Abstract
The curtain of technical limitations impeding rat multichannel non-invasive electroencephalography (EEG) has risen. Given the importance of this preclinical model, development and validation of EEG source imaging (ESI) is essential. We investigate the validity of well-known human ESI methodologies in rats which individual tissue geometries have been approximated by those extracted from an MRI template, leading also to imprecision in electrode localizations. With the half and fifth sensitivity volumes we determine both the theoretical minimum electrode separation for non-redundant scalp EEG measurements and the electrode sensitivity resolution, which vary over the scalp because of the head geometry. According to our results, electrodes should be at least ~3 to 3.5 mm apart for an optimal configuration. The sensitivity resolution is generally worse for electrodes at the boundaries of the scalp measured region, though, by analogy with human montages, concentrates the sensitivity enough to localize sources. Cramér–Rao lower bounds of source localization errors indicate it is theoretically possible to achieve ESI accuracy at the level of anatomical structures, such as the stimulus-specific somatosensory areas, using the template. More validation for this approximation is provided through the comparison between the template and the individual lead field matrices, for several rats. Finally, using well-accepted inverse methods, we demonstrate that somatosensory ESI is not only expected but also allows exploring unknown phenomena related to global sensory integration. Inheriting the advantages and pitfalls of human ESI, rat ESI will boost the understanding of brain pathophysiological mechanisms and the evaluation of ESI methodologies, new pharmacological treatments and ESI-based biomarkers.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Electroencephalography (EEG) is presently an important tool for assisting diagnostics, management and treatment of neurological disorders. In particular, a better understanding of the electrical substrates of pathological events and abnormalities associated with brain network activity has been possible through the use of the estimated EEG sources within the brain, i.e. the EEG source imaging (ESI). When derived from standard EEG high density caps, ESI provides functional images of the whole brain with an exquisite temporal resolution. ESI has been used to determine important features of neuronal connectivity in autism spectrum disorders (Coben et al. 2014); to localize intrinsic sources of either interictal epileptiform discharges in focal epilepsy (Kaiboriboon et al. 2012) or those related to brain lesions (Harmony et al. 1995). It has also been useful in understanding important pathological mechanisms in psychiatric disorders (Hughes et al. 1999) and neurological conditions such as stroke (Bharadwaj and Gofton 2015), attention deficit hyperactivity disorder (Rodrak and Wongsawat 2013) and Alzheimer’s disease (Aghajani et al. 2013).
On the other hand, preclinical models are essential in studying most of these brain disorders. This is due to the possibility of combining invasive with non-invasive techniques, which promises significant improvements to clinical treatments by establishing gold-standards, protocols and useful biomarkers. In particular, the rat is one of the most popular preclinical models, e.g. of Alzheimer’s disease (Do Carmo and Cuello 2013), epilepsy (Löscher 2011), stroke (MacRae 2011), autism (Umeda et al. 2010) and schizophrenia (Shevelkin et al. 2014). There is great interest from both research institutions and the pharmacological industry in developing practical tools for an efficacious evaluation of interventions using these preclinical models. Like in humans, rat ESI—provided that it is based on practical, feasible and exportable methodologies—would provide a promising non-invasive technique for chronic strategies that is needed to understand the evolution of the disease and the responses to specific treatments in this particular species.
Unfortunately, ESI in rodents has been challenging until recently due to the difficulties with obtaining whole scalp high density EEG recordings. The invasiveness of the methods used for electrode fixation poses an additional drawback. Epicranial multi-electrode arrays requires surgical procedures to attach them to the skull or under the skin (Choi et al. 2010; Franceschini et al. 2008; Lee et al. 2011; Mégevand et al. 2008; Quairiaux et al. 2011). We have recently overcome this problem by developing the first EEG mini-cap to non-invasively record high-density (32-channels) scalp EEG in rats (Riera et al. 2012). Preliminary, the methodology for using this mini-cap to perform ESI on a rat preclinical model of focal epilepsy was presented in Bae et al. (2015).
An additional, and very important, practical impediment for ESI is the achievement of realistic MRI-based forward models. This is prohibitive for large preclinical studies, for long EEG recordings precluding rat undergoing MRI, or when MRI scanners suitable for rats are not available. This very reason has already promoted the use of surrogate models in human ESI (Darvas et al. 2006; Valdés-Hernández et al. 2009). For rat ESI, the only precedent is Bae et al. (2015), where an approximate head model—with the consequent approximate electrode positions—was obtained from an MRI rat template. This template—rigorously called minimum deformation template since it minimizes the sum of the template-individual registration deformations across space and individuals—is considered as the most representative among individuals since it is the unbiased shape centroid of an MRI rat database (Valdés-Hernández et al. 2011).
Despite these important advances, rigorous theoretical analysis of the validity of the electrode density proposed by Riera et al. (2012) and of the accuracy in employing a template-based head model approximation is still missing. As a matter of fact, ESI would not be possible in rats if calculations from human data for optimal electrode resolutions (Gevins et al. 1990; Ferree et al. 2001; Malmivuo et al. 1997) and theoretical limits for the dipolar localization errors (Mosher et al. 1993; Beltrachini et al. 2011; von Ellenrieder et al. 2006) still hold for this particular species. Our goal in this paper is to validate practical tools for ESI on rats by evaluating: (1) the electrode sensitivity resolution and (2) the lower bounds for the errors in the estimation of brain source introduced by the use of a template-based LF matrix. Errors from using the template have two sources: one related to the mismatches in the surfaces limiting different brain tissues and another because imprecisions in determining electrode positions in a head model.
Specifically, we firstly establish a reproducible rat EEG forward modelling methodology based on a well-known numerical method for calculating the LF matrix, i.e. the FEM. We validate this methodology by investigating which is the optimal electrode configuration for scalp rat EEG. This is done by evaluating both the theoretical minimum distance at which two electrodes are not measuring redundant information and the electrode sensitivity resolution, using the concepts of half (HSV) and fifth (FSV) sensitivity volumes (Malmivuo et al. 1997; Wendel et al. 2008). Secondly, to theoretically validate the use of the template head model as a surrogate of an individual head model, we investigate if this model misspecification drastically increases the Cramér–Rao lower bound (CRLB) of EEG source estimation errors (Mosher et al. 1993) and provide, at the same time, the maximum possible accuracy of rat ESI. This is tackled with a method based on von Ellenrieder et al. (2006), which incorporates uncertainty in the LF matrix of the model, but deriving the covariance matrix directly from the LF matrices calculated from the individual MRIs of a rat database (individual LF matrices). Thereby this covariance simultaneously accounts for both tissue-limiting surface mismatches and electrode location imprecisions. Since low CRLB values are only necessary conditions for an accurate ESI, we also compare the template LF matrix with the individual LF matrices using both the relative difference measure (RDM) and the magnitude ratio (MAG) (Gramfort et al. 2011; Meijs et al. 1989). This comparison is done for different typical configurations of electrodes to empirically account for the variability in electrode positioning of the rat EEG mini-cap. Finally we demonstrate the scope of our methodology using real ERP data obtained from Wistar rats. A first data set, which comprises simultaneous EEG and fMRI, is used to demonstrate both the accuracy of ESI and of the geometry-electrode template approximation. A second SEP dataset, with different sensory stimulation modalities, is used to illustrate the potential of the technique to extract information about traditional integration of sensory data at a whole brain global scale.
Materials and Methods
Template Set and Individual MRIs
The details of the animal preparation, ethical considerations, MRI acquisition protocol and the template construction can be found in Valdés-Hernández et al. (2011). Briefly, we acquired the MRI of \(N_{K} = 28\) Wistar rats (Charles River Japan, Yokohama, Japan). They consist of respiratory-gated 2D TurboRARE T2 images with parameters: TR/TEeff = 10,971/30, RARE factor = 4, effective SBW = 100 kHz, FOV = 32 × 32 mm2, matrix size = 256 × 256, in-plane resolution = 125 × 125 µm2 and 128 slices with 0.3 mm thickness. The template set was built using these individual MRIs and comprises (1) an unbiased average of the nonlinearly warped MRIs, (2) probabilistic maps of brain, gray matter, white matter and cerebrospinal fluid, and (3) a discrete image labeling 96 structures of the cortex, according to the Paxinos and Watson classification atlas (Paxinos and Watson 2007).
Forward Problem of the EEG
The estimation of the brain electrical sources of EEG is known as the EEG inverse problem. For solving this problem, the generativeEEGmodel must be defined. This supposes the solution of the EEG forward problem, providing the voltage generated at the EEG electrodes by any bioelectrical generator inside the brain. These are conventionally modelled as current dipoles.
For one time instant, let us build the vector comprising \(N_{D}\) dipoles \({\mathbf{j}} \equiv \left[ {\begin{array}{*{20}c} {{\mathbf{j}}_{1}^{\varvec{T}} } & \ldots & {{\mathbf{j}}_{{\varvec{N}_{\varvec{D}} }}^{\varvec{T}} } \\ \end{array} } \right]\), where \({\mathbf{j}}_{\varvec{j}} = Q_{j} {\hat{\mathbf{d}}}_{\varvec{j}}\) is the j-th dipole with orientation \({\hat{\mathbf{d}}}_{\varvec{j}} \in {\mathbb{R}}^{3 \times 1}\), \(\parallel {\hat{\mathbf{d}}}\parallel = 1\). Given the position of \(N_{E}\) scalp EEG electrodes, the forward problem reduces to calculating the LF matrix, which relates these dipoles to the voltage differences \({\mathbf{v}} \in {\mathbb{R}}^{{N_{E} \times 1}}\) they generate at the electrodes with respect to a common referenceFootnote 1:
where \({\mathbf{K}} \in {\mathbb{R}}^{{N_{E} \times 3N_{D} }}\) is the LF matrix, defined as
and the elements of the vector \({\mathbf{k}}_{i,j} \in {\mathbb{R}}^{1 \times 3}\) represent the voltage difference associated with the i-th electrode of an unit amplitude dipole at the j-th location, oriented along the X, Y and Z axes, respectively. The vector \({\varvec{\upvarepsilon}} \sim N\left( {0_{{N_{E} \times 1}} ,\sigma^{2} {\mathbf{I}}_{{N_{E} }} } \right)\) is a multivariate Gaussian white noise.
Head Modelling
We calculated a FEM LF matrix using SimBio, a generic environment for bio-numerical simulations from the SimBio Development Group (online at https://www.mrt.uni-jena.de/simbio) (Vorwerk et al. 2014; Wolters et al. 2004). For that, it is necessary to physically model the head. This requires a model of the possible locations and orientations of current dipoles within the brain (i.e. the source space model) and the spatial dependency of electrical conductivities within the head (i.e. thevolume conductor model). To build a realistic head model, an individual MRI of the rat, or an equivalent representative (i.e. the template MRI), is required.
We considered two types of source space models:
-
(a)
Volumetric model with 3D support either inside the whole brain or only the gray matter. These were discretized into regular grids of points representing possible dipole locations.
-
(b)
Surface model defined as the surface amidst the pial surface and the boundary between the cortical layer and the white matter. This was discretized to a triangular mesh representing possible dipole locations. The dipole orientations were constrained along the normal to the surface, based on the hypothesis that EEG signals are mainly caused by current dipoles in the extracellular space of pyramidal neurons, which are perpendicular to the cortical layer (Riera et al. 2012).
These models and their uses in this paper are detailed in Table 1 and shown in Fig. 1a.
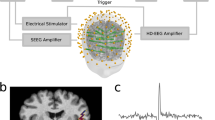
Head models of the rat. a The delineation of the three conductivity compartments is shown in an axial slice: head (white), skull (blue) and brain (magenta). The source space grid points of model Vol3D and VolGM3D are shown as green dots and yellow circles respectively. The intersection between the source model of SurfMid and the slice is shown with a red curve. b Sagittal and axial view of the tetrahedral FEM mesh. This figure evidences the oblong shape of the rat head. c Electrode EEG mini-cap scaffold. The electrodes are labelled with blue numbers and the landmarks used to aid placing the cap in the MRI are labelled in red (Color figure online)
In order to compare the individual LF matrices with the template LF matrix, the points of the discretization of their source space models must be in a one-to-one anatomical correspondence. For this we generated a unique discretization in the template and transformed the points to all the individual spaces. This is related to the notion of the “canonical mesh” used in SPM8Footnote 2 (Litvak and Friston 2008; Mattout et al. 2007) rigorously applied for surfaces in Valdés-Hernández et al. (2009). The LF matrix comparisons were only performed for volumetric models. Thus, the transformations were estimated by spatially normalizing the individual MRIs to our MRI template using the SPM8 “unified segmentation”Footnote 3 (Ashburner and Friston 2005).
Using FreeSurfer,Footnote 4 we divided the template and all individual MRIs into three triangular meshes defining the boundaries of three compartments representing the brain, the skull and the skin, with of homogeneous conductivity of 0.33, 0.0041 and 0.33 S/m, respectively. Figure 1a, b depict this volume conductor model. These compartments were divided into meshes of tetrahedra using Tetgen 1.50 (quality criterion \(q = 1.414\) and volumes no larger than 1, 0.5 and 0.5 mm3 for the skin, the skull and the brain, respectively). This yielded a mesh of about 4 million tetrahedra, shown in Fig. 1b.
Electrode Positioning
The EEG mini-cap scaffold contains 32 electrodes and three jutting circles used as landmarks (Bae et al. 2015), as shown in Fig. 1c. After identifying the latter in the MRI template, the electrode positions are generated in the corresponding head model. The details about this procedure can be found in Bae et al. (2015).
In this paper we generated the electrodes positions on the scalp template using this strategy. Based on our experience, we randomly shifted the landmarks to simulate a small lack of precision in the routinely experimental procedure to place the mini-cap on the rat head. The larger shifts were conferred along the AP direction (maximum ~2 mm) since an experimentalist is less prone to shift the mini-cap to one side of the rat head given its lateral symmetry. Thereby, we produced five different mini-cap configurations. We located the electrodes in each individual of the MRI in a different manner. We first took a photo of the electrodes in the template. Several volunteers, familiarized with both rat anatomy and EEG recordings, were asked to take this photo as a guide to manually place the mini-cap on each individual scalp surface model extracted from MRI the dataset, again using the landmark-based strategy. This second procedure simulates the errors committed by a researcher trying to identify the electrode locations during head modeling based on the experimental documentation. This was done for the five configurations of electrodes.
Electrode Sensitivity and Resolution
A lead is a pair of electrodes on the scalp, say \(i_{E}\) and \(i_{R}\). According to the Helmholtz’s reciprocity theorem the sensitivity for this lead is \({\mathbf{s}} = {\mathbf{k}}_{{i_{R} ,j}} - {\mathbf{k}}_{{i_{E} ,j}}\), for every source point \(j\) (Malmivuo et al. 1997; Rush and Driscoll 1969). The scalar sensitivity is defined as the norm \(S = \parallel {\mathbf{s}}\parallel\). It is desirable to have a sensitivity as concentrated in a small volume as possible, because localization of sources is not possible with leads which sensitivity is homogenous over the entire source space. The concept of HSV is used to quantify the ability of the lead to concentrate its sensitivity in a region (Malmivuo et al. 1997). It is the volume of the source region for which the scalar sensitivity is higher than half of the maximum scalar sensitivity. Likewise, the FSV (Wendel et al. 2008) is the volume of the source region for which the scalar sensitivity is higher than a fifth of the maximum scalar sensitivity. The greater the HSV and the FSV, the more homogenous the sensitivity profile and the worse the ability of the lead to accurately localize sources. Thus, the HSV and FSV provide measures of electrode resolution.
The HSV and FSV have been evaluated in spherical models fitted to human head sizes (Ferree et al. 2001; Malmivuo et al. 1997; Malmivuo and Suihko 2004; Wendel et al. 2008), which is a simplified approximation of the human head geometry. The common practice is to calculate curves of HSV/FSV versus angle or distance separation of the lead. From these curves conclusions regarding electrode resolution of different EEG caps are drawn. The HSV/FSV values for specific lead separations correspond to the resolution of certain electrode cap densities. For example, for a scalp sphere of radius 9.2 cm, the separations 12.4° (2 cm), 15.5° (2.5 cm) and 31° (5 cm) corresponds to the human montages of 256, 128 and 32 electrodes, respectively. Besides, the practical limits for EEG cap density can be identified from the HSV/FSV curves. For increasing cap density (decreasing lead separation) the curve decreases until a baseline is reached. This baseline is the maximum achievable sensitivity resolution and the lead separation at which it starts was called the maximumspatial electrode resolution (Ferree et al. 2001). Electrode caps with densities such that the electrode separation is below the maximum spatial electrode resolution are measuring redundant information.
Due to the rotational invariance of the spherical model, the center of the separating leads can be placed at any point in the scalp. Thus the conclusions drawn from these previous curves are valid for every pair of electrodes in an EEG cap. However, the situation complicates for realistic geometries. Since there is no rotational invariance, we expect the HSV/FSV curves to depend on the location of the lead center. The spherical models may be enough to draw useful conclusions about human settings. However, the rat head is not well approximated by concentric spheres (see Fig. 1b). The scalp is cylindrical-like in the AP direction and both the skull and the brain are ellipsoidal. This implies that the skull-scalp separation is highly variable, steeply increasing along the ML direction. This suggests that the HSV/FSV-based electrode resolution analysis in scalp EEG of rats is scalp position-dependent. We envisaged that different conclusions would be drawn for every pair of electrodes in the rat EEG mini-cap.
To be exhaustive, we propose a novel methodology for realistic geometries: to calculate the HSV/FSV curves for centers located between every pair of electrodes of the rat EEG mini-cap in both the AP and the ML directions. From each curve, we quantified two measures of the sensitivity resolution of the electrode pair. These are the HSV and FSV values in the curves at the separation of the electrode pair. On the other hand, to ensure the electrode pair is not measuring redundant information, we checked if its separation is above the maximum spatial electrode resolution, also identified from the HSV and FSV curves. Since we can derive different maximum electrode resolution for each curve type, we take the maximum among both. The sensitivity was obtained from the LF matrix of the VolB model (Table 1) and the consequent HSV/FSV values were calculated by summing up the number of grid points and multiplying by the grid voxel size. To build the curves, the lead separations ranged from 0.05 to 15 mm at every 0.05 mm. Figure 2 illustrates how the whole procedure was done.
Sensitivity analysis of the rat EEG mini-cap. a A schematic of the position of the centers chosen for calculating the maximum scalar sensitivities, the HSV and FSV for increasing separating leads. The separation is indicated with the gray arrows along the cyan and green cylinders to both sides of the centers. Blue and red dots represent the centers for leads along the ML and AP directions, respectively. The centers were chosen as the middle points between each pair of electrodes of the mini-cap (represented as yellow cylinders). b Curves of S, HSV and FSV versus geodesic distance for two examples. The HSV and FSV are also reported in fractions of the brain volume. In all six curves, the yellow vertical line indicates the electrode separation of the mini-cap (3 mm for ML and 4 mm for AP). In the HSV and FSV curves, the red broken vertical line indicates the maximum spatial electrode resolution (Color figure online)
In spherical models, for small lead separations, the sensitivity profile within the source region contributing to the HSV is predominantly tangential to the electrodes (Malmivuo et al. 1997), though there are still contributions from radial dipoles around the source space points closest to the electrodes (Ferree et al. 2001). For increasing lead separation the radial contribution increases while the tangential decreases until, for very well separated electrodes, the HSV region is split in two identical sub-regions. Each sub-region surrounds the source space below each electrode, with the complete radial sensitivity profile described for one electrode (Ferree et al. 2001; Malmivuo et al. 1997; Malmivuo and Suihko 2004).
For the realistic rat geometry, this might not be straightforward. Thus, we also investigated the sensitivity to the main dipole orientation of the rat EEG mini-cap. For summarizing results in a simple manner, we propose a single vector that averages the orientation sensitivity profile across all the points within the HSV region. Given the sensitivity of a given electrode pair, i.e. \({\mathbf{s}}_{j} = {\mathbf{k}}_{{i_{E} ,j}} - {\mathbf{k}}_{{i_{R} ,j}}\), for which we omit the electrode indices for simplicity, let’s define the matrix:
where \(N_{HSV}\) is the number of source points within the HSV region. Let \(\lambda_{i}\) and \({\varvec{\upupsilon}}_{i}\), for \(i = 1,2,3\), be the eigenvalues and eigenvectors of \({\mathbf{C}}\)(\(\lambda_{1} > \lambda_{2} > \lambda_{3}\)). Since \({\mathbf{C}}\) is positive-definite and symmetric, its eigenvalues are positive and real. Therefore, \({\mathbf{C}}\) can be geometrically represented as an ellipsoid with principal axis \({\varvec{\upupsilon}}_{1}\) denoting the main orientation of the sensitivity profile within the HSV. For non-degenerate ellipsoids eccentricity of the ellipsoid takes values in the interval [0,1) and it is given by
For \(ecc = 0\), the distribution of the sensitivity is isotropic, while for \(ecc \to 1\) the sensitivity is anisotropic. Helmholtz’s theorem allows us to view the sensitivity of a lead as currents flowing from one electrode to the other. Since there is only one source and one sink, which are the electrodes, and these are relatively far from the significant sensitivity region, the LF cannot be isotropic at any region of the source space.Footnote 5 Therefore we do not expect low eccentricity values. However, even with a high eccentricity, the ellipsoid can be either “prolate” (\(\lambda_{1} \gg \lambda_{2} \approx \lambda_{3}\)) or “oblate” (\(\lambda_{1} \approx \lambda_{2} \gg \lambda_{3}\)). In the former case, it is clear that the sensitivity profile is mainly toward the principal eigenvector. However, for the latter case the orientation distribution is shared between the first two eigenvectors. To account for this, we propose the following vector to quantify the main orientations in the sensitivity distribution and also accounts for its variability:
For prolate (oblong) ellipsoids \({\mathbf{c}}\) is approximately collinear with \({\varvec{\upupsilon}}_{1}\) while for oblate ellipsoids is the weighted average of the first two principal directions. The size of this vector, given by \(ecc\), denotes orientation coherency. We calculated \({\mathbf{c}}\) for every ML and AP electrode pair of the EEG mini-cap.
Source Estimation Errors. The Cramér–Rao Lower Bound
Irrespective of the estimation method, the Cramér–Rao theorem provides the CRLB, which is the lower bound for the covariance of the difference between an unbiased estimator and the actual parameter values. Using the LF matrix of a spherical model fitted to the human head size, this theory was firstly applied in Mosher et al. (1993) to assess the CRLB, given the generative Eq. (1), of the noise variance \(\sigma^{2}\), the dipole amplitudes \({\mathbf{j}}\) and the dipoles positions \({\mathbf{r}} \in {\mathbb{R}}^{{3N_{D} \times 1}}\), where \({\mathbf{r}} = \left[ {\begin{array}{*{20}c} {{\mathbf{r}}_{1}^{T} } & \ldots & {{\mathbf{r}}_{{N_{S} }}^{T} } \\ \end{array} } \right]^{T}\) and \({\mathbf{r}}_{k} = \left[ {\begin{array}{*{20}c} {r_{kx} } & {r_{ky} } & {r_{kz} } \\ \end{array} } \right]^{T}\). The latter is explicitly accounted for in Eq. (1) by considering the dependency \({\mathbf{K}}\left( {\mathbf{r}} \right)\)(see Eq. (8) in Appendix 2). In this paper we apply this theory using the rat LF matrix.
In Mosher et al. (1993)’s paper, the only cause of error is the observational noise encoded in the parameter \(\sigma\). Based on the work of von Ellenrieder et al. (2006) we considered an additional cause of error: that of using the template LF matrix as a surrogate of the individual LF matrix. This error accounts for both geometry and electrode misspecifications. In this case the LF matrix is written as \({\mathbf{K}}\left( {\mathbf{r}} \right) = {\mathbf{K}}^{\left( 0 \right)} \left( {\mathbf{r}} \right) + \Delta {\mathbf{K}}\left( {\mathbf{r}} \right)\) where \({\mathbf{K}}^{\left( 0 \right)}\) is the template LF matrix and \(\Delta {\mathbf{K}}\) is a Gaussian random variable which encodes the variability of the LF matrix across different rats.
The Appendix 2 provides the derivation calculation of the CRLB for the errors of estimated dipole locations \({\mathbf{r}}\) and amplitudes \({\mathbf{j}}\), i.e. \({\mathbf{CRLB}}\left( {\mathbf{r}} \right)\) and \({\mathbf{CRLB}}\left( {\mathbf{j}} \right)\) [see Eq. (11)]. These explicitly depend on the covariance matrix of the voltage vector, i.e. \({\mathbf{C}}_{V}\). We propose to calculate this matrix using sample estimators across all individuals and the 5 electrode configurations described in Electrode Positioning” section:
where \(k = 1 \ldots N_{K} ,e = 1,2,3,4,5\), \(\theta_{p}\) is the p-th component of the vector of parameters \({\varvec{\uptheta}} = \left[ {\begin{array}{*{20}c} {\sigma^{2} } & {{\mathbf{j}}^{T} } & {{\mathbf{r}}^{T} } \\ \end{array} } \right]^{T}\). We used the VolB model (see Table 1) to calculate \({\mathbf{K}}^{{\left( {0,e} \right)}} \left( {\mathbf{r}} \right)\), \({\mathbf{K}}^{{\left( {{\text{k}},e} \right)}} \left( {\mathbf{r}} \right)\varvec{ }\) and their spatial derivatives, used to obtain \(\widehat{{\partial {\mathbf{C}}_{V} /\partial \theta_{p} }} = \partial {\hat{\mathbf{C}}}_{V} /\partial \theta_{p}\).
If \(\gamma_{{j^{\prime}}}\), \(j^{\prime} = 1, \ldots ,3N_{D}\) are the diagonal entries of \({\mathbf{CRLB}}\left( {\mathbf{r}} \right)\), a scalar error bound measure for the j-th dipole is defined as the root-mean-square (RMS) of the standard deviations of the dipole locations (Mosher et al. 1993):
On the other hand, is \(\beta_{j^{\prime}}\), \(j^{\prime} = 1, \ldots ,3N_{D}\) are the diagonal entries of \({\mathbf{CRLB}}\left( {\mathbf{j}} \right)\), an analogous measure can be also defined for the amplitude of the j-th dipole. However, we find more informative to define a standardized measure (Z):
If \(Z \le 1\) the estimated amplitude is smaller than its error and it is therefore not significant. We deem the scalar in Eq. (5) as quantitative measures of the upper bounds for the “estimability” or “detectability” of the dipole.
As in Mosher et al. (1993), we consider, for simplicity, that all dipole amplitudes are equal, i.e. \(Q_{j} = Q\) for \(j = 1, \ldots ,N_{D}\). We state, without providing the trivial demonstration, that this assumption allows for the substitutions \(\sigma \to \sigma /Q\) and \({\mathbf{j}}_{j} \to {\hat{\mathbf{d}}}_{j}\). Now both the RMS and Z only depend on the LF matrices, the factor \(f \equiv \sigma /Q\) and the positions and orientation of the dipoles. On the other hand, as expected, in the absence of LF matrix errors, i.e. \(\Delta {\mathbf{\rm K}} = 0\), the formula to obtain the RMS is identical to that derived by Mosher et al. (1993) and linearly proportional to \(f\), while Z is inversely proportional to \(f\).
We calculated the RMS and Z each every dipole oriented along X, Y or Z in the gray matter region defined as the source space model of VolGM (Table 1). Although this restriction was mainly conceived for saving computational cost, it is actually a physiological constraint for the dipole locations. However, the numerical derivatives of the LF matrices and \({\mathbf{C}}_{V}\) were calculated using VolB to avoid undesirable boundary effects (see Appendix 2).
Comparison of the LF Matrices
We compared the individual LF matrices with the template LF matrix using the RDM and the MAG (Gramfort et al. 2011; Meijs et al. 1989). The RDM measures topography errors. It is bounded above by 2 and its optimal value is 0. The MAG measures magnitude errors and its optimal value is 1. These are:
The supra-indices \(\left( {k,e} \right)\) and \(\left( {0,e} \right)\) refer to the k-th individual and the template LF matrices, respectively, for the e-th electrode configuration. Omitting them for simplicity, \({\mathbf{k}}_{j,c} = {\mathbf{K}}_{j} {\hat{\mathbf{r}}}_{c}\), \({\mathbf{K}}_{j} = \left[ {\begin{array}{*{20}c} {{\mathbf{k}}_{1,j}^{T} } & \ldots & {{\mathbf{k}}_{{N_{E} ,j}}^{T} } \\ \end{array} } \right]^{T}\) and \({\hat{\mathbf{r}}}_{c}\), for \(c = x,y,z\), are the unit vectors along the Cartesian axis. The sub-array \({\mathbf{K}}_{j,c}\) is the LF matrix for the j-th source and c component.
Simultaneous EEG-fMRI Experiment
This experiment was performed to:
-
1.
Compare the estimated EEG current sources with the fMRI activation.
-
2.
Compare the EEG inverse solutions obtained with: (a) the individual LF, calculated with actual subject MRI and the exact electrode positions (as identified in the MRI by the marks they leave on the rat scalp) versus (b) the template LF, obtained with the methodology described above.
Animal Preparation
All procedures were performed in agreement with the policies established by the Animal Care Committee at Tohoku University (approval code, 2011AcA-40). The experiment was conducted on a male Wistar rat (Charles River, Yokohama, Japan). The rat was anesthetized with 5 % of isoflurane. The scalp hair was carefully removed and the exposed skin was de-greased with ethanol. The rat was orally intubated and mechanically ventilated (SAR-830/AP, CWE Inc., Ardmore, PA, USA) and a muscle relaxing agent was administered (pancuronium bromide, 2.0 mg/kg/h i.v.). The rat was placed in the prone position on the MRI-bed and the anaesthetic was switched from isoflurane to α-chloralose (80 mg/kg as an initial bolus, followed by a constant infusion of 26.7 mg/kg/h). The core body temperature was carefully regulated during the entire experiment.
Forepaw Electrical Stimulation
A pair of small needle electrodes (NE-224S, Nihon Kohden, Tokyo, Japan) was inserted under the skin of the right forepaw for electrical stimulation. A block-design stimulation paradigm consisting of 10 blocks was employed, where each block comprised of a 30 s forepaw stimulation followed by a 40 s resting condition. A generator (SEN-3401, Nihon Kohden, Tokyo, Japan) and an isolator (SS-203 J, Nihon Kohden, Tokyo, Japan) were utilized to produce the electrical pulses (3 Hz, 3 mA, and 0.3 ms width).
EEG Recordings and Pre-processing
This dataset is actually previous to the 32-electrode mini-cap configuration presented in this paper. It was however acquired with a mini-cap with the sole difference of having 31 electrodes, presented by Sumiyoshi et al. (2011). The data were collected using a 32-channel MR-compatible BrainAmp system (Brain Products, Munich, Germany) with a sampling rate of 5 kHz. We applied both 0.5–250 Hz band-pass and 50 Hz notch filtering. An additional channel was used to record the electrocardiogram through a needle (Model 1025, SA Instruments, Stony Brook, NY, USA) that was inserted into the right hindpaw. To synchronize the EEG data with fMRI scanning, analog outputs (5 V) generated by an MRI console were digitized (16-bit resolution) as triggers to the EEG amplifiers and subsequently utilized for the offline removal of scanning artifacts. The reference and ground electrodes (NE-224S, Nihon Kohden, Tokyo, Japan) were subdermally inserted into the right and left earlobes, respectively.
The Brain Vision Analyzer software (Brain Products, Munich, Germany) was used for the offline correction of fMRI scanning artifacts. This software implements the adaptive artifact subtraction (AAS) method, in which the scanning artifact waveforms are segmented (−10 to 276 ms relative to the scanning trigger), averaged (after baseline correction), and iteratively subtracted from the EEG signals. The data were then down-sampled to 1 kHz, band-pass-filtered from 0.5 Hz to 70 Hz using a Butterworth filter with a 24 dB/oct slope and finally exported into a binary format. The subsequent EEG data analysis was performed using custom-written software in MATLAB (MathWorks, Natick, MA, USA). To reject the ballisto-cardiogram and other undesirable artifacts from the filtered EEG data, an independent component analysis-based subtraction method was employed. The mean ERP for each experiment was computed from 900 individual somatosensory evoked potentials (SEP) (from −50 to 250 ms, time-locked to the stimulus onset).
T1 and T2-Weighted MRI
The MRI data were acquired using a 7.0-T Bruker PharmaScan system (Bruker Biospin, Ettlingen, Germany) with a 38 mm diameter birdcage coil. A T1-weighted image was obtained using a 3D-RARE sequence with the following parameters: TR/TEeff = 300/8.5, RARE factor = 4, SBW = 100 kHz, flip angle = 90°, FOV = 34 × 34 × 30 mm3, matrix size = 272 × 272 × 60 and NAX = 4. T1-weighted images were utilized for the identification of the electrode positions. A T2-weighted anatomical images were obtained using the 2D-RARE sequence with the following parameters: TR/TEeff = 4600/30, RARE factor = 4, SBW = 100 kHz, flip angle = 90°, FOV = 32 × 32 mm, matrix = 256 × 256, voxel size = 125 × 125 μm, 54 slices with 0.5 mm thickness, slice gap = 0 mm and NAX = 24. This image was segmented and normalized using SPM8 (online at http://www.fil.ion.ucl.ac.uk/spm/software/spm8) and the template MRI (Valdés-Hernández et al. 2011). The segmentations were used to define the volumetric gray matter source space. The mid-cortex surface was calculated by shrinking the surface surrounding the brain mask, until reaching the mid layer between the pial and the white/gray interface. The skull boundaries were manually delineated. Using this head model and electrode positions, the LF matrix of the individual versions of the VolGM and SurfMid models were calculated. The resulting SPM spatial normalization transformation was used to warp the fMRI activation to the template space and the atlas digitalization to the individual space.
fMRI Experiment
The fMRI was acquired with the GE-EPI with parameters TR/TE = 2000 ms/15 ms, SBW = 250 kHz, flip angle = 30°, FOV = 25 × 14 mm2 matrix = 125 × 70, voxel size = 200 × 200 μm2, 7 slices with 1.5 mm thickness, slice gap = 0 mm, 370 volumes and 4 dummy scans. The fMRI data analysis was performed using SPM5 (online at http://www.fil.ion.ucl.ac.uk/spm/software/spm5), which consisted of adjustments of the acquisition timing across slices, correction for head movement, and smoothing using a Gaussian kernel of 0.8 mm full width at half maximum. The single-subject analysis of the pre-processed fMRI data was performed also using SPM5 with a critical T-value for each voxel (p < 0.05, FWE corrected).
Further SEP Experiments
We performed additional SEP experiments with forepaw, hindpaw and whisker stimulation on six adult Wistar rats (300–400 g, Charles River, US). This time with no individual MRI. We located the electrodes on the template according to the procedure described above, based on the landmarks in the scaffold, and calculated the template LF matrices for inverse estimation.
Animal Preparation
The study was approved and carried out in full appliance with Animal Care and Use Committee (IACUC) at Florida International University (IACUC 13-004). Anesthesia was induced using 5 % isoflurane and maintained at 1.5–2 % (mixed with 100 % O2 1 L/min) during the fixation of the rat to a stereotaxic apparatus (2 SR-5R, Narishige) and the EEG mini-cap setting. The scalp hair of the rat was trimmed, shaved and rubbed with absolute ethanol (90 %) for both stimulation of blood vessels and depilation of the skin. After the EEG mini-cap was secured on top of the rat head, the anesthesia was switched to dexmedetomidine hydrochloride (i.p 0.25 mg/kg, Dexdomitor, Orion Pharma). The body temperature (ML312 T-type Pod) and heart (FE136 Animal Bio Amp) and respiration (FE221 Bridge Amp) rates were continuously monitored (PL3508/P PowerLab 8/35 with LabChart Pro, AD Instruments) to guarantee a stable level of anesthesia during the entire EEG recording.
Forepaw and Hindpaw Stimulation
A needle electrode was inserted inside each forepaw and hindpaw for electrical stimulation. The stimulation followed a paradigm of 10 blocks with 16 and 44 s for stimulation and resting condition, respectively. The current stimulation was delivered by the isolated pulse stimulator (Model 2100, 110 V, 60 Hz, A-M Systems) with 10 ms pulse-width and 2.0 mA current amplitude at 3 Hz.
Whisker Stimulation
Both sides of whiskers were used to stimulate the vibrissae system of Wistar rats. The whiskers were shortened to 1 cm, and deflections were carried out from the AP direction. Both whiskers were deflected by air puffs of 10 ms duration. The air puffs were generated from a high-pressure air tank controlled by a pneumatic picopump (PV830, World Precision Instruments) at a pressure of 14 psi. A block-design paradigm was applied. Each block consisted of 16 s of stimulation and 44 s of resting period. The air puffs were delivered at 3 Hz within each stimulation period. Each air puff was delivered using a 1 m length tube and produced an inevitable loud sound associated with the switch on/off mechanism. Visual inspection reveal not only movement on the whisker but also on the facial fur.
EEG Recording and Processing
A 32-channel EEG amplifier (PZ3, Tucker-Davis Technology) were used for EEG recordings. EEG signals were amplified, band filtered between 0.5 and 250 Hz, notch filtered at 60 Hz and digitized (5 kHz SR, 0.5 μV resolution). Electrodes on the ears were used as ground (left) and reference (right). The data were then down-sampled to 2034.5 Hz. SEPs were created by averaging the EEG trials (−100 to 300 ms, time-locked to the stimulus onset). These number were more than 400 for all the SEP experiments.
An in-house software (LabVIEW, AD Instruments) was used to both generate the desired triggers for the stimulation and synchronize all the recording devices (PZ3, Model 2100, and PV830) via a multiple input/output analog-to-digital converter (PCI-6259, National Instruments).
EEG Source Imaging Methods
We solved the inverse problem using the LF matrices of models SurfMid and VolGM, and with two inverse methods: (1) exact LORETA (eLORETA) (Pascual-Marqui, 2007), implemented in the fieldtrip software packageFootnote 6 and the (2) LORETA-like implementation of SPM8 (SPM-LOR).Footnote 7 For the cases described in “Simultaneous EEG- fMRI Experiment” section, we also performed a group-based inversion to increase the reliability of detecting systematic responses over subjects (Litvak and Friston 2008).
Results
Electrode Resolution
As we have suggested, among all lead centers, there is a repertoire of different behaviors in the HSV and FSV curves versus lead separation which we shall not describe for brevity. In general, the main difference between these curves and those for spherical models is that the former decrease after a certain lead separation, while the latter saturate. This can be seen in Fig. 2 for the ML and AP directions. A detailed discussion about the reasons for this is beyond the scope of this paper.
The only cases worthy of discussion are those centers near the lateral, the anterior and the posterior boundaries of the source space. In those cases, the decrease, especially of the HSV, starts early in the curve even at lead separations below those of the electrode pairs of the EEG mini-cap. This is because the contribution to the HSV/FSV is mainly from only the electrode closer to the source space, for the other is already too far. This has two implications: (1) the sensitivity resolution, as quantified by the HSV/FSV results better than expected and (2) the sensitivity profile is more radial, or more properly said since we are not dealing with spheres, oriented toward the electrodes. Note that the AP example depicted in Fig. 2 is one of these cases: the lead center is one of the most posterior and the HSV/FSV curves decrease after 4 mm.
Figure 3 illustrates the behavior of the sensitivity for three lead centers. We chose a ML center close to the lateral boundary of the mini-cap (Fig. 3a) and an anterior AP center (Fig. 3b) to demonstrate how they are affected by the aforementioned electrode-source separation effect. For these cases, the sensitivity profile is completely due to one of the electrodes and thus radial at the corresponding separation of the mini-cap. There is another example in Fig. 3c corresponding to a ML center close to the medial plane and central. For this case, the sensitivity profile at the corresponding electrode separation of the mini-cap is still tangential with radial contributions. Figure 3 is also intended to illustrate how the HSV, FSV and sensitivity evolves with lead separation.
Sensitivity behavior for increasing separating leads (green cylinders) for a ML center (black dot). The results for four lead separations are shown, corresponding to the leads represented with the magenta cylinders. To give an idea of the extension and depth of the HSV and FSV regions relative to the brain volume we represent them with dark and light brown surfaces respectively, while the brain volume is represented with a light cyan surface. For each separation case, the scalar sensitivity (values according to the colorbars, units: Ω/m) of the slice containing the leads was projected outside the brain to ease the illustration. The intersections of this slice with the HSV, FSV regions and the brain are represented with a black, light brown and blue curves respectively. To the left, a zoom of the sensitivity profiles for 2 and 15 mm are shown. For the former the lead is more sensitive to tangential dipoles while for the latter the lead is completely sensitive to radial dipoles (Color figure online)
We consider that, even in the presence of these realistic rat geometry effects, the criteria to assess the maximum electrode spatial resolution (Ferree et al. 2001) can remain the same, for differences in the HSV/FSV still mean non-redundant information. We used both HSV and FSV since they provide some complementary information. The former gives a better measure of which source space region is mostly contributing to the sensitivity. The latter is less sensitive to geometry and provides a better measure regarding how much of the whole brain is actually measured by the electrodes. This can be seen in Fig. 3.
Figure 4 summarizes the results for all electrode pairs and corresponding lead centers. Assuming smoothness of the results, we interpolated the values over the whole measurement region to create scalp topographies for better visual understanding. The first conclusion is that all electrode pairs in our rat EEG mini-cap are measuring non-redundant information. A useful information for cap builders is that the limit for cap density in rat scalp EEG is lower in the boundaries of the region, and probably is worse beyond. The electrode resolution, as quantified by the HSV and FSV values, is worse in the boundaries of the mini-cap as well. Although the maximum HSV is higher for the ML topography than for the AP topography, the HSV and FSV resolutions along the AP direction are in general worse than along the ML direction.
Results of the HSV/FSV-based electrode resolution for scalp EEG in rats. The values of the topographies at each location indicates the interpolated result for a pair of electrode at that location. The results are shown only for one hemisphere for obvious symmetry reasons. The first column shows the topographies of the maximum electrode spatial resolution, i.e. the minimum electrode separation for which a pair of electrode is not measuring redundant information. This value is the maximum, i.e. the worst, between those obtained from the HSV and FSV curves. They are also bounded from below by the electrode size of our mini-cap (0.8 mm). Since the separations between AP and ML electrodes pairs of our EEG mini-cap are 3 mm and 4 mm respectively, the cap is not measuring redundant information. The second and third columns report the sensitivity resolution of our rat EEG mini-cap. The fourth column shows the main orientation of the sensitivity. This is represented with the RGB code. Red ML, green AP and blue IS. As expected the ML leads in the middle of the measurement region are mainly sensible to tangential ML oriented dipoles, while in the boundaries to radial dipoles. The central AP leads are more sensitive to tangential AP dipoles, while anterior and posterior AP leads are more sensible to radial dipoles (Color figure online)
The topographies of the average sensitivity orientation, as calculated by Eq. (2), are also shown in Fig. 4. We used the RGB representation for this vector and the intensity of the color represents the LF coherence as quantified between 0 and 1. We note that for electrode pairs far from the boundaries, the main orientation of the sensitivity is a mixture of tangential and radial contributions. This corresponds to magenta (red + blue) and cyan (green + blue) colors for ML and AP orientations, respectively. This is consistent with the LF observed in the medial ML center depicted in Fig. 3c. On the other hand, the main sensitivity orientation for electrode pairs closer to the boundaries is more along the IS direction (bluer color), i.e. they are more radial. This is consistent with the LFs observed in Fig. 3a, b. Note that, for the AP centers in the lateral boundary, the main orientation is a combination of the three ML, AP, and IS directions (whiter color). We point out that this does not mean there is a mixture of sensitivity orientations resulting in an isotropic sensitivity profile. It means that the main orientation is in one of the octants of the 3D space. This is the average of a field that flows from the source electrode along both the ML and IS directions down to the source space, taking the AP direction and returning again along both the ML and IS directions up to the sink electrode. The intensity of all vectors is above 0.7, denoting the expected high coherency in the LF and confirming that we are indeed mapping a coherent “main LF direction” within the HSV.
Mapping the Cramér–Rao Lower Bound
To report RMS and Z [Eqs. (4) and (5)] we chose a value for \(f \equiv \sigma /Q\). We used an estimate of \(\sigma\) obtained from the SEP experiments presented in this paper. After averaging across trials, the absolute value of the signal across the pre-stimulus interval and across experiment is about 2.5 ± 1.0 μV. On the other hand, the equivalent current dipole in whisker stimulation was measured in the barrel cortex (Riera et al. 2012), being \(Q\sim 10 {\text{nAm}}\) at around 50 ms, where the peak occurs. This yields \(f \approx 250\,\varOmega /{\text{m}}\).
We summarize the values of RMS and Z by only reporting their average across X, Y and Z dipole orientations. Figure 5 shows the values of RMS and Z for a single dipole located at each point of the source space model of VolGM. As expected from the high co-planarity in the electrode configuration, and due to the dependency of the values with the dipole-electrode distance, the values of RMS and Z approximately vary in layers from superficial to deep sources, with the latter being very difficult to estimate correctly.
CRLB results for the single dipole case. a–d corresponds to an observational noise level of σ = 2.5 µV. a RMS for the forward model considering both noise and model uncertainties. b RMS of the noise only case. c Z for the noise and model uncertainty case. d Z for the noise only case. Each value in the image corresponds to the results for a dipole located at that point. Only the points in the gray matter source space (VolGM) model are shown since this is usually the only space where dipoles are fitted. The medial sagittal and mid-axial slices are shown. Note that the RMS between a and b look similar, while for Z in d is bigger than in c. e Histograms of the RMS for the noise only case (blue bars) and the noise and model uncertainty case (red bars) for various values of σ (growing from top to bottom). Note that for low σ the RMS is completely due to model uncertainties. For σ = 2.5 μV the RMS distribution is slightly higher for the model uncertainty case. For high σ, both cases are completely dominated by noise, being identical (Color figure online)
In the range of the noise of ERP experiments, the use of the template as a surrogate of the individual implies a slight increase in the lower bound of localization errors with respect to the use of a model only affected by noise. The point-wise difference, averaged across all points, is 0.5 ± 0.2 mm. With the decrease of \(\sigma\), the RMS for the model uncertainty case reaches a limit around 0.05 mm.
With the aid of our digitalization of the Paxinos and Watson atlas, we summarized in Table 2 the mean and standard deviation of RMS and Z across the points belonging to each rat cortical region, for \(\sigma = 2.5\,\mu {\text{V}}\). For brevity, among the 96 structures of the atlas, we only present some of those related to the somatosensory evoked potential (SEP) experiments of this paper. The results for the rest of the cortex is summarized using conglomerates of structures with functional relation. These were also formed attending to closeness and similar depth, based on the fact that depth is probably the most determinant variable on the results. For example, the secondary somatosensory system was left apart from the whole somatosensory system because the former is significantly deeper than the latter. Likewise, we separated the parietal association cortex from the parietal system. On the contrary, other systems like the visual or auditory systems were formed by grouping their corresponding primary and secondary structures since they are all at approximately the same depth.
In order to quantify how large is the RMS compared to the size of the anatomical structure containing the dipole, we defined the relative linear error (RLE). This was calculated for each anatomical structure of the atlas as:
where the denominator is the root-mean-square of linear size of the structure, taken across the AP, ML and IS dimensions. Each linear size is the length of the structure along that dimension. We also report the RLE in Table 2. For SEP experiments, if the dipole would be located at the expected somatosensory region, its RMS would be roughly between 10 and 20 % of the root-mean-square of the linear size of the cortical region.
For brevity we do not show results for more than one dipole. As in Mosher et al. (1993), we also assessed the CRLB for two dipoles, one fixed in space and the other moving across all points of the source space model of VolGM. We omit the results for brevity. We found that two dipoles cannot be estimated for average separations below 3.1 ± 1.4 mm and using the template as a surrogate of the individual model implies an extra localization error of about 0.5 mm that cannot be reduced by decreasing the \(\sigma\). This corresponds to about 3–4 voxels in the template MRI
Comparisons of the LF Matrices
Table 3 shows the values of the RDM and MAG calculated using Eq. (6), for the comparison between the individuals and the template LF matrices. For almost all comparisons, the RDM is very close to 0 and the MAG is very close to 1. These provide evidence for the use of the template LF matrix for ESI in rats. They are not surprising since rat heads are very similar.
Simultaneous EEG-fMRI Experiment
Figure 6 shows where the fMRI activation occurs. The SPM T-map for values above 3 is shown. The activation mostly falls within S1FL, contralateral to the stimulated paw, though a small part is shared with the central region of M1. We shall refer to the locations of the brain as contralateral and ipsilateral to the stimulated side.
Sagittal slices depicting the fMRI activation after the forepaw stimulation in the EEG-fMRI experiment. Red fMRI activation. Green S1FL. Magenta M1. Both the fMRI activation area and atlas structure are contralateral to the stimulated forepaw. It can be seen that the activation is mostly within S1FL but a small part appears in M1 (Color figure online)
The SEP resulting from averaging the 900 trials is shown in Fig. 7. We chose six time instants to show their respective topographies. These instants were chosen after inspecting the forthcoming EEG inverse solutions. We consider them as the instants when the most relevant spatiotemporal features of the solution occur. No wonder that, excepting instant 55 ms, they coincide with the peaks of the SEP. In fact, some of these peaks are well described and reproducible components of SEP in paws stimulations. The first is P1. It is positive and occurs at 15 ms. The second, N1, is negative and occurs at 25 ms. The third is P2 and occurs at 65 ms. They have well localized contralateral topographies. These peaks have been identified for electrical forepaw stimulation elsewhere, e.g. Franceschini et al. (2008). Like in that paper, were could not identify the contralateral N2 component.
Elicited SEP (32 channels) after forepaw stimulation in the EEG-fMRI Experiment. This is the average of 900 trials. Time is locked to the stimulus onset. The topography (in a schematic model of the actual measured region on the rat scalp) of six time instants are shown. These are the same instants chosen for depicting the inverse solutions in the next two figures. As can be seen in the SEP, these time instants were not chosen arbitrarily but coincide with the main maxima and minima of the signals. Three components of the SEP can be identified: P1 (15 ms), N1 (25 ms) and P2 (65 ms)
On the other hand, there is a marked ipsilateral positivity at 25 ms, though its amplitude is lower and its topography is less localized than that of N1. Moreover, there is a localized ipsilateral negative component at 45 ms. It seems as if there would be a translation of the N1 component toward the ipsilateral hemisphere during the interval 25–45 ms. This is suggested by the topography at 35 ms. The inverse solutions for these instants are shown in Figs. 8 and 9.
ESI in the mid-cortex surface model for the forepaw stimulation in the EEG-fMRI experiment. The eLORETA and SPM-LOR methods are presented. The absolute value of the inverse solutions are shown for both the exact individual model (with exact MRI-based electrode positioning) and the template model built as described in this paper. Only the 99 percentile of the values of the inverse solutions are shown. Besides, values below 25 % of the total maxima across surface vertices and time points were also discarded to avoid spurious inverse solutions at instants of no expected ESI activity. The color bars linearly map the values from the lowest to the highest value across all vertices and time instants. The green, red and blue contours delineate the intersection of this surface with the volumes defined by S1FL, S1HL and M1. The black contour is the intersection of the surface model with the volume formed by the T-map activation for values above three. Evidence for the use of the template as a surrogate of the individual is the great similarity between the solutions using the individual and the template models. If we expect that the solutions for the N1 and P2 components of the SEP coincide with the fMRI activation, it seems that the eLORETA outperforms the SPM-LOR. The solutions look in some agreement with what is described in the literature using VSD (Color figure online)
ESI in the volumetric gray matter volume for the forepaw stimulation in the EEG-fMRI experiment using the SPM-LOR. The absolute value of the inverse solutions are depicted for both the exact individual model (with exact MRI-based electrode positioning) and the template model built as described in this paper. Only the 99 percentile of the values of the inverse solutions are shown. Besides, values below 25 % of the total maxima across surface vertices and time points were also discarded to avoid spurious inverse solutions at instants of no expected ESI activity. The color bars linearly map the values from the lowest to the highest value across all vertices and time instants. The light gray surface represents the brain. To the left, the green, red and blue surfaces represent S1FL, S1HL and M1, respectively. The results very much resemble those for the surface model with this inverse method (Color figure online)
By significant ESIactivity, we considered the absolute value of the solution which is above the 99 percentile at each instant and above 25 % of the overall maximum across spatial points and instants. The latter restriction is based on the CRLB results. Table 2 shows that the range of Z for somatosensory regions is ~6 to 8. However these values were calculated assuming the observed maximum dipole amplitude \(Q\) in somatosensory responses. Based on the rough relation \(Z \propto f^{ - 1} \propto Q\), equivalent dipoles with amplitudes 6–8 times smaller than this maximum, cannot be correctly estimated. This does not preclude the estimation of spurious inverse solutions. Based on this rationale, but being cautious, we decided to discard the inverse solutions with an absolute value 4 times (25 %) smaller than the overall maximum. Finally, the overall minimum of the thresholded absolute value was subtracted and the resulting values normalized to 1, by dividing by the overall maximum.
The ESI activity for the SurfMid model are shown in Fig. 8. This figure contains the results for the template and the individual models and for the eLORETA and SPM-LOR methods. For both inverse methods, the ESI activity appears around contralateral S1FL at 14 ms and peaks at 15 ms, disappearing after 16 ms. At 20 ms the ESI activity reappears around contralateral S1FL, with the maximum peak at 25 ms, and lasts until 36 ms. Ipsilateral ESI activity appears at 41 ms in the anterior region of contralateral M1 and in the region occupied by the contralateral secondary somatosensory area which moves toward S1FL until the end of the analyzed interval (5–70 ms).
The main difference between both inverse methods is that while eLORETA localizes the ESI activity almost completely within and centered in the S1FL region, SPM-LOR shifts the center toward the central part of M1. The second main difference between both methods is that the ipsilateral ESI activity at 45 ms is more extended and anterior for SPM-LOR than for eLORETA.
Figure 9 shows the solution for the volumetric model (VolGM, see Table 1) using SPM-LOR. The results essentially resemble those obtained using SurfMid in Fig. 8. We do not show the eLORETA solution for the volumetric model because we consider the solution was plainly non-sense, for the ESI activity appeared at too deep regions and spread into exaggerated volumes toward the olfactory bulb. This might be a consequence of this type of standardized LORETA methods, which only guarantee zero localization error of the maximum of the solution, ignoring the whole spatial distribution (Pascual-Marqui 2007).
There is a great similarity between the ESI activity estimated using the template and the individual LF matrices in Figs. 8 and 9. We remind the reader that the individual solutions were estimated using the LF matrix calculated from the head model extracted from the individual MRI; after accurately identifying the electrodes in the MRI by the marks they leave on the rat scalp. On the other hand, the template solution was calculated using the template LF matrix calculated with the strategy described in this paper and in Bae et al. (2015).
The polarity of the SEP components P1, N1 and P2 is mirrored by the corresponding ESI activity using both methods. For the surface model, the ESI activity is negative for 15 ms and positive for 25 and 65 ms. For the volumetric model, the estimated distributed dipoles corresponding to the ESI activity are oriented inward for 15 ms and outward for 25 and 65 ms, almost all perpendicular to the cortical layers.
Further SEP Experiments
Due to electrolytic shortcut provoked by the smearing of the conductive gel, we discarded two rats from this dataset. Figure 10 shows the average across rats and hemispheres of the EEG inverse solutions after the forepaw, hindpaw and whisker. For obtaining these group-based inverse solutions, we used the method described in Litvak and Friston (2008) and implemented in SPM8. For brevity, we shall only depict the systematic effects in the sample, and not the individual solutions. However the latter very much resemble their averages. We have also restricted the presentation of the results for the volumetric model since both volumetric and surface solutions using SPM-LOR are alike, as we could verify in the EEG-fMRI experiment. In this respect we also deem the volumetric solution as more descriptive of the whole process, allowing for the estimation of both unconstrained dipole orientations and deep sources.
ESI in the volumetric gray matter model for forepaw, hindpaw and whisker SEPs, estimated using the template LF matrix. The absolute value of the average of the inverse solutions with group constraints using SPM-LOR is shown. Only the 99 percentile of the values of the inverse solutions are shown. Besides, values below 25 % of the total maxima across surface vertices and time points were also discarded to avoid spurious inverse solutions at instants of no expected ESI activity. The color bars linearly map the values from the lowest to the highest value across all vertices and time instants. The light gray surface represents the brain. To the left, the green, red, blue, orange, yellow and black surfaces (these two appear only in the bottom row) represent S1FL, S1HL, M1, SLBF, Pt and M2, respectively. Notice that forepaw ESI activations in this figure are different from to the one in Fig. 10. This is because the results in this figure is an average after a grouped estimation and the result in Fig. 10 is the estimation of an individual case (Color figure online)
Similar to the EEG-fMRI experiment, the forepaw ESI activity appears at 14 ms in both contralateral S1FL and in a small portion in the central M1. The ESI activity reappears later and peaks at 26 ms, centered at the S1FL. Then it attenuates, spreads and splits toward anterior and posterior regions. The ESI activity resurges during 50–70 ms. These patterns resemble those obtained from the EEG-fMRI experiments. For the hindpaw stimulation, the ESI activity also appears at 14 ms within the contralateral S1HL and remains concentrated in this region until it splits toward M1 after 23 ms. The maximum peak is reached at 33 ms mainly at both the contralateral S1HL and M1. The ESI activity continues and spread until 55 ms. For the whisker stimulation, the first activation appears at 15 ms in the contralateral Pt. It continues and spreads toward the contralateral S1BF, peaking at 48 ms. After this instant the ESI activity is shared by both structures until the end of the interval, but mainly within Pt.
Discussion
Interpretation of the Results from Human to Rat Scales
It is the usual practice in rodent Neuroimaging to multiply the dimensions of the voxels by 10, to be dealt with by the usual programs and algorithms used in human Neuroimaging, such as tissue segmentation, spatial normalization and fMRI analysis (Ashburner and Friston 2000, 2005). This is justified by a rough scaling of one order of magnitude between human and rat head sizes. Indeed the linear sizes can be easily obtained from an MRI. For the adult human brain, they are about 13, 13 and 18 for the ML, IS and AP directions, respectively. For an adult rat brain they are about 16, 11 and 24 mm, respectively.
Downscaling a head model by 10 only implies multiplying the LF matrix by 10−2. Thus, the relative spatial distribution of electrode sensitivity, and the resulting HSV and FSV do not change. Besides, the RMS is downscaled by 10 and the Z remains unchanged, yielding exactly the same conclusion regarding EEG and ESI. However, the rat head is not a plain 10-downscaled version of the human head. In fact, the analysis done for spheres fitted to human heads does not apply for rats, for the rat head and its conductivity compartments are very unlike to those of the spherical model, as can be seen in Fig. 1b. Besides, the area covered by the rat EEG mini-cap is more reduced and less curved than that of standard human head EEG montages. Furthermore, the relative skin/skull thickness is not the same between adult humans and rats, and does not distribute similarly and evenly across the head. These are the reasons that led us in this paper (a) to evaluate the electrode resolution for our rat EEG mini-cap, (b) to investigate the sensitivity profile of the electrodes of the mini-cap, and (c) to calculate the lower bounds for the source estimation errors under the template-based forward models in Eq. (1); all this after realistically modeling the rat head with the three conductivity compartments of brain, skull and skin. Nevertheless, the 10-scale rule can be used to approximately validate the range of the results of this paper. The first check is with the values of the LF matrix. The maximum value for the rat is ~33,000 Ω/m while for the sphere model fitted to human head (brain sphere 8 cm) it is about two orders of magnitude smaller, i.e. ~460 Ω/m.
An important question is whether our rat EEG mini-cap is measuring signals from the source space with enough sensitivity resolution. Figure 4 shows that the range of the HSV of our cap is ~2 to 23 mm3. Since there is no reference to compare but the reports using spheres fitted to the human head, we shall appeal to the aforementioned 10-scale rule. For separations equivalent to a human 129 channel cap there are reports of HSV = 2.8 cm3 (Malmivuo et al. 1997) or HSV ~6 to 8 cm3 (Ferree et al. 2001). For low density montages, such as the 19 channel cap, HSV ~22 to 37 cm3 (Ferree et al. 2001). According to this, we could conclude that our rat EEG mini-cap has a variable sensitivity resolution spanning from low to high density human EEG caps, depending on the measurement site.
Alternatively, the sensitivity resolution can be also reported independent of the scale of the model, i.e. in terms of the volume fractions relative to the volume of the brain. The rat brain volume is ~1950 mm3 (Valdés-Hernández et al. 2011), while the spherical brain volume fitted to a human head is ~683 cm3 (Malmivuo et al. 1997). Following again the 10-scale rule, given two HSV with the same values, but one in mm3 for the rat and the other in cm3 for the human head, the volume fraction relative to the source space of the former is about 2.8 times smaller than that of the latter and so the sensitivity resolution of the former is 2.8 better than the latter. But using volume fractions as measures of the relative sensitivity resolution tend to overestimate the resolution of the mini-cap, as compared to that of the human spherical model. This is because the elongated shape of the rat head model, which provides a large contribution to the source space model. We propose a different relative measure, more independent of this anisotropic shape. This is the ratio of the depth of the HSV region to the IS distance of the brain. The average maximum depth of the HSV region in our study is ~1.5 mm, thus this ratio is ~1.5/11 ≈ 0.14. A similar value is obtained for the spherical human head, i.e. ~2.5/2 × 8 ≈ 0.156. For this math, we’ve used the maximum depth of 2.5 cm reported by Ferree et al. (2001) and the diameter of the sphere (radius = 8 cm) as the IS distance.
A similar comparative analysis can be applied to the CRLB results. An exhaustive evaluation of the CRLB for different electrode configurations in the spherical model was described by Mosher et al. (1993). Among these, the so-called high density configuration of 37 electrodes, all grouped in the top of the sphere, is the one that mostly resembles our rat EEG mini-cap. Like with our mini-cap, the way in which the electrodes are spatially distributed furnishes a laminar configuration of increasing RMS values for increasing depth. We suggest that the reader compare our results for the noise only model, presented in Fig. 5b, with the Fig. 10 of that paper. Our value \(f = 250\,\varOmega /{\text{m}}\) is 6 times higher than that used in Mosher et al. (1993). Their suggestions were \(\sigma = 0.4\;\mu {\text{V}}\), \(Q = 10\,{\text{nAm }}\) and thus \(f = 40\,\,\varOmega /{\text{m}}\). If we use this value in our noise only model, the RMS must be multiplied by 6. This yields RMS similar to those in the aforementioned electrode configuration presented by Mosher et al. (1993), if we appeal to the 10-scale rule for comparisons.
Using Sample Estimators of the Covariance Matrix
The assessment of the CRLB for EEG forward models was firstly used to investigate the performance of MUSIC and the Maximum Likelihood estimator (Stoica and Nehorai 1989). Also, an exhaustive evaluation of the CRLB for different electrode configurations in the spherical model was described by Mosher et al. (1993). Yet, these initial approaches consider that the model is correct and only corrupted by unknown noise at the sensors. More recent papers investigated the effect of errors in the forward model on the CRLB, as well as on the source estimation error. For the generative Eq. (8), these model errors are as many as the variables that determine the LF matrix. For example, uncertainty of the skull conductivity was considered by Plis et al. (2007). They also evaluated the CRLB when this conductivity is also an unknown parameter to be estimated. On the other hand, the effect of incorrect electrode position was tackled by Beltrachini et al. (2011). Uncertainty of the parameters that characterize the surfaces defining the conductivity compartments was the subject of the CRLB analysis in von Ellenrieder et al. (2006). In those papers, the uncertainties (or errors) of the parameters that determine the LF matrix were theoretically modelled and propagated to the covariance matrix through the forward model. On the contrary, in the present paper, we use the actual variability of the LF matrices present in the sample to directly estimate the data covariance matrix, using Eq. (3). This approach could be biased by the lack of enough samples to provide an asymptotic estimate. However, the CRLB results in the aforementioned papers could be also biased because the parameters and functions chosen to model the uncertainties could be wrong. We think that a correct strategy might be estimating the data covariance matrix by characterizing the variability of either the LF matrix or the parameters that determine it, using a large enough sample. For too small samples, shrinkage estimators of the covariance matrix must be employed (Chen et al. 2011; Ledoit and Wolf 2004; Wiesel and Hero 2010). In this case, the way the estimated uncertainty of the parameters is propagated to the covariance matrix must be tackled carefully, for the variability might not be small enough to consider a linear relation between the LF matrix and the parameters, as was done by von Ellenrieder et al. (2006).
EEG Source Imaging of SEP in Rats
Figures 8 and 9 show that the ESI activation in the EEG-fMRI experiment is appreciably different between eLORETA and SPM-LOR. While for the former the ESI activations at 15 ms, 25 ms and 65 ms are almost completely localized within the S1FL region, for the latter the ESI activations are shifted toward the central part of M1. Actually, we cannot ascertain which method is outperforming the other since we don’t know exactly where the ESI activity should actually be. If we follow the classical somatotopic criterion (Brecht et al. 2004; Chapin et al. 1987), the ESI activity should be always at S1FL, favoring the eLORETA. In this case, the displacement of the SPM-LOR solutions would be simply the consequence of the known bias in localization error of LORETA methods (Pascual-Marqui 2007). However, recent functional studies using voltage sensitive dye (VSD) revokes this anatomical constraint (Ghosh et al. 2009; Morales-Botello et al. 2012). Electrical excitation results not only in sensory but also in proprioceptive input due to feeble muscle and joint movements. On the other hand, if we consider that the ESI activity correlates with the fMRI activation, then a small shift of the former toward M1 is possible, especially around 25 ms and 65 ms, as previous studies suggest (Franceschini et al. 2010, 2008). These reveal that the components of the SEPs that most correlate with diffuse optical imaging (DOI) of hemodynamic responses are N1 and P2, rather than P1.
Let us compare our ESI results for the stimulations of the paws with those reported using VSD in rats (Ghosh et al. 2009; Morales-Botello et al. 2012). VSD maps the neuronal activity of the supra-granular 2/3 layer over large spatial scales with very high temporal resolution. It is highly correlated with the local field potentials (LFP) (Ferezou et al. 2007; Petersen et al. 2003). In a VSD study, the instant of appearance of the first activity (called “amplitude latency”) and the peak latency were found to be dependent on the intensity of the current applied to the paws (Morales-Botello et al. 2012). The mean and standard deviation across different rats at these latencies were higher for 0.6 mA than for 6 mA. We have not found experimental reports of the latencies for the 2–3 mA range we use in our experiments. Thus, we applied an inverse linear rule to provide a guess of the range in which our latencies might be. We stress that these are not real experimental values. We estimated that the contralateral amplitude latency and peak latency of the forepaw stimulation might be around ~14 ± 5 ms and ~24 ± 6 ms, respectively. On the other hand, our forepaw ESI amplitude latency is 15 ms for the EEG-fMRI experiment and 14 ms for the other forepaw experiment, while the peak latencies are 25 ms and26 ms, respectively. In the hindpaw stimulation, our estimated ranges are ~19 ± 4 ms for the amplitude latency and ~29 ± 6 ms for the peak latency; while our ESI amplitude and peak latencies are 14 and 33 ms, respectively. In summary, although the hindpaw amplitude latency of our experiment was low, the latencies are in general within the rough guessed ranges.
In our forepaw experiments, the spread and split of the ESI activity after the amplitude latency toward posterior or anterior regions is expected according to VSD results (Ghosh et al. 2009; Morales-Botello et al. 2012). Besides, the later spread toward motor regions in the contralateral and ipsilateral hemispheres of our EEG-fMRI ESI activations resembles the VSD images. The spread of contralateral VSD activity after the amplitude latency is more pronounced for the forepaw stimulation than for the hindpaw stimulation (Morales-Botello et al. 2012). This is consistent with the higher spread of ESI activation in our forepaw experiments, as compared with the hindpaw experiment (see Fig. 10).
On the other hand, our results for the whisker stimulation are somewhat arguable and provocative. Based on VSD images and LFP recordings (Petersen et al. 2003), the first activation is expected to be localized within S1BF. During a later time interval, the VSD activity spreads toward S2, the contralateral and ipsilateral M1 and anteriorly to the Agranular Medial Motor cortex (Agm) (Ferezou et al. 2007; Matyas et al. 2010; Tsytsarev et al. 2010), which is the central and anterior part of the secondary motor cortex (M2) as defined in the Paxinos & Watson atlas.Footnote 8 This spatiotemporal behavior of the activity is in agreement with Current Source Density (CSD) maps in layer 2/3 based on LFP recordings in both contralateral and ipsilateral S1, S2 and Agm (Mégevand et al. 2008). However, in our experiments the ESI activity appears at Pt and only moves toward the barrel field after 40 ms. There might be some explanations to this clear disagreement. First, our experimental setup has several differences with that of the aforementioned studies, e.g. whole-whisker air puff versus selected-whisker piezoelectric/tube, analgesia versus anesthesia, scalp electrodes versus subcutaneous/epicranial, and rats versus mice. But nevertheless, we believe that the fundamental cause is the careless inclusion of multimodal stimulation. First, the use of a 10 ms-duration air puff was visibly stimulating both the rat face and part of the body. The somatosensory representation of the body is the most posterior part of the somatosensory cortex, colliding with the parietal cortex (Brecht et al. 2004). This, combined with a possible localization bias of the LORETA, might be the reason of the ESI localization in Pt. But second, and probably more likely, the picopump used to create the air puffs produces a sharp sound (a click) that cannot be ignored since in this experimental protocol the rat is drugged medetomidine, i.e. not anesthetized. Thus, there might be a simultaneous somatosensory and auditory stimulation. If so, the activation at the posterior parietal cortex and its surroundings is expected, since the parietal cortex is the epicenter of the multisensory integration in the rat (Lippert et al. 2013). Instead of discarding this data, given the inconsistency with classical results using other modalities, we decided to present the experiment to illustrate the scope of ESI in rats to deal with complex inquiries regarding global integration/processing of information across sensory and motor areas where consciousness is not compromised. This type of map of brain activity is not possible using other non-invasive modalities like fMRI, which do not have the temporal resolution needed to separate the different components of the sensorial responses.
In general, the significant ipsilateral ESI activity is difficult to detect when there is a high contralateral ESI activity. This is because, in that case, the ipsilateral is not above the 99 % chosen for thresholding the inverse solutions. Note that, as suggested by VSD studies, the ipsilateral maximum is 40–50 % of the contralateral one (Ghosh et al. 2009; Morales-Botello et al. 2012). Besides, averaging the inverse solutions of the second set of SEP experiments cancels the ipsilateral ESI activity that appears in the individual solutions. This is because its spatial and temporal variability is much higher than that of the contralateral ESI activity, consistent with VSD-based results in paw stimulations (Morales-Botello et al. 2012). On the other hand, the ipsilateral ESI activity is more pronounced in the EEG-fMRI experiment than in the individual solutions of the second set of SEP experiments. This might be a consequence of the different sedation drugs used. In a previous study, inter-hemispheric BOLD correlations were lower when using medetomidine (Domitor, a sedative very similar to Dexdomitor) than when using α-chloralose (Williams et al. 2010). We used α-chloralose anesthesia in our EEG-fMRI experiment, while the dexdomitor was used in the second set of experiments. This protocol difference owes to the datasets were acquired in different laboratories as part of disparate main research projects. While the α-chloralose anesthesia might be more suitable for fMRI localization (Williams et al. 2010), the dexdomitor analgesic is preferred for epilepsy studies (Bae et al. 2015). Although this heterogeneity in drug protocols does not affect the main goal of this paper, it could be account for in future studies. Nevertheless, there is no consensus regarding which drug is the gold standard for general stimulation protocols (Williams et al. 2010).
In general, the 99 percentile threshold does not correctly detect the ESI activity, at a given instant, in different spatial regions with large differences in amplitude. However, decreasing this threshold introduces spurious inverse solutions. We consider that proper statistical confidence intervals, such as the desparsification methods (van de Geer et al. 2014; Zhang and Zhang 2014), must be used in future studies to correctly estimate the significant activity. This is still an unsolved problem in ESI nowadays.
Conclusions
In this paper, we evaluated methodological and theoretical aspects related to the EEG forward modelling of rats. We demonstrated that a valid ESI can be achieved using non-invasive scalp EEG recordings in rats. We also demonstrated that the LF matrix built from the minimum deformation MRI template can be used as a surrogate of the individual LF matrix without drastically compromising the accuracy of ESI.
By means of the HSV/FSV we demonstrated that the rat EEG mini-cap does not measure redundant information. Our results suggests that, for the scalp measured region, 32 electrodes is optimal. In order to increase the number of electrodes, the measurement region must be extended. However this might not represent a significant improvement given the significant decrease in the sensitivity resolution and the consequent redefinition of the whole cap density. We also demonstrated that for electrode pairs in the boundary of the mini-cap, the main contribution to the sensitivity comes from radial dipoles and the sensitivity resolution is worse. Appealing to “up-scaled” comparisons with human EEG caps, we conclude that the mini-cap concentrates the sensitivity enough to obtain a valid rat ESI.
We calculated the Cramér–Rao lower bounds of the error of any unbiased estimator of dipole location, orientation and amplitude. We model the use of a template LF by incorporating uncertainty in the LF matrix to account for imprecisions in the electrode locations and mismatches in the tissue-limiting surfaces. In contrast to previous approaches (Beltrachini et al. 2011; von Ellenrieder et al. 2006), we estimated the data covariance matrix of the model using sample estimators using the individual LF matrices of the database.
Our CRLB results suggest that the localization error of a dipole located at a primary somatosensory structure is such that the dipole is very likely estimated within the structure. Moreover, two equivalent dipoles can be estimated only if they are within different Paxinos & Watson structures. Besides, for the typical range of noise level in ERP experiments, the use of the LF template with approximated electrode positions yields only a slightly higher localization error than using the actual individual LF matrix. We provide further evidence by demonstrating a high similarity in topography (RDM) and magnitude (MAG) between the template and the individual LF matrices.
Finally, we performed ESI in real SEP experiments to demonstrate the accuracy of the whole methodology. We first estimated the ESI activity of a joint EEG-fMRI experiment after forepaw electrical stimulation. Face validity for the use of the template was evidenced by similarities of both individual-template ESI and ESI-fMRI activations. A second set of SEP experiments, consisting on forepaw, hindpaw and whisker stimulations, carved the initial path of the non-invasive scalp ESI-based exploration of rodent global sensory integration.
Notes
See notation in Appendix 1.
See (Valdés-Hernández et al. 2011) for details of the procedure with rats and our Template Set.
online at http://surfer.nmr.mgh.harvard.edu.
Note that \(\nabla {\mathbf{k}} = 0\). Thus the “volumetric LF flux density” is zero, i.e. \(1/G\mathop \smallint \limits_{G} {\mathbf{k}} \cdot d{\mathbf{G}} = 0\) for an arbitrarily small enough volume \(G\). This means that lead field lines do not diverge to form isotropic profiles.
online at http://www.fieldtriptoolbox.org/.
To avoid confusion: the Agm is considered by some authors as part of the primary motor cortex and M1 is called the Agranular Lateral Motor cortex (Agl) (Brecht et al. 2004). The whisker somatosensory motor region is suggested to be either at the Agm (Brecht et al. 2004) or in the boundary between and Agm and Agl (Smith and Alloway 2013).
Matrix \({\mathbf{A}}\) “no lower” than \({\mathbf{B}}\) means that \({\mathbf{A}} - {\mathbf{B}}\) is a semi-positive matrix.
Abbreviations
- EEG:
-
Electroencephalography
- ESI:
-
EEG source imaging
- MRI:
-
Magnetic resonance imaging (or magnetic resonance image)
- fMRI:
-
Functional MRI
- FEM:
-
Finite element method
- LF:
-
Lead field
- RDM:
-
Relative difference measure
- MAG:
-
Magnitude ratio
- AP:
-
Anterior–posterior (rostro-caudal)
- ML:
-
Mediolateral
- IS:
-
Inferior–superior (ventro-dorsal)
- HSV:
-
Half sensitive volume
- FSV:
-
Fifth sensitive volume
- CRLB:
-
Cramér–Rao lower bound
- RMS:
-
Root-mean-square of the CRLB of the localization error
- S1FL:
-
Forelimb region of the primary somatosensory (We’ll use the anatomical structures of the Paxinos and Watson atlas (Paxinos and Watson 2007).
- S1HL:
-
Hindlimb region of the primary somatosensory
- S1BF:
-
Barrel field of the primary somatosensory
- M1:
-
Primary motor cortex
- M2:
-
Secondary motor cortex
- Pt:
-
Posterior parietal cortex
- ERP:
-
Event related potential
- SEP:
-
Somatosensory evoked potential (somatosensory ERP)
- RGB:
-
Red–green–blue color code
References
Aghajani H, Zahedi E, Jalili M, Keikhosravi A, Vahdat BV (2013) Diagnosis of early Alzheimer’s disease based on EEG source localization and a standardized realistic head model. IEEE J Biomed Health Inform 17:1039–1045. doi:10.1109/JBHI.2013.2253326
Ashburner J, Friston KJ (2000) Voxel-based morphometry–the methods. Neuroimage 11:805–821. doi:10.1006/nimg.2000.0582
Ashburner J, Friston KJ (2005) Unified segmentation. Neuroimage 26:839–851. doi:10.1016/j.neuroimage.2005.02.018
Bae J, Deshmukh A, Song Y, Riera J (2015) Brain source imaging in preclinical rat models of focal epilepsy using high-resolution EEG recordings. J Vis Exp 100:e52700. doi:10.3791/52700
Beltrachini L, von Ellenrieder N, Muravchik CH (2011) General bounds for electrode mislocation on the EEG inverse problem. Comput Methods Programs Biomed 103:1–9. doi:10.1016/j.cmpb.2010.05.008
Bharadwaj S, Gofton TE (2015) EEG as a surrogate to brain imaging for diagnosing stroke in morbidly obese patients. J Neurosurg Anesthesiol 27:77–78. doi:10.1097/ANA.0000000000000074
Brecht M, Krauss A, Muhammad S, Sinai-Esfahani L, Bellanca S, Margrie TW (2004) Organization of rat vibrissa motor cortex and adjacent areas according to cytoarchitectonics, microstimulation, and intracellular stimulation of identified cells. J. Comp Neurol 479:360–373. doi:10.1002/cne.20306
Chapin JK, Sadeq M, Guise JL (1987) Corticocortical connections within the primary somatosensory cortex of the rat. J Comp Neurol 263:326–346. doi:10.1002/cne.902630303
Chen Y, Wiesel A, Hero AO (2011) Robust shrinkage estimation of high-dimensional covariance matrices. IEEE Trans Signal Process 59:4097–4107. doi:10.1109/TSP.2011.2138698
Choi JH, Koch KP, Poppendieck W, Lee M, Shin H-S (2010) High resolution electroencephalography in freely moving mice. J Neurophysiol 104:1825–1834. doi:10.1152/jn.00188.2010
Coben R, Mohammad-Rezazadeh I, Cannon RL (2014) Using quantitative and analytic EEG methods in the understanding of connectivity in autism spectrum disorders: a theory of mixed over- and under-connectivity. Front Hum Neurosci 8:45. doi:10.3389/fnhum.2014.00045
Darvas F, Ermer JJ, Mosher JC, Leahy RM (2006) Generic head models for atlas-based EEG source analysis. Hum Brain Mapp 27:129–143. doi:10.1002/hbm.20171
Do Carmo S, Cuello A (2013) Modeling Alzheimer’s disease in transgenic rats. Mol Neurodegener 8:37. doi:10.1186/1750-1326-8-37
Ferezou I, Haiss F, Gentet LJ, Aronoff R, Weber B, Petersen CCH (2007) Spatiotemporal dynamics of cortical sensorimotor integration in behaving mice. Neuron 56:907–923. doi:10.1016/j.neuron.2007.10.007
Ferree TC, Clay MT, Tucker DM (2001) The spatial resolution of scalp EEG. Neurocomputing 38–40:1209–1216. doi:10.1016/S0925-2312(01)00568-9
Franceschini MA, Nissilä I, Wu W, Diamond SG, Bonmassar G, Boas DA (2008) Coupling between somatosensory evoked potentials and hemodynamic response in the rat. Neuroimage 41:189–203. doi:10.1016/j.neuroimage.2008.02.061
Franceschini MA, Radhakrishnan H, Thakur K, Wu W, Ruvinskaya S, Carp S, Boas DA (2010) The effect of different anesthetics on neurovascular coupling. Neuroimage 51:1367–1377. doi:10.1016/j.neuroimage.2010.03.060
Gevins A, Brickett P, Costales B, Le J, Reutter B (1990) Beyond topographic mapping: towards functional-anatomical imaging with 124-channel EEGs and 3-D MRIs. Brain Topogr. 3:53–64. doi:10.1007/BF01128862
Ghosh A, Sydekum E, Haiss F, Peduzzi S, Zorner B, Schneider R, Baltes C, Rudin M, Weber B, Schwab ME (2009) Functional and anatomical reorganization of the sensory-motor cortex after incomplete spinal cord injury in adult rats. J Neurosci 29:12210–12219. doi:10.1523/JNEUROSCI.1828-09.2009
Gramfort A, Papadopoulo T, Olivi E, Clerc M (2011) Forward field computation with OpenMEEG. Comput Intell Neurosci 2011:1–13. doi:10.1155/2011/923703
Harmony T, Fernández-Bouzas A, Marosi E, Fernández T, Valdés P, Bosch J, Riera J, Bernal J, Rodríguez M, Reyes A (1995) Frequency source analysis in patients with brain lesions. Brain Topogr 8:109–117
Hughes JR, Ph D, John ER (1999) Conventional and quantitative electroencephalography in psychiatry. J Neuropsychiatry Clin Neurosci 11:190–208
Kaiboriboon K, Lüders HO, Hamaneh M, Turnbull J, Lhatoo SD (2012) EEG source imaging in epilepsy—practicalities and pitfalls. Nat Rev Neurol 8:498–507. doi:10.1038/nrneurol.2012.150
Kay SM (1993) Fundamentals of Statistical Signal Processing: Estimation Theory, ser. Signal Processing
Ledoit O, Wolf M (2004) Honey, I shrunk the sample covariance matrix. J Portf Manag 30:110–119. doi:10.3905/jpm.2004.110
Lee M, Kim D, Shin H, Sung H, Choi JH (2011) High-density EEG recordings of the freely moving mice using polyimide-based microelectrode. J Vis Exp. doi:10.3791/2562
Lippert MT, Takagaki K, Kayser C, Ohl FW (2013) Asymmetric multisensory interactions of visual and somatosensory responses in a region of the rat parietal cortex. PLoS One. doi:10.1371/journal.pone.0063631
Litvak V, Friston K (2008) Electromagnetic source reconstruction for group studies. Neuroimage 42:1490–1498. doi:10.1016/j.neuroimage.2008.06.022
Löscher W (2011) Critical review of current animal models of seizures and epilepsy used in the discovery and development of new antiepileptic drugs. Seizure 20:359–368. doi:10.1016/j.seizure.2011.01.003
MacRae I (2011) Preclinical stroke research—advantages and disadvantages of the most common rodent models of focal ischaemia. Br J Pharmacol 164:1062–1078. doi:10.1111/j.1476-5381.2011.01398.x
Malmivuo JA, Suihko VE (2004) Effect of skull resistivity on the spatial resolutions of EEG and MEG. IEEE Trans Biomed Eng 51:1276–1280. doi:10.1109/TBME.2004.827255
Malmivuo J, Suihko V, Eskola H (1997) Sensitivity distributions of EEG and MEG measurements. IEEE Trans Biomed Eng 44:196–208. doi:10.1109/10.554766
Mattout J, Henson RN, Friston KJ (2007) Canonical source reconstruction for MEG. Comput Intell Neurosci 2007:67613
Matyas F, Sreenivasan V, Marbach F, Wacongne C, Barsy B, Mateo C, Aronoff R, Petersen CCH (2010) Motor control by sensory cortex. Science 330:1240–1243. doi:10.1126/science.1195797
Mégevand P, Quairiaux C, Lascano AM, Kiss JZ, Michel CM (2008) A mouse model for studying large-scale neuronal networks using EEG mapping techniques. Neuroimage 42:591–602. doi:10.1016/j.neuroimage.2008.05.016
Meijs JWH, Weier OW, Peters MJ, Van Oosterom A (1989) On the numerical accuracy of the boundary element method. IEEE Trans Biomed Eng 36:1038–1049. doi:10.1109/10.40805
Morales-Botello ML, Aguilar J, Foffani G (2012) Imaging the spatio-temporal dynamics of supragranular activity in the rat somatosensory cortex in response to stimulation of the paws. PLoS One 7:e40174. doi:10.1371/journal.pone.0040174
Mosher JC, Spencer ME, Leahy RM, Lewis PS (1993) Error bounds for EEG and MEG dipole source localization. Electroencephalogr Clin Neurophysiol 86:303–321. doi:10.1016/0013-4694(93)90043-U
Pascual-Marqui RD (2007) Discrete, 3D distributed, linear imaging methods of electric neuronal activity. Part 1: exact, zero error localization. Math Phys. doi:10.1186/1744-9081-4-27
Paxinos G, Watson C (2007) The rat brain in stereotaxic coordinates, 6th edn. Elsevier, Amsterdam
Petersen CCH, Grinvald A, Sakmann B (2003) Spatiotemporal dynamics of sensory responses in layer 2/3 of rat barrel cortex measured in vivo by voltage-sensitive dye imaging combined with whole-cell voltage recordings and neuron reconstructions. J Neurosci 23:1298–1309
Plis SM, George JS, Jun SC, Ranken DM, Volegov PL, Schmidt DM (2007) Probabilistic forward model for electroencephalography source analysis. Phys Med Biol 52:5309–5327. doi:10.1088/0031-9155/52/17/014
Quairiaux C, Mégevand P, Kiss JZ, Michel CM (2011) Functional development of large-scale sensorimotor cortical networks in the brain. J Neurosci 31:9574–9584. doi:10.1523/JNEUROSCI.5995-10.2011
Riera J, Ogawa T, Goto T, Sumiyoshi A, Nonaka H, Evans A, Miyakawa H, Kawashima R (2012) Pitfalls in the dipolar model for the neocortical EEG sources. J Neurophysiol 108:956–975. doi:10.1152/jn.00098.2011
Rodrak S, Wongsawat Y (2013) EEG brain mapping and brain connectivity index for subtypes classification of attention deficit hyperactivity disorder children during the eye-opened period. Proc Annu Int Conf IEEE Eng Med Biol Soc EMBS. doi:10.1109/EMBC.2013.6611268
Rush S, Driscoll DA (1969) EEG electrode sensitivity–an application of reciprocity. IEEE Trans Biomed Eng 16:15–22. doi:10.1109/TBME.1969.4502598
Shevelkin AV, Ihenatu C, Pletnikov MV (2014) Pre-clinical models of neurodevelopmental disorders: focus on the cerebellum. Rev Neurosci 25:177–194. doi:10.1515/revneuro-2013-0049
Smith JB, Alloway KD (2013) Rat whisker motor cortex is subdivided into sensory-input and motor-output areas. Front Neural Circuits 7:4. doi:10.3389/fncir.2013.00004
Stoica P, Nehorai A (1989) Music, maximum likelihood, and cramer-rao bound. IEEE Trans Acoust Speech Signal Process 37:720–741
Sumiyoshi A, Riera JJ, Ogawa T, Kawashima R (2011) A mini-cap for simultaneous EEG and fMRI recording in rodents. Neuroimage 54:1951–1965. doi:10.1016/j.neuroimage.2010.09.056
Tsytsarev V, Pope D, Pumbo E, Yablonskii A, Hofmann M (2010) Study of the cortical representation of whisker directional deflection using voltage-sensitive dye optical imaging. Neuroimage 53:233–238. doi:10.1016/j.neuroimage.2010.06.022
Umeda T, Takashima N, Nakagawa R, Maekawa M, Ikegami S, Yoshikawa T, Kobayashi K, Okanoya K, Inokuchi K, Osumi N (2010) Evaluation of Pax6 mutant rat as a model for autism. PLoS One 5:e15500. doi:10.1371/journal.pone.0015500
Valdés-Hernández PA, von Ellenrieder N, Ojeda-Gonzalez A, Kochen S, Alemán-Gómez Y, Muravchik C, Valdés-Sosa PA (2009) Approximate average head models for EEG source imaging. J Neurosci Methods 185:125–132. doi:10.1016/j.jneumeth.2009.09.005
Valdés-Hernández PA, Sumiyoshi A, Nonaka H, Haga R, Aubert-Vázquez E, Ogawa T, Iturria-Medina Y, Riera JJ, Kawashima R (2011) An in vivo MRI template set for morphometry, tissue segmentation, and fMRI localization in rats. Front Neuroinform 5:1. doi:10.3389/fninf.2011.00026
van de Geer S, Bühlmann P, Ritov Y, Dezeure R (2014) On asymptotically optimal confidence regions and tests for high-dimensional models. Ann Stat 42:1166–1202. doi:10.1214/14-AOS1221
von Ellenrieder N, Muravchik CH, Nehorai A (2006) Effects of geometric head model perturbations on the EEG forward and inverse problems. IEEE Trans Biomed Eng 53:421–429. doi:10.1109/TBME.2005.869769
Vorwerk J, Cho JH, Rampp S, Hamer H, Knösche TR, Wolters CH (2014) A guideline for head volume conductor modeling in EEG and MEG. Neuroimage 100:590–607. doi:10.1016/j.neuroimage.2014.06.040
Wendel K, Narra NG, Hannula M, Kauppinen P, Malmivuo J (2008) The influence of CSF on EEG sensitivity distributions of multilayered head models. IEEE Trans Biomed Eng 55:1454–1456. doi:10.1109/TBME.2007.912427
Wiesel A, Hero AO (2010) Robust shrinkage estimation of high-dimensional covariance matrices 189–192
Williams KA, Magnuson M, Majeed W, LaConte SM, Peltier SJ, Hu X, Keilholz SD (2010) Comparison of α-chloralose, medetomidine and isoflurane anesthesia for functional connectivity mapping in the rat. Magn Reson Imaging 28:995–1003. doi:10.1016/j.mri.2010.03.007
Wolters CH, Grasedyck L, Hackbusch W (2004) Efficient computation of lead field bases and influence matrix for the FEM-based EEG and MEG inverse problem. Inverse Probl 20:1099–1116. doi:10.1088/0266-5611/20/4/007
Zhang C-H, Zhang SS (2014) Confidence intervals for low dimensional parameters in high dimensional linear models. J R Stat Soc Ser B 76:217–242. doi:10.1111/rssb.12026
Acknowledgments
We thank Lloyd Smith for his help on typos and English grammar; Dr. Eduardo Martinez Montes for his revision and suggestions and Joe Michel Lopez Inguanso for some useful searches in the Internet. We also thank Professor Pedro A. Valdés-Sosa for useful advices in the organization of the paper and the presentation of the main ideas.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
Cortech Solution’s has licensed a patent (US 9,078,584 B2) for the EEG mini-cap named “The Riera High-Density Non-Invasive EEG Mini-Cap” after its principal inventor Jorge Riera, the corresponding author of this paper. The patent was licensed from Tohoku University (Sendai, Japan). Authors Jorge Riera and Akira Sumiyoshi might respectively receive 10 and 5 % of the patent-generated royalties due to intellectual property rights. All other co-authors have nothing to declare.
Additional information
This is one of several papers published together in Brain Topography on the “Special Issue: Controversies in EEG Source Analysis”.
Appendices
Appendix 1: Notation
Lowercase bold letter are column vectors, uppercase bold letters are matrices and non-bold italic letters are scalars. The matrix \({\mathbf{I}}_{N}\) is the N-th order identity matrix and \(0_{N \times M}\) is the \(N \times M\) matrix of zero elements. The operator \(\otimes\) is the Kronecker product, \(\parallel \cdot \parallel\) is the \(\ell_{2} - {\text{norm}}\) and \({\text{diag}}\left( {{\mathbf{A}}_{1} ,{\mathbf{A}}_{2} , \ldots } \right)\) denotes a block-diagonal matrix.
Appendix 2: Cramér–Rao Lower Bounds for the Dipoles Estimation Error in the Presence of Noise and Uncertainties of the LF Matrix
The EEG generative model is that of Eq. (1), which we rewrite accounting for the dependency of the LF matrix on the dipole locations:
where, \({\mathbf{v}}\) is the voltage, \({\mathbf{j}}\) is the vector of dipoles, \({\varvec{\upvarepsilon}}\) is the observational noise and \({\mathbf{r}} \in {\mathbb{R}}^{{3N_{D} \times 1}}\), with \({\mathbf{r}} = \left[ {\begin{array}{*{20}c} {{\mathbf{r}}_{1}^{T} } & \ldots & {{\mathbf{r}}_{{N_{D} }}^{T} } \\ \end{array} } \right]^{T}\) and \({\mathbf{r}}_{j} = \left[ {\begin{array}{*{20}c} {r_{jx} } & {r_{jy} } & {r_{jz} } \\ \end{array} } \right]^{T}\) being the position of the j-th dipole.
The error due to model uncertainty is \(\Delta {\mathbf{K}}\left( {\mathbf{r}} \right) = {\mathbf{K}}\left( {\mathbf{r}} \right) - {\mathbf{K}}^{\left( 0 \right)} \left( {\mathbf{r}} \right)\), where \({\mathbf{K}}\) and \({\mathbf{K}}^{\left( 0 \right)}\) are the individual and template LF matrices, respectively. If the parameters that contribute to the calculation of \({\mathbf{K}}\) (e.g. shape of the surfaces, conductivities, etc.) have a normal variability and the variations are small enough, \(\Delta {\mathbf{K}}\) is also multivariate normal (von Ellenrieder et al. 2006). We expected small variations in our case given the usual similarity of the rats. This can be in fact evidenced by the high similarity between the individual and template LF matrices, as quantified by the RDM and the MAG measures (see “Source estimation errors. The Cramér–Rao Lower Bound and Comparisons of the LF matrices” sections). To ensure the approximation, we directly performed an element-wise Kolmogorov–Smirnov test under the null hypothesis of zero-mean normality. Figure 11 in Appendix demonstrates that only the 0.17 % of the elements of the LF are non-Gaussian. Therefore, we consider the zero-mean normality assumption of the LF elements justified. Consequently the voltage is also multivariate normal:
Demonstration of the Gaussian behavior of the elements of the LF matrices across individuals and electrode configurations. a Matrix of the same size of the LF matrix with the result of the Smirnov–Kolmogorov test under the null hypothesis of Gaussianity (p < 0.05). The red dots (which were augmented for visualization purposes) identify the elements where the null hypothesis was rejected. We note that the Gaussianity is violated more toward deep sources and anterior electrodes. This might imply an overestimation of the CRLB for deep and anterior dipoles. However non-Gaussian elements are barely the insignificant 0.17 % of the total number of elements. b Histograms of randomly chosen elements which are identified with blue dots in the matrix above. The histograms follow the same order, from left to right, of their corresponding blue dots (Color figure online)
For a vector of parameters to be estimated \({\varvec{\uptheta}}\), the Cramér–Rao theorem states that the covariance of the errors of the estimators \({\hat{\mathbf{\theta }}}\) cannot be lower than the inverse of the Fisher information matrix,Footnote 9 i.e.
For Gaussian likelihoods the Fisher information matrix is (Kay, 1993)
Thus, if \({\varvec{\uptheta}} = \left[ {\begin{array}{*{20}c} {\sigma^{2} } & {{\vec{\mathbf{J}}}^{T} } & {{\mathbf{r}}^{T} } \\ \end{array} } \right]^{T}\) the Fisher information matrix is then:
where \({\mathbf{X}} = {\text{diag}}\left( {{\mathbf{I}}_{3} \otimes {\mathbf{j}}_{1} , \ldots ,{\mathbf{I}}_{3} \otimes {\mathbf{j}}_{{N_{D} }} } \right)\) and \({\mathbf{D}} = \left[ {\begin{array}{*{20}c} {{\mathbf{d}}\left( {{\mathbf{r}}_{1} } \right)} & \ldots & {{\mathbf{d}}\left( {{\mathbf{r}}_{{N_{D} }} } \right)} \\ \end{array} } \right]\) stores the derivatives of the LF matrix with respect to the positions, being \({\mathbf{d}}\left( {{\mathbf{r}}_{k} } \right) \equiv \left[ {\begin{array}{*{20}c} {\partial {\mathbf{K}}_{0} \left( {{\mathbf{r}}_{k} } \right)/\partial r_{xk} } & {\partial {\mathbf{K}}_{0} \left( {{\mathbf{r}}_{k} } \right)/\partial r_{yk} } & {\partial {\mathbf{K}}_{0} \left( {{\mathbf{r}}_{k} } \right)/\partial r_{zk} } \\ \end{array} } \right]\) For \(\Delta {\mathbf{K}} = 0\) the first terms of the sub-matrices in Eq. (10) are identical to those in Mosher et al. (1993) while the second terms become zero. The latter are:
We have proposed in this paper to calculate \({\mathbf{C}}_{V}\) using sample estimators (see Eq. (3) in “Source Estimation Errors. The Cramér–Rao Lower Bound” section). Besides, the dependence of the LF matrix (and thus of CV) on dipole positions is smooth enough to approximate their values as piece-wise linear functions with a high accuracy. Therefore we use finite-difference operators for estimate D and \(\partial {\mathbf{C}}_{V} /\partial r_{p}\).
From Eq. (10) the CRLB for the location and amplitude of the dipoles are:
Rights and permissions
About this article
Cite this article
Valdés-Hernández, P.A., Bae, J., Song, Y. et al. Validating Non-invasive EEG Source Imaging Using Optimal Electrode Configurations on a Representative Rat Head Model. Brain Topogr 32, 599–624 (2019). https://doi.org/10.1007/s10548-016-0484-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10548-016-0484-4