Abstract
Glucose and urea enzymatic biosensors were fabricated. One-step electrochemical immobilization process was used to produce thin polyaniline films with entrapped enzymes. Chronopotentiometric analysis, scanning electron microscopy, electrochemical impedance spectroscopy and optical reflectance spectroscopy were used to determine the structure-property relationship of the functionalized polymeric thin films. The device has a recognition stage connected to a potentiometric field-effect-transistor stage and is based on the measurement of microenvironment pH variation or locally produced ions. Optimization of biosensor fabrication and effective measurement conditions were performed. The optimized films presented sensitivity, linearity and detection range to glucose of 14.6 ± 0.4 mV/decade, 99.8% and from 10−4 M to 10−1 mol/L. Two different biosensors were produced based on the enzymatic reaction of urea with selectivity to ammonium or hydroxyl ions. For ammonium ion selective film, the sensor’s parameters were 14.7 ± 0.9 mV/decade, 98.2% and from 10−5 to 10−1 mol/L. For the hydroxyl ion selective film, the same parameters were 7.4 ± 0.5 mV/decade, 98.1% and from 10−5 to 10−1 mol/L. The change in the oxidation state of the polymeric matrix explains: i) the large loss of functionality of glucose biosensor in time, ii) the conservation of functionality to the hydroxyl ions for urea biosensor and iii) the selectivity variation of the ammonium ion selective urea biosensor. The results indicate that the polymeric matrix has indeed changeable selectivity, what can be applied in different situations for biosensors production.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Biosensors are chemical sensors whose bio-analyte recognition system uses a biologically sensitive receptor, such as enzyme, living cell, DNA, antibody, and/or others (Gründler 2007; Stetter et al. 2003). The various components that make up a biosensor are the biologically sensitive element or biocatalyst, transducer and signal amplifiers/processors. For the specific case of enzymatic biosensors, the chosen enzyme is immobilized near the surface of the electrode, which allows the reduction of material needed to carry out an analysis. Among these, we can mention the enzymatic biosensors used to detect urea and glucose (Gürel et al. 1997). The enzymes used as the biological recognition element are urease (Ur) (Lakard et al. 2011) and glucose oxidase (GOx) (Liao et al. 2007), respectively. The most common immobilization techniques are: physical adsorption, covalent bonding, entrapment and crosslinking (Subramanian et al. 1999). Regardless of the method of immobilization, it must be simple to perform, highly reproducible (to favour the large-scale production of the biosensor) and to avoid non-specific binding and extreme environmental conditions. In addition, the biomolecule to be immobilized must be easily accessible after immobilization and chemically inert to the host structure.
Conducting polymers are of interest for biosensors because they function as the enzyme immobilization matrix. The conducting polymers are used to increase the speed, sensitivity and versatility of biosensors (Gerard and Malhotra 2005; Piao et al. 2015). Its use allows the immobilization of molecules of varying sizes and geometry and is particularly suitable for the manufacture of miniaturized biosensors. As for its use in conjunction with different transduction systems, in the specific case of potentiometric biosensors, the sensitivity of the conducting polymers to the pH variation or the presence of specific ions, results in a change in the energy levels, band levels, or work function of the polymer, which is consequently transduced by the potentiometric system.
Polyaniline (PANI) has been extensively used in the development of enzymatic biosensors, mainly due to its relative low cost and easy synthesis from aniline monomer in aqueous solution, easy doping/de-doping process, which can remarkably change its electronic structure, electrical conductivity and others. In addition, it presents high surface area and chemical specificities, redox conductivity and polyelectrolyte characteristics, direct and easy deposition on electrodes, biocompatibility, excellent environmental stability and effective anti-interferences property. The ability to control PANI properties by varying electrochemical polarization conditions and its ability to be not only the immobilization matrix but also the electron mediator in a redox or enzymatic reaction due to its excellent conductivity and inherent electroactivity are also highlighted (Lai et al. 2016; Teles and Fonseca 2008). As example, PANI and its derivatives have been deposited by cyclic voltammetry and then, functionalized by a layer of glucose-oxidase and chitosan to produce a chronoamperometry dose-response to glucose device (Mousa et al. 2018). A PANI-PEO (polyaniline-poly (ethylene oxide)) based glucose biosensor was also fabricated without the need for complex and expensive materials presenting an efficient and reproducible biosensor (Hansen et al. 2016).
This work describes the use of functionalized PANI thin films with glucose oxidase and urease enzymes through the one-step electrochemical immobilization process into potentiometric enzymatic field-effect-transistor (EnFET) biosensors. The structure-property relationship of the immobilization matrix is discussed concerning the properties presented by the biosensor such as sensitivity, linearity, detection range, long term stability, repeatability and selectivity.
2 Materials and methods
2.1 Reagents
Fluorine-doped tin oxide (FTO) thin films deposited on glass (Sigma-Aldrich) were used as substrates. The samples were cleaned with de-ionized water followed by acetone using the ultra-sonication method (15 min each) to remove contaminants from the surface. The other chemicals were aniline (C6H5NH2) (Vetec Brazil, > 99.5%), potassium chloride (KCl) (Cinética, Brazil, 99%), hydrochloric acid (HCl) (Sigma Aldrich, 99%), glucose oxidase (GOx) (Sigma Aldrich, EC 1.1.3.4), urease (Ur) (Sigma Aldrich, EC 3.5.1.5), glucose (anhydrous d-glucose P.A. A.C.S., Synth), urea (urea P.A. A.C.S., Vetec Brazil), anhydrous bibasic sodium phosphate (P.A.-A.C.S. Synth, 99.5%) and sodium phosphate monobasic (P.A.-A.C.S. Synth, 99.5%).
2.2 Functionalized PANI thin films fabrication
The functionalized PANI thin films were obtained by the galvanostatic entrapment of enzymes in a one-step process, by applying a desirable current density for a specific deposition time. The electropolymerization was carried out in a two-electrode system in a cell at 25 °C (Mello and Mulato 2018). The chronopotentiometric curve was recorded with a Data Acquisition model 34970A (HP). A platinum inert electrode was used as the counter electrode and FTO was used as the working electrode. The aqueous polymerization solution contained aniline monomer (0.1 mol/L), KCl (0.1 mol/L), de-ionized water and GOx or Ur, respectively for each kind of biosensor, in specific concentrations, namely 0.25, 0.50 and 1.00 mg/mL for GOx and 0.15, 0.30 and 0.60 mg/mL for Ur. The current density was 0.5 mA/cm2 and three deposition times of 300, 600 and 1200 s were used to obtain total deposition charges of 150, 300 and 600 mC/cm2, respectively.
The PANI thin films functionalized with urease were produced in two batches of three samples each, which selectivity to ammonium ions and hydroxyl ions, by changing the PANI oxidation state through chemical oxidation. After the electropolymerization process, the samples were dried during 10 min under flow in the exhaust hood prior to use. The produced glucose biosensor was labelled as GOx/PANI, while the urease biosensor selective to ammonium ions was labelled as Ur/PANI (NH4) and the one selective to hydroxyl ion was labelled as Ur/PANI (OH).
2.3 Sample characterization
The morphology of PANI films was studied using scanning electron microscopy (SEM). A JEOL microscope JSM-6610 model operating at 20 kV was used. A thin gold coating (≈20 Å) was applied to the samples prior to measurement. The average surface roughness analysis was performed using ImageJ (NIH) software. Electrochemical impedance spectroscopy (EIS) was used to analyse the conductivity of the materials and to verify whether they were altered due to the immobilization process. An AUTOLAB (Metrohm) with FRA module potentiostat controlled by the software NOVA was used. A conventional three-electrode cell system was used. The functionalized and non-functionalized PANI thin films were used as working electrodes, Ag/AgCl electrode was used as the reference electrode and the counter electrode was made of platinum. The measurements were performed at 25 °C in a 0.1 mol/L sodium phosphate buffer solution (optimum pH of each functionalized samples) under open circuit potential (OCP) vs. Ag/AgCl in the ac frequency ranged from 10 mHz to 100 kHz with ac voltage amplitude equal to 10 mV (rms). OCP was recorded prior of each experiment.
Visible reflectance spectroscopy was obtained non-destructively in the 400 to 700 nm range using a portable spectrophotometer model Colour-Guide, BYK-Gardner (Columbia, USA). The data were recorded in 20 nm step with a 20 mm diameter circular aperture and a 65/10° optical geometry. The device displayed the respective International Commission on Illumination (CIE) colour values: luminance (L*), the position between red and green (a*), and the position between yellow and blue (b*) (International Commission on Illumination 2004).
2.4 Biosensor measurements
The biosensor tests occurred in buffer solutions containing the analyte to be measured. We used 0.1 mol/L sodium phosphate buffer solutions prepared with anhydrous bibasic sodium phosphate and sodium phosphate monobasic in specific proportion. Once the buffer solutions are ready, we added glucose and urea to prepare the analytes solutions in the concentration range from 10−6 to 10−1 mol/L.
The EnFET potentiometric biosensor is based on the potentiometric IA-EGFET sensor (Mello et al. 2015; Mello and Mulato 2016). The EnFET biosensor is a potentiometric chemical sensor with high input impedance due to field effect transistors (FET) devices (van der Spiegel et al. 1983) having its recognition stage made of functionalized polymeric thin films with the enzymes responsible for the biological recognition of bio-analytes in solution. The IA-EGFET differential mode of operation uses a contrast film in the reference input of the IA circuit. The EnFET configuration presented here used a grounded reference input for preliminary studies. The potential data were recorded during 180 s for each analyte solution. The functionalized PANI thin films were carefully washed in de-ionized water in between each measurement.
The sensitivity (S, from the sensors’ calibration curve, units of millivolts per decade) and linearity (L, from the coefficient of determination R2 times 100) of the biosensors were measured for each specific detection range depending on the analysed bio-element (glucose or urea).
3 Results and discussion
3.1 Sample fabrication, characterization and optimization
The chronopotentiometric curves for GOx/PANI and Ur/PANI biosensors are shown in Fig. 1 (a) and (b), respectively, for varied enzyme concentration and total deposited charge. The deposition is characterized by an initial coating period, related to the initial deposition of PANI, followed by a process of smooth potential increase due to continuous increase in the film impedance caused by electrode passivation (Cheraghi et al. 2009). The chronopotentiometric curves for the pure PANI thin film and for the functionalized PANI thin films with GOx have similarity. The same occurring for the functionalized PANI thin films with Ur enzymes. The chronopotentiometric curves comparing the two distinct types of biosensors also presented similarity. These results indicate that the variation of the type of enzyme used and its concentration does not significantly affect the deposition process, which could be affected by variation of pH, temperature and other parameters concerning the electrochemical immobilization of enzymes in polymeric matrices (Uang and Chou 2003). The curves differ mainly in relation to the total deposition time, which was changed to allow varied deposited charges.
Functionalized PANI thin films characterization. The chronopotentiometric curves for GOx/PANI and Ur/PANI biosensors in (a) and (b), respectively, showing no main differences between the samples. The SEM morphology images for PANI, GOx/PANI and Ur/PANI thin films prepared under the same experimental conditions are shown in (c), (d) and (e), respectively. The RQ surface roughness parameter increased 12.5% by incorporating GOx into PANI and decreased 9.6% by incorporating Ur
The surface morphology for PANI, GOx/PANI and Ur/PANI thin films prepared under the same experimental conditions can be seen by the SEM images in Fig. 1 (c), (d) and (e), respectively. PANI thin film shows a typical granular morphology (Iroh and Rajagopalan 2000). After enzymes were immobilized, the original morphology of the thin film was slightly changed. The analysis of RQ (root mean square deviation) showed that by incorporating GOx into PANI the surface roughness increased 12.5% and by incorporating Ur it decreased 9.6%. This result indicates that addition of enzymes into PANI thin films has almost no effect in the sample morphology. No big difference seems to exist in the growth processes of GOx/PANI and Ur/PANI thin films, as previously indicated.
The investigated electrodeposition parameters were the enzyme concentration and deposited charge, obtained directly by variation of the deposition time. For each batch of produced samples with a specific enzyme concentration and deposited charge (see materials and methods section), the potentiometric response was evaluated from pH 5.6 to 7.8, with analyte concentration of 1 mmol/L for both glucose and urea. The final analysis was made using the difference between the output potential measured for the functionalized film and the pure PANI film, the latter usually having a larger value when compared to the biosensor. The functionalized films presenting the larger differential response were chosen as the optimized one. The optimized glucose biosensor was produced with 0.25 mg/mL of glucose oxidase and 600 mC/cm2 of deposited charge, while the optimized urea biosensor was produced with 0.60 mg/mL of urease and 150 mC/cm2 of deposited charge. These are the enzyme functionalized PANI thin film used in the presented EnFET biosensor.
The optimal buffer pH analysis was made based on the data previously described. It is shown in Fig. 2(a). The working pH range for the glucose biosensor is between 6.2 and 6.5. A less acidic optimum pH for glucose biosensor when compared to the optimum pH of the free enzyme (5.5) was already reported in the literature, for a glucose biosensor made with an organically modified sol-gel/chitosan composite, which is used to make covalent bonding of the enzyme that naturally changes its conformation and microenvironment (Chen et al. 2003). This effect can be less pronounced if other techniques are used for the immobilization. Ekanayake and co-workers presented a glucose oxidase based biosensor using entrapment by physical adsorption in polypyrrole with optimum pH of 6.5 (Ekanayake et al. 2007) while Kausaite-Minkstimiene and co-workers presented a glucose oxidase based biosensor with the enzyme entrapment within formed PANI films with optimum pH of 6.0 (Kausaite-Minkstimiene et al. 2010). In both cases, the optimum pH for entrapped enzymes in polymeric films are closer to the pH for the free enzyme than biosensors using other immobilization techniques, such as covalent binding. The results agree with the ones found in the present work.
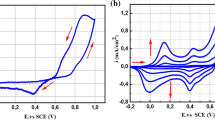
Analysis of the optimal electrodeposition conditions and optimum pH for bio-sensing of glucose and urea. The functionalized PANI thin films for the biosensor obtained with the optimal parameters, as well as the optimal pH range in (a). The EIS Nyquist graphs for GOx/PANI, Ur/PANI and the PANI samples without enzymes for both biosensors in (b). The inset shows more detailed graphs for the high frequency region. Analysis of potentiometric EnFET biosensor. The calibration curve with the sensitivity and linearity parameters for the GOx/PANI, Ur/PANI (NH4) and Ur/PANI (OH) biosensors in (c)
The analysis of the optimal conditions for the urea biosensor is also shown in Fig. 2(a). Its optimal working pH range is around 7.4. For the urea biosensor it was obtained an optimum pH around the reported optimum pH for the same kind of urease enzyme (around 7.4) (Cesareo and Langton 1992). However, for biosensors based on covalent immobilized urease enzymes, a slight decrease in the optimum pH can be achieved, as reported by Saeedfar and co-workers. For a covalent binding of the enzyme in a modified fullerene nanomaterial, the optimal pH decreased to 7.0 (Saeedfar et al. 2013). It was reported a pH range from 7.25 to 7.4 by Busono using an entrapped urease in polypyrrole film, (Busono 2015). The entrapment of enzymes by the one-step electrochemical immobilization technique produces functionalized films presenting optimal working pH close to that of free enzymes due to the low degree of modification in the conformation and microenvironment of the enzymes.
The Nyquist plot from EIS measurements for the two types of enzymatic biosensors are shown in Fig. 2(b). Typically, the Nyquist plot of electrochemical impedance spectra consists of two sections: a semicircle in high frequencies which reflects charge transfer resistance (RCT) at electrode surface and a linear part in low frequencies corresponding to diffusion limited process. Usually, it is expected the slope of each linear section in Nyquist plots to be approximately 1 in diffusional related processes, which is not the case for the spectra of the analysed films, indicating that, possibly, other phenomena are occurring in the system. Also, the characteristic semicircle is only present in the non-functionalized PANI thin films (samples deposited with 600 and 150 mC, as optimized parameters for glucose and urea biosensors, respectively). The inset in Fig. 2(b) shows a more detailed Nyquist plot illustrating changes in the electrical properties of the PANI thin films due to enzyme immobilization. After GOx and Ur were immobilized into PANI thin films, the corresponding RCT, obtained from the equivalent circuit (a R(RC)(RC) circuit, (Yang et al. 2018)) fitting of each electrochemical spectra, increased, starting in 30 and 16 kΩ for the non-functionalized PANI thin films matrix of GOx and Ur, respectively, and growing up to 427 and 147 kΩ for GOx/PANI and Ur/PANI, respectively, while the corresponding final slopes did not change. This indicates that the enzymes were well immobilized into the matrix, leading to a slight hindrance in the charge transfer process (Feng et al. 2015).
3.2 Biosensor measurements
Potential data were recorded during 180 s (for a stable response) for each analyte solution. A calibration curve was obtained for each biosensor, from which sensitivity and linearity parameters were extracted. The calibration curve with the sensitivity and linearity parameters for the biosensors are shown in Fig. 2 (c). The glucose biosensor, GOx/PANI, presented sensitivity of 14.6 ± 0.4 mV/decade, linearity of 99.8% and detection range from 10−4 to 10−1 mol/L of glucose. These values agree with other potentiometric biosensors. Khun and colleagues (Khun et al. 2012) described a biosensor with a sensitivity of 27.3 ± 0.8 mV/decade with linearity of 99%, in a range from 10−6 to 10−2 mol/L of glucose. However, this biosensor used a gold-coated glass electrode with glucose oxidase immobilized in a compound of iron and chitosan magnetic nanoparticles, which was a time-consuming and complex process with expensive materials when compared to the GOx/PANI biosensor reported in this work. Other reported biosensors present similar sensitivity with a much lower detection range or more complex fabrication processes (Table 1).
The urease biosensor selective to ammonium ions, Ur/PANI (NH4), presented sensitivity of 14.7 ± 0.9 mV/decade, linearity of 98.2% and detection range from 10−5 to 10−1 mol/L of urea. The Ur/PANI (NH4) selectivity to the NH4+ ion is evidenced by the ascending character of the calibration curve. The polymeric matrix measures the variation of ammonium ions, NH4+, which increases with the increase of urea. The PANI matrix works as an ion-selective film detecting the ammonium ion, as shown by Zhybak and colleagues (Zhybak et al. 2016). Urea biosensors measuring ammonium ions are faster and function in a broader concentration range (Chou et al. 2008). The urease biosensor selectivity to the hydroxyl ion, Ur/PANI (OH), presented sensitivity of 7.4 ± 0.5 mV/decade, linearity of 98.1% and detection range from 10−5 to 10−1 mol/L of urea. These values show that the proper function of the biosensor with selectivity to OH− ions is related to the variation of the local pH caused by the enzyme, which increase with increasing urea concentration, causing a decrease in potential, leading to the descending behaviour presented by the film (Lakard et al. 2011). As for the GOx/PANI biosensor, there are reported urea biosensors in the literature with comparable sensitivity, although some presented higher sensitivity. However, these biosensors operate at a much lower detection range than the biosensors described in this paper (Table 1).
The sensitivity of GOx/PANI and Ur/PANI (NH4) biosensors are similar, around 14.5 mV/decade, while the sensitivity of the Ur/PANI (OH) biosensor are approximately half of the other two, around 7.4 mV/decade. This can be explained by the catalyse process that occurs with each enzyme and the proportion between the bio-analytes, glucose and urea, and the ions produced, H+, OH− and NH4+, that will interact with the PANI immobilization matrix that possess ion-selectivity characteristic. For each mole of glucose and urea, two moles of H+ and NH4+ ions are produced, respectively, but one mole of OH− ions are produced, explaining the ratios between the enzymatic biosensor described here. This proportion reinforces the bio-sensing mechanism of the biosensors produced.
3.3 Stability and repeatability analysis
The stability, as evolution over five weeks, and repeatability, as evolution over five consecutive experiments, of sensitivity and linearity of the PANI based enzymatic biosensors are shown in Fig. 3. The samples were storage in petri dishes, in atmospheric air, inside the refrigerator at −4 °C. The stability and repeatability of GOx/PANI biosensors are shown in Fig. 3(a). The GOx/PANI biosensor rapidly loses its functionality, with a decrease in the values of both parameters. The distinct effect of time and reuse can be clearly seen for the GOx/PANI biosensor, where the effect of the five weeks increased a sensitivity loss of about 20% compared to five sequential experiments. The linearity presented the same comparative behaviour. The differences between stability and repeatability may be related to the distinct wear modes of the biosensor, i.e. from the detach and inactivation of the enzyme which take longer than the polymeric oxidation of the immobilization matrix. The latter seems the main responsible for the quality decline of the biosensor. However, in a long-term analysis after five weeks, the complete linearity loss indicates a non-functional biosensor, probably with the absence of the bio-recognition element (the enzyme), while in the other case, the residual linearity indicates that the bio-functionality of the biosensor remains in a certain level and that the PANI oxidation, causing the loss of pH sensitivity, is the main cause of biosensor decline.
The repeatability and stability of both sensitivity and linearity for Ur/PANI (OH) biosensors are shown in Fig. 3(b). The analysis indicates that the Ur/PANI (OH) biosensor does not lose its functionality over five weeks or after five sequential experiments, presenting good stability and repeatability. This is different to what previously occurred to the GOx/PANI biosensor. The behaviour of biosensor parameters for five weeks or five sequential experiments does not make it unfeasible to be used. Although it is also subject to the same inherent effects of biosensors with immobilized enzymes, the Ur/PANI (OH) biosensor does not have diminished functionality, which can be attributed to the immobilization matrix that retains the enzyme within its structure, mainly because urease have a size of 540 kDa, more than three times greater than the glucose oxidase, with 160 kDa. The electrochemical polymerization techniques, such galvanostatic one, causes the formation of porous PANI structures, as reported by Das and Sarkar (Das and Sarkar 2016) and as shown in this work (Fig. 1). In this context, such enzyme size difference is important to evaluate the detachment of entrapped bio-elements from PANI immobilization matrix. Besides that, the PANI oxidation effect does not interfere in the Ur/PANI (OH) biosensor operation, once that in this case the polymer is selective to hydroxyl ions, and the sensitivity to this ion is not affected by the polymer oxidation.
The repeatability and stability of sensitivity and linearity for Ur/PANI (NH4) biosensors are shown in Fig. 3(c). The variation in sensitivity and linearity is caused by the change in the selectivity of the Ur/PANI (NH4) biosensor, from ammonium ions, NH4+, to hydroxyl ions, OH−. This variation in selectivity is verified by the change in the character of the calibration curve, changing from ascending to descending with increasing urea concentration. This change can be detailed seen in Fig. 4.
Figure 4 indicates that a gradual change in the character of the calibration curve occurred. The behaviour of the calibration curves along the five weeks justifies the variation in sensitivity and linearity. The selectivity profile of the curve in the fifth week is the same of the urea biosensor selective to hydroxyl ions as shown in Fig. 3(b). As already shown and discussed in Fig. 3(b), the detachment and inactivation of urease enzyme for urea biosensor is not pronounced for the one-step functionalized PANI thin film. For the described EnFET biosensor the variation in the selectivity must be related to the variation of the polymeric immobilization matrix. The PANI film changed its oxidation state, changing its selectivity from positive ions (NH4+) to negative ones (OH−). The previous results indicate that it is possible to obtain two urea biosensors with immobilized urease in PANI matrix.
3.4 Selectivity analysis
The analysis of the oxidation states of PANI by means of reflectance spectroscopy gives information about the selectivity of the immobilization matrix. Figure 5(a) shows the reflectance spectrum for four different samples: unfunctionalized PANI, GOx/PANI, Ur/PANI (NH4) and Ur/PANI (OH) thin films. In general, the reflectance spectra for thin films of PANI having a higher intensity between 550 and 700 nm and lower intensity between 400 and 550 nm, relatively to each other, indicate a more oxidized material, whereas the opposite indicates a smaller oxidized material. In between, it presents a half-oxidation state such as PANI emeraldine base (PANI-EB), as seen by Albuquerque and co-workers (Albuquerque et al. 2000). In addition, PANI in a higher or lower oxidation state is insulator and only remains stably protonated under extreme conditions, such as pH close to zero. This occurs because the completely oxidized and reduced forms of PANI have structure with a strong localization of charges, which indicates that only PANI in a state of half-oxidation can be protonated and become more conductive (D’Aprano et al. 1992). The functionalized PANI thin films presenting a higher oxidation level have lower potential to be protonated, consequently presenting low capacity for the detection of NH4+ and H+ ions, as in the case of Ur/PANI (OH) biosensor and in this way, it becomes a selective sensing material for OH− (or negative) ions. On the other hand, functionalized films in a half-oxidation level present higher potential of protonation and selectivity to NH4+ and H+ (or positive) ions, as the Ur/PANI (NH4) and the GOx/PANI biosensor, respectively. There is a possibility of altering the selectivity of Ur/PANI (NH4) biosensor through the action of time and environment on the PANI oxidation, which would cause not only the change in the selectivity of Ur/PANI (NH4) biosensor (Fig. 3(b) and 4) but also the decrease in sensitivity of the GOx/PANI biosensor.
Analysis of the oxidation states of PANI by means of reflectance spectroscopy to determine the selectivity of the immobilization matrix. Reflectance spectrum for four different samples: unfunctionalized PANI, GOx/PANI, Ur/PANI (NH4) and Ur/PANI (OH) thin films in (a). Evaluation of the oxidation state variation using the difference between the fifth- and first-week spectra of the films for GOx/PANI and both Ur/PANI (NH4) and (OH) in (b)
The evaluation of the oxidation state variation of the functionalized PANI thin film through the analysis of the reflectance spectra is shown in Fig. 5(b) for the GOx/PANI and both Ur/PANI (NH4) and (OH) biosensors using the difference between the spectra of the films for the fifth- and first-week experiments. A general behaviour of the three functionalized PANI thin films is the increase in reflectance intensity between 600 and 700 nm, approximately, and reduction of the reflectance intensity between 400 and 550 nm, approximately. This behaviour, as discussed before, is characteristic of thin films of PANI that change to a state of greater oxidation. This corroborates the loss of sensitivity to glucose, maintenance of sensitivity to urea for a biosensor with selectivity to hydroxyl ions, Ur/PANI (OH), and alteration of the selectivity to urea for a biosensor initially selective to ammonium ions, Ur/PANI (NH4). This indicates that the Ur/PANI (OH) produced so far is more appropriated to be used in the next stage of tests. The GOx/PANI biosensor proved to be robust with an important sensitivity and selectivity but need improvements in order to become a fully useful device.
4 Conclusions
Rapidly and easily fabricated glucose and urea biosensors using the one-step electrochemical immobilization technique by galvanostatic method to entrap glucose oxidase and urease in PANI immobilization matrix presented changeable selectivity. Functionalized PANI thin films were used as recognition stage connected to a potentiometric transducer stage based on the IA-EGFET system, originating the EnFET biosensor. The optimized biosensors were produced with 0.25 mg/mL of glucose oxidase and 600 mC/cm2 deposited charge for the GOx/PANI biosensor and 0.60 mg/mL of urease and 150 mC/cm2 deposited charge for both Ur/PANI (NH4) and (OH) biosensors. The optimum pH range for the glucose biosensor was around 6.2 to 6.6 and for the urea biosensor the optimum pH was around 7.4. The sensitivity, linearity and detection range were 14.6 ± 0.4 mV/decade, 99.8% and from 10−4 to 10−1 mol/L for GOx/PANI, 14.7 ± 0.9 mV/decade, 98.2% and from 10−5 to 10−1 mol/L for Ur/PANI (NH4) and 7.4 ± 0.5 mV/decade, 98.1% and from 10−5 to 10−1 mol/L for Ur/PANI (OH), respectively. These biosensors presented similar sensitivity to other reported devices, with a much simpler and faster fabrication method and a broad detection range. The time evolution analysis of stability and repeatability showed that the glucose biosensor response presented a fast decline, that the urea biosensor selective to hydroxyl ions could maintain its proper function over all the tested period and that the ammonium selective urea biosensor presented a changeable selectivity with sensitivity recovery after change of selectivity from ammonium to hydroxyl ions. The characteristics depended on the enzyme behaviour and mainly on the oxidation variation of the PANI immobilization matrix, which was responsible to the selectivity variation of Ur/PANI (NH4) biosensor and great functionality loss of GOx/PANI biosensor. The EnFET system is reliable and the functionalized thin films can be easily produced with controllable selectivity.
References
J.E. Albuquerque, L.H.C. Mattoso, D.T. Balogh, R.M. Faria, J.G. Masters, A.G. MacDiarmid, Synth. Met. 113, 19 (2000)
P. Busono, Development of amperometric biosensor immobilized by entrapment of urease enzyme in polypiroll film for the determination of blood urea. (Atlantis Press, 2015). https://doi.org/10.2991/icopia-14.2015.23
S.D. Cesareo, S.R. Langton, FEMS Microbiol. Lett. 99, 15 (1992)
X. Chen, J. Jia, S. Dong, Electroanalysis 15, 608 (2003)
B. Cheraghi, A.R. Fakhari, S. Borhani, A.A. Entezami, J. Electroanal. Chem. 626, 116 (2009)
N.H. Chou, J.C. Chou, T.P. Sun, S.K. Hsiung, Sens. Actuators B Chem. 130, 359 (2008)
G. D’Aprano, M. Leclerc, G. Zotti, Macromolecules 25, 2145 (1992)
J. Das, P. Sarkar, Enzymatic electrochemical biosensor for urea with polyaniline grafted conducting hydrogel composite modified electrode. RSC. Adv. 6, 92520–92533 (2016). https://doi.org/10.1039/C6RA12159D
E.M.I.M. Ekanayake, D.M.G. Preethichandra, K. Kaneto, Biosens. Bioelectron. 23, 107 (2007)
X. Feng, H. Cheng, Y. Pan, H. Zheng, Biosens. Bioelectron. 70, 411 (2015)
M. Gerard, B.D. Malhotra, Curr. Appl. Phys. 5, 174 (2005)
P. Gründler, Chemical Sensors an Introduction for Scientists and Engineers (Springer, Berlin, 2007)
I. Gürel, M.Y. Arica, V. Hasirci, Turk. J. Chem. 21, 387 (1997)
B. Hansen, M.A. Hocevar, C.A. Ferreira, Synth. Met. 222, 224 (2016)
International Commission on Illumination, CIE 15: Technical Report: Colorimetry (Viena, Austria, 2004), p. 72
J.O. Iroh, R. Rajagopalan, Surf. Eng. 16, 321 (2000)
E. Jaworska, K. Maksymiuk, A. Michalska, Chemosensors 3, 200 (2015)
A. Kausaite-Minkstimiene, V. Mazeiko, A. Ramanaviciene, A. Ramanavicius, Biosens. Bioelectron. 26, 790 (2010)
K. Khun, Z.H. Ibupoto, J. Lu, M.S. AlSalhi, M. Atif, A.A. Ansari, M. Willander, Sens. Actuators B Chem. 173, 698 (2012)
J. Lai, Y. Yi, P. Zhu, J. Shen, K. Wu, L. Zhang, J. Liu, J. Electroanal. Chem. 782, 138 (2016)
B. Lakard, D. Magnin, O. Deschaume, G. Vanlancker, K. Glinel, S. Demoustier-Champagne, B. Nysten, A.M. Jonas, P. Bertrand, S. Yunus, Biosens. Bioelectron. 26, 4139 (2011)
C.-W. Liao, J.-C. Chou, T.-P. Sun, S.-K. Hsiung, J.-H. Hsieh, Sens. Actuators B Chem. 123, 720 (2007)
Y.-H. Lin, C.-P. Chu, C.-F. Lin, H.-H. Liao, H.-H. Tsai, Y.-Z. Juang, Biomed. Microdevices 17, 1 (2015)
H.J.N.P.D. Mello, T. Heimfarth, M. Mulato, Mater. Chem. Phys. 160, 257 (2015)
H.J.N.P.D. Mello, M. Mulato, Influence of galvanostatic electrodeposition parameters on the structure-property relationships of polyaniline thin films and their use as potentiometric and optical pH sensors. Thin. Solid. Films. 656, 14–21 (2018). https://doi.org/10.1016/j.tsf.2018.04.022
H. J. N. P. D. Mello and M. Mulato, Thin Solid Films 656, 14 (2018)
H.M. Mousa, J.R. Aggas, A. Guiseppi-Elie, Electropolymerization of Aniline and (N-Phenyl-O-phenylenediamine) for Glucose Biosensor Application. Mater. Lett. (2018). https://doi.org/10.1016/j.matlet.2018.12.012
Y. Piao, D.J. Han, M.R. Azad, M. Park, T.S. Seo, Biosens. Bioelectron. 65, 220 (2015)
A.S. Poghossian, Sens. Actuators B Chem. 44, 361 (1997)
K. Saeedfar, L.Y. Heng, T.L. Ling, M. Rezayi, Sensors 13, 16851 (2013)
A. Seki, S. Ikeda, I. Kubo, I. Karube, Anal. Chim. Acta 373, 9 (1998)
J. van der Spiegel, I. Lauks, P. Chan, D. Babic, Sensors Actuators 4, 291 (1983)
J.R. Stetter, W.R. Penrose, S. Yao, J. Electrochem. Soc. 150, S11 (2003)
A. Subramanian, S.J. Kennel, P.I. Oden, K.B. Jacobson, J. Woodward, M.J. Doktycz, Enzym. Microb. Technol. 24, 26 (1999)
F.R.R. Teles, L.P. Fonseca, Mater. Sci. Eng. C 28, 1530 (2008)
Y.-M. Uang, T.-C. Chou, Biosens. Bioelectron. 19, 141 (2003)
F. Yang, T. Liu, J. Li, S. Qiu, H. Zhao, RSC Adv. 8, 13237 (2018)
M. Zhybak, V. Beni, M.Y. Vagin, E. Dempsey, A.P.F. Turner, Y. Korpan, Biosens. Bioelectron. 77, 505 (2016)
Acknowledgements
This work was funded by FAPESP – Fundação de Amparo à Pesquisa do Estado de São Paulo (2014/24559-0, 2017/24201-6, 2014/09562-4) and CNPq – Conselho Nacional de Desenvolvimento Científico e Tecnológico (30538/2015-0) Brazilian agencies.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Mello, H.J.N.P.D., Mulato, M. Enzymatically functionalized polyaniline thin films produced with one-step electrochemical immobilization and its application in glucose and urea potentiometric biosensors. Biomed Microdevices 22, 22 (2020). https://doi.org/10.1007/s10544-020-00478-4
Published:
DOI: https://doi.org/10.1007/s10544-020-00478-4