Abstract
Triple-negative breast cancer (TNBC) is an aggressive cancer with limited treatment options. Dual specificity phosphatase 4 (DUSP4) has recently been suggested as a potential marker of chemotherapy resistance for TNBC. DUSP4 gene expression levels were measured in breast cancer tissue from 469 TNBC patients aged 20–75 years who participated in the Shanghai Breast Cancer Survival Study, and their association with recurrence/breast cancer mortality and total mortality was evaluated. Information on breast cancer diagnosis, treatment, and disease progression was collected via medical chart review and multiple in-person follow-up surveys. A Cox regression model was applied in the data analyses. Over a median follow-up of 5.3 years (range: 0.7–8.9 years), 100 deaths and 92 recurrences/breast cancer deaths were documented. Expression levels of transcript variant 1 (NM_001394) and transcript variant 2 (NM_057158) of the DUSP4 gene were studied and were highly correlated (r = 0.76). Low DUSP4 expression levels, particularly of variant 1, were associated with both increased recurrence/breast cancer mortality and increased overall mortality. Hazard ratios with adjustment for age at diagnosis and TNM stage associated with below versus above the median expression level were 1.97 (95 % confidence interval (CI): 1.27–3.05) for recurrence/breast cancer mortality and 2.09 (95 % CI: 1.38–3.17) for overall mortality. Additional adjustment for expression levels of MKI67 and TP53, common treatment types, breast cancer subtype, and grade did not materially alter the observed associations. Low DUSP4 expression levels predict recurrence and mortality in TNBC patients independently from known clinical and molecular predictors.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Approximately 15–20 % of breast cancers in the United States are classified as triple-negative breast cancer (TNBC), characterized by minimal or no expression of estrogen receptors (ER) and progesterone receptors (PR) and the absence of overexpression of human epidermal growth factor 2 (HER2) [1]. TNBC is typically an aggressive cancer type displaying high rates of proliferation and metastasis and having generally poorer prognosis, particularly among those with advanced disease, as evidenced by increased recurrence and mortality compared with other breast cancer types [2]. Due to the lack of known therapeutic targets, such as ER, PR, and HER2 receptors, there are no specific therapies for TNBC tumors [3]. Thus, treatment options are limited. Chemotherapy is commonly used to treat TNBC, but a pathological complete response only occurs in approximately 30 % of cases; therefore, residual disease following treatment is common and accounts for the increased risk of metastatic recurrence and poorer outcomes [4].
Dual specificity phosphatase 4 (DUSP4) negatively regulates members of the mitogen-activated protein kinase (MAPK) superfamily, specifically extracellular-regulated kinase (ERK) activity, which is involved in cellular proliferation and survival [4–6]. Previous studies have suggested that DUSP4 inactivation and subsequent activation of the ERK pathway may play a role in tumor cell proliferation and growth [7, 8]. A recent study showed that low levels of DUSP4 gene expression was associated with basal-like breast cancer (BLBC), high tumor cell proliferation after chemotherapy, dampened response to chemotherapy, and shorter recurrence-free survival [4]. Conversely, overexpression was correlated with increased chemotherapy-induced apoptosis. The authors suggested that the response to MEK inhibitors may be indicated by low DUSP4 expression [9]. This finding was based on 111 total TNBC cases after neoadjuvant chemotherapy recruited from a single institute.
In the present study, we evaluated the association between DUSP4 expression and overall and disease-free survival (DFS) in a cohort of 469 TNBC patients recruited into a population-based cohort study, the Shanghai Breast Cancer Survival Study (SBCSS).
Materials and methods
Study population
The SBCSS, described elsewhere previously, is a longitudinal, population-based cohort study of breast cancer survival in Shanghai, China [10]. Between March 2002 and April 2006, 5,042 women with incident breast cancer were identified from the population-based Shanghai Cancer Registry (response rate: 80 %). All participants were between 20 and 75 years of age at cancer diagnosis and were permanent residents of Shanghai. The study protocol was approved by the institutional review boards of Vanderbilt University and the Shanghai Municipal Center for Disease Control and Prevention, and all participants provided written informed consent.
Data collection
Participants were recruited to the study approximately 6 months following breast cancer diagnosis [mean (standard deviation):6.6 (0.7) months] [10]. In-person interviews, with trained interviewers, were conducted and information obtained included demographic characteristics, selected lifestyle factors, and clinical and treatment factors. Medical charts from the initial diagnostic hospital were reviewed to obtain clinical information, i.e., tumor characteristics, first-line treatments, and ER/PR status. HER2 status was measured in the Vanderbilt Molecular Epidemiology Lab using rabbit polyclonal antibody recognizing the HER2 cytoplasmic domain (DAKO, Cat# A0485, 1:100) following the DAKO EnvisionTM kit protocol (DAKO, Cat# K4011) (details published elsewhere) [11]. Ten tumor sections (nine tumor sections (slides) in 5 micron and one section (slide) in 15 micron) were collected from the diagnostic hospitals for 4,036 SBCSS participants (80 %). Among these subjects, 525 were diagnosed with TNBC and included in the current study. Participants were followed up at 18, 36, and 60 months after breast cancer diagnosis to obtain information on disease progression and survival status. Survival information for participants lost to follow up was obtained using annual record linkage with the Shanghai Vital Statistics Registry.
Gene expression levels were measured in total RNA isolated from formalin-fixed paraffin-embedded (FFPE) breast cancer tissues using Nanostring technology. FFPE tissue sections and the H&E slides were reviewed by a study pathologist. Tumor tissue was selected and dissected to ensure that tissues contained >80 % tumor cells for total RNA extraction. Total RNA were isolated and purified using Qiagen’s miRNeasy FFPE Kit (Qiagen, Valencia, CA), a kit specifically for purification of total RNA including miRNA from FFPE tissue sections, following the manufacturer’s instructions. The quantity and quality of the RNA samples were checked by Nanodrop (E260, E260/E280 ratio, spectrum 220–320 nm) and by separation on an Agilent BioAnalyzer. We excluded 29 samples from the gene expression assay either due to the fact that tumor tissue was too small for RNA extraction (n = 19) or low RNA concentrations (n = 10), leaving 496 samples for expression assay.
Expression levels of transcript variant 1 (NM_001394) and transcript variant 2 (NM_057158) of the DUSP4 gene were measured as a part of large gene expression effort. A custom-designed nCounter Gene Expression CodeSet profiling of 311 selected gene targets using the NanoString nCounter technology was performed following the NanoString standard protocol [12]. The selected gene panel included PAM50 genes, drug metabolism genes, reference genes, and a set of targeted genes based on review and analysis of published data. The assay CV for the 2 DUSP4 isoforms was 7.84 and 10.43 % based on eight QC samples (duplicated measurements of seven individual QC samples and ten repeated measurements of a pooled QC sample). The R package NanoStringNorm (version 1.1.16), developed for normalization, diagnostics, and visualization of NanoString nCounter data, was used for quality assurance and data normalization. Specifically, quality control and raw data normalization were completed as follows: (1) adjust each sample’s counts based on its relative value to the geometric mean of all samples to reduce technical variation; (2) correct background count level by subtracting each sample’s count value from the mean +2 standard deviations of counts of the negatives controls; and (3) normalize for sample RNA content, i.e., “pipetting” fluctuations using the geometric mean of the expression levels of 5 pre-specified housekeeping genes (ACTB, RPLP0, MRPL19, SF3A1, and PSMC4). From 496 samples assayed, samples were excluded from statistical analysis if they met at least one of the following conditions: (1) positive normalization factor was outside of the range (0.3, 3) (N = 6); (2) samples with a background level > 3 standard deviations from the mean (N = 4); (3) samples with >90 % missing after background level correction (N = 11); and (4) samples with RNA content >3 standard deviations from the mean (N = 6), leaving a total of 469 samples with valid data for analysis. We applied the calling algorithm developed by Parker et al. to classify tumors into subgroups most resembling Basal-like, Luminal A, Luminal B, HER2-enriched, or Normal-like breast cancer based on PAM50 genes [13]. Briefly, for each sample, we calculated the Spearman’s rank correlations between the gene set and the centroids of the five intrinsic subtypes: Luminal A, Luminal B, Basal-like, HER2-enriched, or Normal-like. These correlations were used as the distance metric, and all samples were then assigned to the subtype that had the minimum distance to the sample. To maintain the subtype structure in our study cohort, we did not apply the gene-wise centering during this calling process.
For the 469 samples included in the study, 418 were collected prior to chemotherapy [or the patient did not receive chemotherapy (N = 28)], 34 were collected after chemotherapy, and for the remaining 17 samples, the time line relevant to chemotherapy or radiotherapy could not be determined due to the overlapping of month of surgery and initiation of chemo- or radiotherapy (information on specific day of initiation was not collected).
Statistical analysis
The major endpoints for the study were any death for overall survival (OS) and cancer recurrence/metastasis or death related to breast cancer for DFS. The date of last in-person contact or June 2011 (6-months prior to date of latest record linkage), whichever was more recent, was used as the censor date for event-free subjects.
Clinical, treatment, demographic, and lifestyle factor variables were evaluated for their association with DUSP4 expression levels (split at median expression level) using generalized linear models for continuous variables and χ2 tests for categorical variables. Information on TNM stage was missing for 16 participants.
Using Cox proportional hazards models, the associations between DUSP4 expression levels, analyzed both as a continuous variable and using median cutpoints, and recurrence and mortality were evaluated. The multivariate analyses were adjusted for age at diagnosis and TNM stage (five levels: stage 0–I (reference), IIA, IIB, III–IV, and missing). The associations were also examined by tumor molecular subtype.
Additional analyses stratified by chemotherapy (yes/no), type of chemotherapeutic drug (for the four most commonly used drugs), radiotherapy (yes/no), tamoxifen use, TNM stage, tumor grade, or use of vitamins C or E were performed to evaluate whether the relationship between DUSP4 expression and recurrence and overall mortality was modified by these factors.
Statistical analyses were performed using SAS software (version 9.3; SAS Institute, Inc., Cary, NC). The REMARK guidelines were followed in reporting the results of this study [14].
Results
The expression levels for the two isoforms for DUSP4 were highly correlated (r = 0.76, P < 0.0001); the DUSP4 variant 1 (NM_001394) was more predictive of outcome in this study and was chosen for more in-depth analyses as presented below. Over a median follow-up of 5.3 years (range: 0.7–8.9 years), 100 deaths and 92 recurrences/breast cancer mortalities were documented in this cohort of TNBC patients from the SBCSS cohort. DFS rate and OS were inversely associated with advanced stage disease and receiving radiotherapy (Table 1). Additionally, OS was inversely associated with menopausal status.
Participants with DUSP4 (variant 1) expression levels below the median were significantly younger at diagnosis and were more likely to have a family history of breast cancer. No association was observed between DUSP4 expression and other demographic and lifestyle factors. Breast cancer cases with DUSP4 expression levels below the median were significantly more likely to have a higher grade tumor as compared to those with DUSP4 expression levels above the median (P < 0.0001) (Table 2). Tumors with low DUSP4 expression level were more likely to possess markers of the BLBC subtype (P < 0.0001).
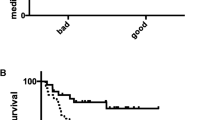
Overall, the unadjusted 5-year disease-free and OS rates were 79.3 and 81.9 %, respectively, for this group of TNBC cases. DUSP4 expression was inversely associated with the risk of recurrence/breast cancer mortality and total mortality (HR associated with per unit decrease of log2 transformed DUSP4 expression level = 1.16, 95 % confidence interval (CI): 1.08, 1.25 and HR = 1.19, 95 % CI: 1.10, 1.28, respectively) (Table 3). Categorization of DUSP4 expression into quartiles suggested that the median split could serve as an indicator of threshold effect. Compared to those with DUSP4 expression above the median, those with DUSP4 expression levels below the median had a 1.97-fold (95 % CI: 1.27, 3.05) increased risk of recurrence/breast cancer mortality and a 2.09-fold (95 % CI: 1.38, 3.17) increased risk of mortality. When the lower median was further categorized into two categories and compared to the upper median, the association was the strongest between those with the lowest DUSP4 expression levels and the upper median for recurrence/breast cancer mortality (HR = 2.28, 95 % CI: 1.40, 3.72) and overall mortality (HR = 2.40, 95 % CI: 1.50, 3.84); the HRs for the upper half of the lower median were 1.66 (95 % CI: 0.97, 2.81) for recurrence/breast cancer mortality and 1.79 (95 % CI: 1.08, 2.95) for overall mortality as compared to the upper median. Those with the lowest DUSP4 expression levels also had the lowest disease-free and OS rates (69.6 and 72.6 %, respectively). Participants with stage 0 tumors (n = 7) had DUSP4 expression levels greater than the median (mean = 9.72), and the participant with a stage IV tumor (n = 1) had a DUSP4 expression level below the median (DUSP4 expression = 6.69). Excluding participants with stage 0 or stage IV tumors did not materially change the observed results (HR for the upper median compared to the lower median was 1.88, 95 % CI: 1.22, 2.90 for recurrence/breast cancer mortality and 1.99, 95 % CI: 1.31, 3.01 for overall mortality; HR for the continuous DUSP4 measure was 1.16, 95 % CI: 1.08, 1.25 for recurrence/breast cancer mortality and 1.19, 95 % CI: 1.10, 1.28 for overall mortality). Figure 1 shows the Kaplan–Meier DFS curve by the lower and upper median of DUSP4 expression.
Survival time since diagnosis in years is on the x-axis and disease-free survival as a percentage is on the y-axis. Kaplan–Meier curves, comparing those with DUSP4 (variant 1) expression levels above (n = 234) and below (n = 235) the median, show a significantly worse prognosis for patients with DUSP4 expression levels below the median (P = 0.002)
DUSP4 expression was inversely correlated with MKI67 gene expression (r = −0.24, P < 0.0001), an indicator for higher tumor cell proliferation [15]. However, controlling for MKI67 gene expression level did not alter the association between DUSP4 and recurrence and mortality (lower median compared to upper: HR = 2.00, 95 % CI: 1.28, 3.15 and HR = 2.13, 95 % CI: 1.38, 3.28, respectively). Furthermore, DUSP4 expression was positively associated with expression levels of ESR1, PGR, and ERBB2. Controlling for ESR1 expression changed the association with DUSP4 expression for recurrence/breast cancer mortality to HR = 2.21 (95 % CI: 1.37, 3.55). The DUSP4 association was not materially changed when controlling for expression levels of PRG and ERRB2. DUSP4 expression was inversely associated with expression level of the MYC (r = −0.19, P < 0.0001) and positively associated with expression level of the MAPT (r = 0.37, P < 0.0001), genes which are both in the ERK pathway; controlling for these genes did not alter the results materially (data not shown). No correlation between DUSP4 and TP53 expression was observed, and controlling for TP53 did not alter the observed results.
Despite that tumors with markers of the basal-like subtype were more likely to have low DUSP4 expression, analysis stratified by molecular subtype based on PAM50 genes showed that DUSP4 expression was associated with overall mortality and recurrence/breast cancer mortality in basal-like (HR for continuous DUSP4 measure = 1.22, 95 % CI: 1.05, 1.41 for overall mortality and HR = 1.16, 95 % CI: 1.01, 1.33 for recurrence/breast cancer mortality) and non-basal-like breast cancer subtypes (HR = 1.16, 95 % CI: 1.05, 1.30 for overall mortality and HR = 1.14, 95 % CI: 1.02, 1.28 for recurrence/breast cancer mortality).
No interactions with TNM stage (0-IIA vs IIB-IV), tumor grade (1 and 2 vs 3), chemotherapy, radiotherapy, tamoxifen use, and use of vitamin C or E (ever vs never) were observed for DUSP4 expression. We evaluated the most commonly used chemotherapy drugs (5-fluorouracil, cyclophosphamide, methotrexate, and epirubicin) as potential effect measure modifiers (Table 4); DUSP4 expression, analyzed continuously, was more predictive of DFS in patients who received methotrexate (HR for continuous DUSP4 measure = 1.32, 95 % CI: 1.11, 1.58) than those who did not (HR = 1.10, 95 % CI: 1.01, 1.21) (P = 0.02 for interaction). We did not find that the DUSP4 associations varied by other common chemotherapy drugs or radiotherapy.
Discussion
The results from this large, population-based cohort study show that low DUSP4 gene expression is associated with increased mortality and breast cancer recurrence among patients with TNBC. Although low DUSP4 expression was more common in tumors with BLBC markers and was inversely associated with MKI67 and MYC gene expressions, its association with breast cancer outcomes was independent of these factors.
A member of the dual specificity protein phosphatase family, DUSP4 dephosphorylates the phosphoserine/threonine and phosphotyrosine residues of its target kinase, resulting in an inactivation of the kinase [16]. DUSP4 regulates cell proliferation and survival by negatively regulating members of the MAPK superfamily through inactivation of ERK1, ERK2, and JNK [6]. DUSP4 downregulation results in Ras–ERK pathway activation. The Ras signaling pathways play a substantial role in controlling normal cell growth and, when activated, lead to uncontrolled cell growth [17]. Activation of this pathway may in turn lead to reduced response to chemotherapy [4, 9]. If this hypothesis is correct, DUSP4 gene expression quantification may be useful in predicting activity of the Ras–ERK pathway [9]. MEK inhibitors could be used in combination with chemotherapy agents to potentially improve response to chemotherapy [9].
In a recent study using Nanostring technology to profile 49 breast cancer tissue samples, primarily TNBC, surgically resected after neoadjuvant chemotherapy, Balko et al. found that low DUSP4 expression correlated with high tumor cell proliferation following neoadjuvant chemotherapy and BLBC tumors [4]. Pathological response to chemotherapy was inversely associated with DUSP4 expression, and the authors concluded that the Ras–ERK pathway may be activated by downregulation of DUSP4 in BLBC, resulting in an attenuated response to chemotherapy. The authors further demonstrated that treatment with a MEK inhibitor enhanced sensitivity to the chemotherapeutic drug, docetaxel, in vivo. The authors concluded that DUSP4 may be a potential marker of resistance to chemotherapy drugs.
Similarly, in our study, low DUSP4 expression was more common in BLBC tumors and was associated with increased recurrence/breast cancer mortality and increased total mortality. Tumors with BLBC subtype markers had increased recurrence/breast cancer mortality and total mortality compared with tumors without these markers. However, adjustment for DUSP4 expression attenuated, but did not completely eliminate, the survival disadvantage for BLBC tumors, suggesting that DUSP4 down regulation may only be one of the mechanisms responsible for the aggressive clinical behavior of BLBC. In our study, DUSP4 expression was inversely related to MKI67 and MYC expression and positively associated with ERS1, PR, ERBB2, and MAPT expression, but not TP53 expression. Additionally, adjustment for TP53, MKI67, ESR1, PGR, and ERBB2 gene expression level did not change the association. These results suggest that the association of DUSP4 with recurrence/survival is independent of the molecular subtypes of breast cancer as defined by PAM50 and the known molecular prognostic markers for TNBC.
While our study only involved TNBC, deregulation of DUSP4 on cancer prognosis may not be confined to TNBC. Aberrations in many of the above-mentioned DUSP4-correlated genes are commonly seen in other types of breast cancer [18–20]. In our study, DUSP4 expression was associated with breast cancer outcomes for non-basal-like TNBC, including those that expressed luminal A or B signature genes. Using the publicly available Dutch Cancer Institute (NKI) breast cancer dataset (https://www.synapse.org/#!Synapse:syn4517), we found that DUSP4 expression was associated with outcomes of ER + tumors (HR associated with per unit decrease of log2 transformed DUSP4 expression level = 1.45, 95 % CI: 1.12, 1.86 for recurrence and HR = 1.57, 95 % CI: 1.13, 2.18 for overall mortality, after adjustment for age at diagnosis and tumor grade) [21]. These results call for more investigation on the role of DUSP4 in other major breast cancer subtypes.
It is important to note the differences in populations used in our study compared with the previous study by Balko et al. The previous findings were observed in breast cancer tissue taken from European-ancestry women, whereas our finding of a similar association in an Asian-ancestry population suggests that the DUSP4 association is independent of race and ethnicity. Furthermore, all patients in the study by Balko et al. were treated with the chemotherapy drug, docetaxel, before tumor resection. The percentage of patients who received docetaxel in our population was low (3.4 %), and the majority of patients received other chemotherapy agents or combinations of agents, including 5-fluorouracil (75.5 %), cyclophosphamide (71.9 %), epirubicin (54.2 %), and methotrexate (21.3 %). Additionally, the majority of participants in our study had not received chemotherapy prior to surgery when the tissue sample for gene expression quantification was resected.
Our data show that DUSP4 expression was predictive of outcomes among many different groups, including those whose DUSP4 expression level was quantified prior to chemotherapy treatment and those who did not receive chemotherapy treatment at all (although the sample size for the latter group was small). There were some indications that DUSP4 expression was more predictive of outcome among those who received certain types of chemotherapy; however, the interaction was only significant for those who took methotrexate. Taken together, our study suggests that DUSP4 expression may predict breast cancer outcomes beyond its association with response to chemotherapy as previously suggested.
The present study has several strengths. The data come from a large, population-based cohort study with comprehensive information collected on potential covariates. Information on clinical and disease factors was verified through medical chart review, which increased the validity of the data. A limitation of our study is that the timing of biological sampling with respect to chemotherapy treatment could not be determined for 17 patients. However, the significant association between DUSP4 gene expression and recurrence/breast cancer mortality and overall mortality was similar when only those with tumor tissue samples taken prior to chemotherapy initiation were included in the analyses.
Further research is needed to better understand the biological mechanism(s) underlying the association of DUSP4 expression with TNBC outcomes and to investigate the role of DUSP4 in the prognosis of various molecular subtypes of breast cancer. The gene–drug interaction, in particular the role of DUSP4 expression on chemotherapy resistance, may be better evaluated in a randomized clinical trial.
In summary, our study confirmed that low DUSP4 expression was associated with increased recurrence/breast cancer mortality and increased total mortality among TNBC patients. This association was independent of markers of BLBC and other known clinical and molecular predictors.
Abbreviations
- DUSP4:
-
Dual specificity phosphatase 4
- TNBC:
-
Triple-negative breast cancer
- ER:
-
Estrogen receptor
- PR:
-
Progesterone receptor
- HER2:
-
Human epidermal growth factor receptor 2
References
Pal SK, Childs BH, Pegram M (2011) Triple negative breast cancer: unmet medical needs. Breast Cancer Res Treat 125(3):627–636. doi:10.1007/s10549-010-1293-1
Ossovskaya V, Wang Y, Budoff A, Xu Q, Lituev A, Potapova O, Vansant G, Monforte J, Daraselia N (2011) Exploring molecular pathways of triple-negative breast cancer. Genes cancer 2(9):870–879. doi:10.1177/1947601911432496
Dent R, Trudeau M, Pritchard KI, Hanna WM, Kahn HK, Sawka CA, Lickley LA, Rawlinson E, Sun P, Narod SA (2007) Triple-negative breast cancer: clinical features and patterns of recurrence. Clin Cancer Res 13(15 Pt 1):4429–4434. doi:10.1158/1078-0432.CCR-06-3045
Balko JM, Cook RS, Vaught DB, Kuba MG, Miller TW, Bhola NE, Sanders ME, Granja-Ingram NM, Smith JJ, Meszoely IM, Salter J, Dowsett M, Stemke-Hale K, Gonzalez-Angulo AM, Mills GB, Pinto JA, Gomez HL, Arteaga CL (2012) Profiling of residual breast cancers after neoadjuvant chemotherapy identifies DUSP4 deficiency as a mechanism of drug resistance. Nat Med 18(7):1052–1059. doi:10.1038/nm.2795
Jeffrey KL, Camps M, Rommel C, Mackay CR (2007) Targeting dual-specificity phosphatases: manipulating MAP kinase signalling and immune responses. Nat Rev Drug Discovery 6(5):391–403. doi:10.1038/nrd2289
Lawan A, Al-Harthi S, Cadalbert L, McCluskey AG, Shweash M, Grassia G, Grant A, Boyd M, Currie S, Plevin R (2011) Deletion of the dual specific phosphatase-4 (DUSP-4) gene reveals an essential non-redundant role for MAP kinase phosphatase-2 (MKP-2) in proliferation and cell survival. J Biol Chem 286(15):12933–12943. doi:10.1074/jbc.M110.181370
Waha A, Felsberg J, Hartmann W, von dem Knesebeck A, Mikeska T, Joos S, Wolter M, Koch A, Yan PS, Endl E, Wiestler OD, Reifenberger G, Pietsch T (2010) Epigenetic downregulation of mitogen-activated protein kinase phosphatase MKP-2 relieves its growth suppressive activity in glioma cells. Cancer Res 70(4):1689–1699. doi:10.1158/0008-5472.CAN-09-3218
Cagnol S, Rivard N (2013) Oncogenic KRAS and BRAF activation of the MEK/ERK signaling pathway promotes expression of dual-specificity phosphatase 4 (DUSP4/MKP2) resulting in nuclear ERK1/2 inhibition. Oncogene 32(5):564–576. doi:10.1038/onc.2012.88
Rottenberg S, Jonkers J (2012) MEK inhibition as a strategy for targeting residual breast cancer cells with low DUSP4 expression. Breast cancer res 14(6):324. doi:10.1186/bcr3327
Shu XO, Zheng Y, Cai H, Gu K, Chen Z, Zheng W, Lu W (2009) Soy food intake and breast cancer survival. JAMA 302(22):2437–2443. doi:10.1001/jama.2009.1783
Su Y, Zheng Y, Zheng W, Gu K, Chen Z, Li G, Cai Q, Lu W, Shu XO (2011) Distinct distribution and prognostic significance of molecular subtypes of breast cancer in Chinese women: a population-based cohort study. BMC Cancer 11:292. doi:10.1186/1471-2407-11-292
Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, Fell HP, Ferree S, George RD, Grogan T, James JJ, Maysuria M, Mitton JD, Oliveri P, Osborn JL, Peng T, Ratcliffe AL, Webster PJ, Davidson EH, Hood L, Dimitrov K (2008) Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol 26(3):317–325. doi:10.1038/nbt1385
Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T, Davies S, Fauron C, He X, Hu Z, Quackenbush JF, Stijleman IJ, Palazzo J, Marron JS, Nobel AB, Mardis E, Nielsen TO, Ellis MJ, Perou CM, Bernard PS (2009) Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol 27(8):1160–1167. doi:10.1200/JCO.2008.18.1370
McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM (2006) REporting recommendations for tumor MARKer prognostic studies (REMARK). Breast Cancer Res Treat 100(2):229–235. doi:10.1007/s10549-006-9242-8
Guarneri V, Piacentini F, Ficarra G, Frassoldati A, D’Amico R, Giovannelli S, Maiorana A, Jovic G, Conte P (2009) A prognostic model based on nodal status and Ki-67 predicts the risk of recurrence and death in breast cancer patients with residual disease after preoperative chemotherapy. Ann Oncol Off J European Soc Med Oncol 20(7):1193–1198. doi:10.1093/annonc/mdn761
Peng DJ, Zhou JY, Wu GS (2010) Post-translational regulation of mitogen-activated protein kinase phosphatase-2 (MKP-2) by ERK. Cell Cycle 9(23):4650–4655
Downward J (2003) Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer 3(1):11–22. doi:10.1038/nrc969
Byler S, Goldgar S, Heerboth S, Leary M, Housman G, Moulton K, Sarkar S (2014) Genetic and epigenetic aspects of breast cancer progression and therapy. Anticancer Res 34(3):1071–1077
Sabatier R, Goncalves A, Bertucci F (2014) Personalized medicine: present and future of breast cancer management. Crit Rev Oncol Hematol 91(3):223–233. doi:10.1016/j.critrevonc.2014.03.002
Baquero MT, Lostritto K, Gustavson MD, Bassi KA, Appia F, Camp RL, Molinaro AM, Harris LN, Rimm DL (2011) Evaluation of prognostic and predictive value of microtubule associated protein tau in two independent cohorts. Breast Cancer Res 13(5):R85. doi:10.1186/bcr2937
van de Vijver MJ, He YD, van’t Veer LJ, Dai H, Hart AA, Voskuil DW, Schreiber GJ, Peterse JL, Roberts C, Marton MJ, Parrish M, Atsma D, Witteveen A, Glas A, Delahaye L, van der Velde T, Bartelink H, Rodenhuis S, Rutgers ET, Friend SH, Bernards R (2002) A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med 347(25):1999–2009. doi:10.1056/NEJMoa021967
Acknowledgments
We thank Regina Courtney and Bo Huang for their help with RNA sample preparation. Sample preparation was conducted at the Survey and Biospecimen Shared Resources, which is supported in part by the Vanderbilt-Ingram Cancer Center (P30CA068485). NanoString nCounter assays were conducted at the University of Miami Sylvester Comprehensive Cancer Center. This study was supported by grants from the US Department of Defense (DOD) Breast Cancer Research Program (DAMD 17-02-1-0607 to X.-O. Shu) and the National Institutes of Health (NIH; R01 CA118229 to X.-O. Shu, P50CA098131 to C. Arteaga). Michelle Baglia is funded by a Clinical and Translational Science Award (CTSA) TL1 fellowship.
Ethical Standards
This study complies with the current laws of the country in which they were performed.
Conflict of interest
The authors have no conflicts of interest to disclose.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Baglia, M.L., Cai, Q., Zheng, Y. et al. Dual specificity phosphatase 4 gene expression in association with triple-negative breast cancer outcome. Breast Cancer Res Treat 148, 211–220 (2014). https://doi.org/10.1007/s10549-014-3127-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-014-3127-z