Abstract
Epithelial–mesenchymal transition (EMT) is a crucial step in epithelial cancer invasion and metastasis. miR-153 has been identified as a key EMT suppressor. Accordingly, this study aimed to determine the possible relation of miR-153 downregulation to EMT through MTDH modulation. The miR-153 and MTDH expression profiles of human breast cancer specimen were determined by qPCR and evaluated by correlation analysis. Cell viability and clonogenic assays were applied to explore the impact of miR-153 on suppression of proliferation and oncogenic potential of breast cancer cells. Cell migration and invasion assays were used for the functional analysis of miR-153 in MCF-7 and MDA-MB-231 cells. Luciferase assay was adopted to identify MTDH as a new direct target of miR-153. Ectopic expression of miR-153 could significantly inhibit tumor growth and impair the migration and invasion of breast cancer cells. Overexpression of miR-153 simultaneously increased E-cadherin, decreased vimentin expression, and downmodulated EMT-associated transcription factors. miR-153 was negatively correlated with MTDH in cell lines and clinical samples. Overexpression of miR-153 significantly suppressed MTDH, as demonstrated by in vitro MTDH 3′-untranslated region luciferase report assay. MTDH is a direct downstream target of miR-153 and is involved in the miR-153-induced suppression of the migration and invasion of breast cancer cells. Our findings indicate that miR-153 functions as a tumor suppressor and miR-153/MTDH link is a promising therapeutic target for breast cancer.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Breast cancer is the most common malignancy in women around the world. Invasion and metastasis remain the main obstacles in the effective treatment of this disease. Thus, research on the molecular mechanisms leading to invasive and metastatic dissemination of carcinoma cells is receiving increased interest. Recent evidence indicates that epithelial–mesenchymal transition (EMT) is a critical step in cancer cell invasion and metastasis. EMT has been found to be positively correlated with poor prognosis in breast cancer patients [1, 2].
Metadherin (MTDH) is originally identified as an oncogene and its abnormalities have been identified in many types of human malignancy [3, 4]. MTDH could enhance invasion ability by inducing EMT in human breast cancer cells through the NF-kappaB, Ha-ras, PI3K/Akt, and Wnt/beta-catenin signaling pathways [5, 6].
miRNAs reportedly play an important role in tumorigenesis and metastasis in human cancer [7, 8]. Recently, miR-153 has been found to be markedly downregulated in cells when EMT occurs [9]. More data have demonstrated that miR-153 is a novel regulator of EMT by targeting SNAI1 and ZEB2 [9]. MTDH has further been identified as a direct target of miR-145 and miR-22 in ovarian and gastric cancer [10, 11]. Given that miR-153 has been marked as a key EMT suppressor, close attention is paid to the modulation of EMT through the crosstalk between miR-153 and MTDH.
Materials and methods
Patients and tissue samples
Invasive breast carcinoma and the adjacent normal tissues were collected in The Affiliated Hospital of Weifang Medical University from April 2008 to July 2012. The specimens were obtained from primary breast cancer patients receiving no surgery or chemotherapy previously. Written informed consent was obtained from all participants, as delineated by the protocol (approved by the Ethical Committee of Weifang Medical University).
Cell lines and reagents
Breast cancer cell lines MCF-7 and MDA-MB-231 were obtained from American Type Culture Collection (Rockville, MD, USA). They were routinely cultured in DMEM supplemented with 10 % FBS at 37 °C with 5 % CO2. Rabbit anti-E-cadherin, anti-vimentin antibodies were purchased from Cell Signaling Technology (Beverly, MA, USA) and rabbit anti-MTDH antibody was purchased from Invitrogen (Carlsbad, CA, USA).
Plasmid construction and transfection
For MTDH overexpression, the cDNA of MTDH was cloned into the multiple cloning site of the pcDNA3.1 as previously described [12]. The sequences of miR-153 mimic were as follows: 5′-UUGCAUAGUCACAAAAGUGAUC-3′/5′-GAUCACUUUUGUGACUAUGCAA-3′. The sequence of miR-153 inhibitor was as follows: 5′-AUCACUU UUGUGACUAUGCA-3′ [9]. Cells were seeded at 2 × 105 cells per well in a six-well plate and transfected using Lipofectamine according to the manufacturer’s instructions. Total RNA and protein were collected for assay 2 days post-transfection.
Cell proliferation and clonogenic assay
Cell proliferation was assessed by MTT assay as previously described [10]. To determine clonogenic ability, cells were allowed to grow for 14 days to form colonies, which were then stained with crystal violet (2 %, w/v; Sigma, St Louis, MO, USA).
Scratch assay
For the scratch assay, cells were grown to confluence in a 24-well plate, and a “w“unding” ”ine was scratched into the cell monolayer with a sterile 200 μL pipette tip. The width of the wound was measured under microscope at 0 and 24 h after the scratch to assess the cell migration ability.
Transwell invasive assay
Invasion assay was performed using a Transwell system according to manuals. Chambers were incubated at 37 °C for 24 h, and three duplicates were prepared for each group. Successfully translocated cells were fixed and stained with 0.2 % crystal violet. The total cell numbers of five visual fields at the center and in the surrounding areas were counted, and the average was calculated.
RNA extraction and qPCR
Total RNA from tissues and cultured cells was extracted using Trizol (Invitrogen) according to the manufacturer’s instructions. The specific primers were designed as follows: SNAI1: 5′-GCTGCAGGACTCTAATCCAGA-3′/5′-ATCTCCGGAGGTGGGATG-3′, ZEB2: 5′-AGGAGCTGTCTCGCCTTG-3′/5′-GGCAAAAGCATCTGGAGTTC-3′ and GAPDH: 5′-CATGAGAAGTATGACAACAGCCT-3′/5′-AGTCCTTCCACGATACCAAAGT-3′. GAPDH was used as the internal control. mRNA expression was quantified using the \(\Delta \Delta C_{\text{t}}\) method. For miRNA analysis, qPCR was performed using PrimeScript® miRNA RT-PCR Kit (Takara) according to the manufacturer’s instructions. The entire sequences of miR-153 (TTGCATAGTCACAAAAGTGATC), or U6 (CGCAAGGATGACACG CAAATTCGT) were used as specific forward primers in combination with the universal reverse quantitative PCR primers provided in the kit. All miRNA data were expressed relative to a U6 small nuclear RNA from the same sample.
Immunohistochemistry
The resected specimens were paraffin embedded and stored at 4 °C. The expression of MTDH was detected through immunohistochemistry and the intensity of positive staining was measured with integrated optical density (IOD). The intensity of MTDH was classified as either high or low expression (median value of IOD (mIOD) was used as the cutoff value; MTDHhigh:IOD > mIOD, and MTDHlow:IOD ≤ mIOD).
E-cadherin and vimentin immunofluorescence
Cells were grown on coverslips, fixed in 4 % paraformaldehyde for 20 min, incubated in a blocking buffer (1 % BSA and 0.25 % Triton X-100 in PBS; pH 7.4), and then probed with an E-cadherin antibody or vimentin antibody. To detect nuclei, cells were costained with DAPI.
MTDH 3′-untranslated region (UTR) reporter analysis
The 3′-UTR of MTDH containing a putative miR-153 binding site was amplified and cloned into pGL3 vector to generate a wild-type construct. For mutant plasmid, overlap extension PCR assay was used as previously described [13]. Cells were cultured in 24-well plates, for a transfection complex. About 2 µl of 20 µM chemically synthesized miR-153 mimic, 150 ng of pGL3 reporter plasmid and 50 ng of pRL-TK plasmid were mixed with Lipofectamine 2000. Luciferase activities were measured according to the manufacturer’s instructions (Dual-Luciferase Assay System; Promega). Renilla luciferase activity was normalized to corresponding firefly luciferase activity and plotted as a percentage.
Western blot analysis
Cells were lysed in RIPA buffer, and protein was separated by SDS-PAGE and then transferred to nitrocellulose. The blots were probed with primary antibodies and then incubated with 1:5000 secondary antibody. Signals were detected with enhanced chemiluminescence, and resulting images were analyzed with Gel-Pro Analyzer software. The membranes were stripped and probed with monoclonal antibody for β-actin as loading control.
Statistical analysis
SPSS V18.0 was used for statistical analysis. Student′s t test and one-way ANOVA were used to determine the significance of two groups and multiple groups, respectively. Spearman′s correlation was used to identify the correlation between the expression of miR-153 and MTDH. All error bars represent the SE of three experiments. Differences with P < 0.05 were considered significant.
Results
Downregulation of miR-153 in breast cancer tissues and cell lines
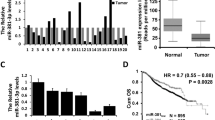
The expression of miR-153 in tumor samples was significantly lower than that in benign tissues (Fig. 1a). Clinicopathological analysis further revealed that the downregulation of miR-153 was significantly associated with the molecular-based classification and lymph node metastasis (Table 1). The expression of miR-153 in patients without lymph node metastasis was significantly higher than in those who had at least one positive lymph node (Fig. 1b). We also found that the expression levels of miR-153 in breast cancer cell lines were much lower than in MCF-10A (Fig. 1c). Moreover, miR-153 was revealed to be a potential participant miRNA in TGF-β-mediated-EMT by qPCR (Fig. 1d).
miR-153 expression is downregulated in breast cancer. a Expression of miR-153 in each sample was quantitatively analyzed using qPCR and normalized with U6 snRNA. b Validation of miR-153 in fresh primary breast cancer specimens with or without lymph node metastasis. c Relative miR-153 expression levels of breast cancer cell lines and a normal mammary epithelial cell line were examined with qPCR assay. d Expression of miR-153 in breast cancer cell lines with or without TGF-β treatment. *P < 0.05, **P < 0.01
Ectopic expression of miR-153 suppresses migration and invasion of breast cancer cells in vitro
To determine whether miR-153 functionally behaved as a tumor suppressor, we overexpressed miR-153 in MDA-MB-231 and MCF-7 cells (Fig. 2a). As shown in Fig. 2b, cells overexpressing miR-153 exhibited epithelial morphology. We found that miR-153 could significantly suppress invasion by Transwell assay (Fig. 2c), and that MCF-7 cells overexpressing miR-153 migrated much more slowly than the control cells by wound scratch assay (Fig. 2d). Overexpression of miR-153 significantly suppressed the proliferation of breast cancer cells (Fig. 2e). We also adopted the clonogenic assay to evaluate the oncogenic potential of miR-153, as shown in Fig. 2f. Colony-forming efficiency was found to be significantly reduced in miR-153 overexpressed cells.
Overexpression of miR-153 suppressed cell proliferation, migration, and invasion ability in vitro. a Overexpression of miR-153 was validated with qPCR. b Morphological changes in miR-153 overexpressed cells. c Reduction of invasion caused by expression of miR-153. d Overexpression of miR-153 resulted in a significant decrease in migratory ability of MCF-7 and MDA-MB-231 cells. e Ectopic expression of miR-153 on the proliferation in MCF-7 (left panel) and MDA-MB-231 (right panel) cells is shown as growth curve. f Overexpression of miR-153 on the clonogenic ability of MCF-7 cells. The upper panel illustrates the number of colonies formed, and the lower panel represents dishes by colony formation assay. *P < 0.05, **P < 0.01
miR-153 inhibited the EMT process of breast cancer cells
To determine whether the inhibitory effect of miR-153 on migration and invasion was mediated by EMT, we examined the expression of several EMT markers. As expected, miR-153 overexpression increased the expression level of E-cadherin and decreased the level of vimentin (Fig. 3a, b). The majority of the EMT-associated genes tested were downmodulated by miR-153 overexpression, and SNAI1 was the most severely affected (Fig. 3c).
Ectopic expression of miR-153 induced mesenchymal-to-epithelial transformation. a E-cadherin and vimentin immunofluorescence in MCF-7 cells. b Expression of E-cadherin and vimentin in MCF-7 and MDA-MB-231 cells by Western blot. c Ectopic expression of miR-153 inhibited the expression of EMT-associated genes. *P < 0.05, **P < 0.01
miR-153 was negatively correlated with MTDH in cell lines and clinical samples and can function as a tumor suppressor
To further validate that miR-153 suppressed invasion and metastasis by targeting MTDH, the expression levels of miR-153 and MTDH were detected in a variety of breast cancer cell lines. As indicated in Fig. 4a, the expression levels of miR-153 and MTDH were inversely correlated with each other. Afterward, the clinical relevance of the above findings was evaluated. We measured the expression of MTDH in a cohort of breast cancer samples (Fig. 4b), and the relative expression level of MTDH was plotted against that of miR-153 in each sample. Lower miR-153 expression was observed in breast cancer tissues with higher immunostaining of MTDH (Fig. 4c), and a moderate negative correlation of MTDH with miR-153 was observed (Fig. 4d).
miR-153 was negatively correlated with MTDH in cell lines and clinical samples. a Diagram represents the relative miR-153 and MTDH mRNA expression levels in breast cancer cells. b 1 Expression of MTDH in MCF-7 cells, 2 negative normal sample for MTDH expression, 3 weak positive tumor sample for MTDH, and 4 example of strong immunoreactivity for MTDH. c miR-153 was downregulated in breast cancer tissues (MTDHlow vs. normal, P < 0.01; MTDHhigh vs. normal, P < 0.01; MTDHlow vs. MTDHhigh, P < 0.05). d In breast cancer tissues, lower miR-153 expression was accompanied with higher immunostaining of MTDH
miR-153 directly targeted the MTDH 3′ 3′-UTR)
Computational prediction with TargetScan software revealed that an evolutionarily conserved region in the 3′-UTR of MTDH had a perfect complementary matching region to the seed sequence of miR-153 (Fig. 5a). As shown in Fig. 5b, the ectopic overexpression of miR-153 led to the inhibition of MTDH expression. By contrast, miR-153 inhibitor dose-dependently restored MTDH expression (Fig. 5c). To confirm that MTDH was the direct target of miR-153, we introduced MTDH 3′-UTR and corresponding mutant counterparts into pGL3 luciferase reporter vector (Fig. 5d). miR-153 overexpression reduced luciferase activity in cells transfected with the wild-type 3′-UTR of MTDH but not in cells with mutant 3′-UTR in MCF-7 cells (Fig. 5e).
Oncogene MTDH was specifically targeted by miR-153. a Alignment of miR-153-targeting sequences located in the 3′-UTR of the MTDH genes from 15 organisms; evolutionarily conserved nucleotides are highlighted in white. b miR-153 overexpression reduced MTDH protein expression in MCF-7 and MDA-MB-231 cells. c Different concentrations of miR-153 inhibitor transfection gradually increased MTDH expression in MCF-7 cells. d Sequences of wild type and mutant 3′-UTR showed the segments cloned into luciferase reporter plasmid. e Overexpression of miR-153 suppressed MTDH 3′-UTR luciferase activity. **P < 0.01
Exogenous MTDH overturned the inhibitory effects of miR-153 on breast cancer cells
We established four stable subgroups of cells and measured the expression levels of miR-153 and MTDH by qPCR. As shown in Fig. 6a, we successfully established these four subgroups of cells for rescue experiment. We then attempted to test whether restoration of MTDH could reverse the miR-153-mediated inhibition of invasion ability of breast cancer cells. As shown in Fig. 6b, overexpression of MTDH with a cDNA without 3′-UTR could partially abrogate the miR-153-mediated suppression of the migration and invasion of breast cancer cells.
Overexpression of MTDH reversed the inhibitory effects of miR-153 on breast cancer cells. a qPCR analysis showed MTDH expression in cells transfected with different vectors. b Transwell assay revealed that the reduction of migration and invasion caused by the overexpression of miR-153 could be reversed by the introduction of MTDH. *P < 0.05, **P < 0.01
Discussion
Accumulating studies have reported that miRNAs are aberrantly expressed in many types of human cancers. miRNAs are emerging as master regulators of tumorigenesis and have also been linked to the control of EMT in the development of cancer through regulation of transcription network [14, 15]. For example, members of the miR-200 family play key roles in mediating the effects of TGF-β and other EMT regulators by targeting ZEB1 and ZEB2 [16, 17].
In a previous study, a comprehensive screening approach combined the analysis of the dysregulation of miRNAs in a natural epithelial–mesenchymal phenotype pair of cell lines and TGF-β-induced EMT models. Results indicated that aberrantly expressed miR-153 played an important role in obtaining or maintaining EMT status [18]. In another report, the downregulation of miR-153 is shown to be a frequent event in certain types of tumors [19]. In the present study, the introduction of miR-153 overexpression was able to significantly suppress cell proliferation and reduce clonogenic ability in two breast cancer cell lines. Moreover, the overexpression of miR-153 in MCF-7 cells resulted in markedly decreased invasive behavior. All these observations demonstrated that ectopic expression of miR-153 could significantly inhibit tumor growth and impair the migration and invasion of breast cancer cells.
Studies have revealed a tendency toward the downregulation of miR-153 in relation to lymph node metastasis in ovarian epithelial tumors [19]; thus, downregulated miR-153 may be a useful marker for the differentiation of malignant serous tumors from serous borderline tumors [19, 20]. Nevertheless, the role of miR-153 in EMT is less investigated. In the present work, miR-153 overexpressed MCF-7 cells exhibited a more epithelial-shaped phenotype, increased E-cadherin expression, and decreased vimentin expression. EMT-associated transcription factors tested, including TWIST, ZEB2, ERG, Notch1, and SNAI1, were also downmodulated by miR-153 overexpression,. These data provided new evidence that miR-153 inhibited EMT breast cancer cells.
Overexpression of MTDH promoted metastatic seeding as well as the chemoresistance of breast tumors and was correlated with poor prognosis in breast cancer patients. miR-375 and miR-145 could concurrently regulate the expression level of MTDH [21]. Thus, we speculated that the downregulation of miR-153 may account for the overexpression of MTDH in breast cancer as an alternative mechanism distinct from 8q22 gain. In the current study, we found that overexpression of miR-153 in breast cancer cells significantly suppressed MTDH in vitro, so we used bioinformatic strategy to reveal that MTDH was a particular candidate target of miR-153. miR-153 overexpression reduced luciferase activity in cells transfected with the wild-type 3′-UTR of MTDH. By contrast, overexpression of MTDH with a cDNA without 3′-UTR could partially abrogate miR-153-mediated suppression of the migration and invasion of breast cancer cells.
Taken together, our results indicated that MTDH was a direct downstream target of miR-153 and was involved in the miR-153-induced suppression of the migration and invasion in breast cancer cell. The miR-153/MTDH link may possibly play roles in breast cancer as markers of metastasis and prognostic factors. Furthermore, these results provided insight into the potential therapeutic value of miR-153 in reducing cancer invasion and metastasis. Nevertheless, these data should be further validated by independent cohorts and prospective trials.
References
Creighton CJ, Gibbons DL, Kurie JM (2013) The role of epithelial–mesenchymal transition programming in invasion and metastasis: a clinical perspective. Cancer Manag Res 5:187–195
Brabletz T (2012) To differentiate or not–routes towards metastasis. Nat Rev Cancer 12(6):425–436
Zhang B, Liu XX, He JR, Zhou CX, Guo M, He M, Li MF, Chen GQ, Zhao Q (2011) Pathologically decreased miR-26a antagonizes apoptosis and facilitates carcinogenesis by targeting MTDH and EZH2 in breast cancer. Carcinogenesis 32(1):2–9
Xia X, Du R, Zhao L, Sun W, Wang X (2014) Expression of AEG-1 and microvessel density correlates with metastasis and prognosis of oral squamous cell carcinoma. Hum Pathol 45(4):858–865
Lee SG, Su ZZ, Emdad L, Sarkar D, Franke TF, Fisher PB (2008) Astrocyte elevated gene-1 activates cell survival pathways through PI3K-Akt signaling. Oncogene 27(8):1114–1121
Yoo BK, Emdad L, Su ZZ, Villanueva A, Chiang DY, Mukhopadhyay ND, Mills AS, Waxman S, Fisher RA, Llovet JM et al (2009) Astrocyte elevated gene-1 regulates hepatocellular carcinoma development and progression. J Clin Investig 119(3):465–477
Di Leva G, Croce CM (2013) The role of microRNAs in the tumorigenesis of ovarian cancer. Front Oncol 3:153
Korpal M, Ell BJ, Buffa FM, Ibrahim T, Blanco MA, Celia-Terrassa T, Mercatali L, Khan Z, Goodarzi H, Hua Y et al (2011) Direct targeting of Sec23a by miR-200s influences cancer cell secretome and promotes metastatic colonization. Nat Med 17(9):1101–1108
Xu Q, Sun Q, Zhang J, Yu J, Chen W, Zhang Z (2013) Downregulation of miR-153 contributes to epithelial–mesenchymal transition and tumor metastasis in human epithelial cancer. Carcinogenesis 34(3):539–549
Dong R, Liu X, Zhang Q, Jiang Z, Li Y, Wei Y, Yang Q, Liu J, Wei JJ, Shao C et al (2014) miR-145 inhibits tumor growth and metastasis by targeting metadherin in high-grade serous ovarian carcinoma. Oncotarget 5(21):10816–10829
Tang Y, Liu X, Su B, Zhang Z, Zeng X, Lei Y, Shan J, Wu Y, Tang H, Su Q (2014) microRNA-22 acts as a metastasis suppressor by targeting metadherin in gastric cancer. Mol Med Rep 11(1):454–460
Hu G, Chong RA, Yang Q, Wei Y, Blanco MA, Li F, Reiss M, Au JL, Haffty BG, Kang Y (2009) MTDH activation by 8q22 genomic gain promotes chemoresistance and metastasis of poor-prognosis breast cancer. Cancer Cell 15(1):9–20
Heckman KL, Pease LR (2007) Gene splicing and mutagenesis by PCR-driven overlap extension. Nat Protoc 2(4):924–932
Bullock MD, Sayan AE, Packham GK, Mirnezami AH (2012) MicroRNAs: critical regulators of epithelial to mesenchymal (EMT) and mesenchymal to epithelial transition (MET) in cancer progression. Biology of the cell/under the auspices of the European Cell Biology Organization 104(1):3–12
Gregory PA, Bracken CP, Bert AG, Goodall GJ (2008) MicroRNAs as regulators of epithelial–mesenchymal transition. Cell Cycle 7(20):3112–3118
Mongroo PS, Rustgi AK (2010) The role of the miR-200 family in epithelial–mesenchymal transition. Cancer Biol Ther 10(3):219–222
Peter ME (2009) Let-7 and miR-200 microRNAs: guardians against pluripotency and cancer progression. Cell Cycle 8(6):843–852
Zhang N, Wang X, Huo Q, Sun M, Cai C, Liu Z, Hu G, Yang Q (2014) MicroRNA-30a suppresses breast tumor growth and metastasis by targeting metadherin. Oncogene 33(24):3119–3128
Kim TH, Kim YK, Kwon Y, Heo JH, Kang H, Kim G, An HJ (2010) Deregulation of miR-519a, 153, and 485-5p and its clinicopathological relevance in ovarian epithelial tumours. Histopathology 57(5):734–743
Xu J, Liao X, Wong C (2010) Downregulations of B-cell lymphoma 2 and myeloid cell leukemia sequence 1 by microRNA 153 induce apoptosis in a glioblastoma cell line DBTRG-05MG. Int J Cancer 126(4):1029–1035
Nohata N, Hanazawa T, Kikkawa N, Mutallip M, Sakurai D, Fujimura L, Kawakami K, Chiyomaru T, Yoshino H, Enokida H et al (2011) Tumor suppressive microRNA-375 regulates oncogene AEG-1/MTDH in head and neck squamous cell carcinoma (HNSCC). J Hum Genet 56(8):595–601
Acknowledgments
This study was supported by grants from National Nature Scientific Foundation of China (30901779, 81072068) and The Natural Science Foundation of Shandong Province (BS2011YY060, ZR2009CM047).
Conflict of interest
All authors of this paper have no financial and personal relationships with other people or organizations that could inappropriately influence this work.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Li, W., Zhai, L., Zhao, C. et al. miR-153 inhibits epithelial–mesenchymal transition by targeting metadherin in human breast cancer. Breast Cancer Res Treat 150, 501–509 (2015). https://doi.org/10.1007/s10549-015-3346-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-015-3346-y