research Program W81XWH-11-2-0135).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Rummel, S.K., Lovejoy, L., Shriver, C.D. et al. Contribution of germline mutations in cancer predisposition genes to tumor etiology in young women diagnosed with invasive breast cancer. Breast Cancer Res Treat 164, 593–601 (2017). https://doi.org/10.1007/s10549-017-4291-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-017-4291-8