Abstract
Maintenance of phenotypic and genotypic diversity within and across species and populations is critical for their capacity to survive and adapt to changing environments. Climate change potentially puts cryptic diversity and populations at increased risk, highlighting the importance of quantifying and understanding this diversity before it is lost. This study focuses on Bombus lapponicus sylvicola, a bumble bee species that has undergone recent taxonomic additions and revisions. We tested the null hypotheses that B. l. sylvicola over a 40,000 km2 geographic range and climatic gradient in the Canadian Rocky Mountains represented a single genetic population. Furthermore, we evaluated predictions for mechanisms behind genomic divergence among groups under this framework. We sampled bumble bees from 69 sites and used DNA from two different species (131 B. l. sylvicola and 435 individuals of the closely related B. melanopygus as an outgroup) to characterize 20,000 SNPs and measure relatedness and gene flow. We collected phenotypic data on color patterns and mapped population distribution based on environmental variables. We found evidence of two phenotypically and genetically distinct parapatric populations of B. l. sylvicola that appear to have diverged under conditions of gene flow and differential recombination. Our models suggest that these populations occupy distinct climatic regions, with a newly described cryptic population found in locations reaching a lower minimum temperature. This research presents evidence for the role of adaptative evolution in response to different climate conditions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Biological diversity is important for the persistence of populations, species and ecosystems in the face of changing environmental conditions (Chapin et al. 1998; Tilman et al. 2006; Cortés and López-Hernández 2021). Biodiversity should be considered both for its intrinsic value and for the ecosystem services and other extrinsic value that it provides, including the maintenance of genotypic and phenotypic diversity which can help buffer against the impacts of climate change by increasing the potential for future adaptation (Johnson et al. 1996; Loreau 2000; Oliver et al. 2015; Obura et al. 2022). However, biodiversity is considered at risk globally, and can also decline in rapidly changing environments as individuals and species are lost (Bellard et al. 2012; Garcia et al. 2014). It is therefore important to understand biodiversity in the context of both past events (e.g. Riddle 2019) and future resilience (Bellard et al. 2012; Garcia et al. 2014).
While diversity may be evident and more readily quantified at higher taxonomic levels (e.g., by comparing species richness among communities), other types of diversity can be cryptic and therefore could be overlooked. This cryptic diversity may be at the genetic level, it may manifest in physiological responses, or constitute distinct behaviours or geographic ranges among morphologically similar groups (e.g., ecotypes) (Bickford et al. 2007; Trontelj and Fišer 2009). Characterizing this cryptic diversity is important for explaining spatial patterns of genetic structure, understanding speciation, in evaluating the relationship between communities and past, present and future environments, and in planning conservation strategies (Bickford et al. 2007; Pearman et al. 2010; Marske et al. 2013; Vodă et al. 2015; Schön et al. 2017; Chen et al. 2022). Cryptic populations are becoming more easily identified as genomic methods continue to become more widely available. The mechanisms driving and maintaining cryptic diversity remain unknown for many species, however, and may be elucidated through the evaluation of a priori assumptions about the processes behind cryptic speciation.
Speciation and the associated genetic divergence between populations plays an integral role in the creation and maintenance of biodiversity (Nosil and Feder 2012; Schluter and Pennell 2017). In understanding how and why the process of genetic divergence may be occurring, one of the first steps is to test predictions associated with the levels of gene flow between populations (Feder et al. 2012; Sousa and Hey 2013). Gene flow can be restricted between populations due to geographic separation with barriers to movement (allopatry) or partial separation, for example, due to distances among individuals across a geographically widespread population (parapatry). Gene flow between populations can also be restricted by phenological and habitat barriers or by assortative mating and/or barriers to fertilization (Seehausen et al. 2014). When gene flow is restricted between populations, they can experience genetic divergence due to selection and/or genetic drift (Feder et al. 2012).
Selective pressure is one commonly hypothesized mechanism that is often involved in population divergence, especially in conditions of gene flow (Feder et al. 2012). Population divergence under selective pressure can sometimes be detected based on patterns in the genome, however patterns of genomic divergence can also be influenced by other processes such as differential recombination across the genome, and the interactions between selection, recombination and gene flow (Butlin 2005; Nosil and Feder 2012; Wolf and Ellegren 2017). Together, gene flow, selective pressure and genome recombination are important factors in population divergence and the evolution of diversity. Current ecological genomic methods make it increasingly possible to test hypotheses about these factors, while also examining cryptic population divergence (Ungerer et al. 2008).
Bumble bees (Bombus sp.) are important pollinators in many natural systems, especially in the northern temperate regions to which they are particularly well adapted (Ollerton 2017). They are also an interesting group in which to test for evidence of cryptic diversity—especially in natural populations occupying heterogeneous mountain habitats. More than a dozen bumble bee species may occur in a single region (e.g., Clake et al. 2022), with taxa sharing many morphological characteristics while exhibiting variation in color patterns and behavioural adaptations (Cameron et al. 2007). Bumble bee taxonomy has also been the subject of several recent updates with the advent of genomic data, including both the grouping of species previously considered to be separate (e.g. Bombus melanopygus Nylander and Bombus edwardsii Cresson; Owen et al. 2010) and the splitting of what was previously one species into two (e.g. Bombus bifarius Cresson and Bombus vancouverensis Cresson; Ghisbain et al. 2020). In particular, the Bombus lapponicus/Bombus sylvicola species complex has undergone several recent taxonomic revisions and additions, including evidence suggesting that B. sylvicola is most likely a subspecies of B. lapponicus (Martinet et al. 2019; hereafter referred to as B. l. sylvicola). A cryptic population originally thought to belong to B. l. sylvicola in Colorado, USA was described as a new species Bombus incognitus (Christmas et al. 2021). A new closely related species, Bombus interacti (Martinet et al. 2019), was also described in Alaska, USA before it was found to be synonymous with the previously described species Bombus johanseni (Sladen 1919; Sheffield et al. 2020). While much remains to be learned about these newly described species, it is clear that the B. lapponicus/B. sylvicola species complex has high potential for cryptic genetic diversity. To our knowledge there have not been evaluations of genetic or phenotypic diversity in this species in the Canadian Rocky Mountains, leaving open an important area for assessment.
Here, we test predictions of adaptive divergence and cryptic diversity in bumble bees sampled in a region adjacent to where B. interacti/B. johanseni (Martinet et al. 2019; Sheffield et al. 2020) and B. incognitus (Christmas et al. 2021) have been documented. The objective of this study is to investigate mechanisms driving cryptic population divergence over a large area, with reference to the B. l. sylvicola species complex. Specifically, we: (A) test the hypothesis that B. l. sylvicola populations in the Canadian Rocky Mountains represent a single genetic population and (B) examine predictions for mechanisms behind genomic divergence between groups. We evaluate evidence for three non-mutually exclusive predictions, i.e., that population genetic structure may be created by: (A) reduced gene flow between populations; (B) regions of differentiated recombination and (C) selection based on adaptation to different environmental conditions.
This study uses samples from an under-studied area in the Canadian Rocky Mountains with locations covering a range of environmental variability, selected to minimize the potential for spatial autocorrelation (see Clake et al. 2022). The sampling design, therefore, has potential to offer novel insight into the interaction between speciation and environmental conditions.
Materials and methods
Sampling
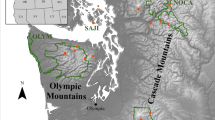
We sampled bumble bees in June–August 2017 and July–August 2019 in the Rocky Mountain and Columbia Mountain Ranges in Alberta and British Columbia, Canada (Fig. 1). Sampling occurred at 69 unique sites clustered in 17 broad sampling locales across roughly 40,000 km2, with two locales sampled in both 2017 and 2019, and the remainder in one of the 2 years (Fig. 1). Locales generally corresponded to established hiking paths in protected areas. Sites were initially selected to be roughly 2 km apart to minimize the chances of capturing individuals from the same nest at multiple sites. In some cases, our original target locations were not accessible due to safety constraints and were modified in the field. Bumble bees were collected using blue vane traps filled with 100% propylene glycol with 3 or 6 traps at each site (based on requirements for a parallel study), for a total of 260 unique trap locations. Each trap was deployed for five weeks, with samples collected every two or three weeks and immediately transferred to 95% ethanol for transportation. While this method of sample collection resulted in DNA degradation in some cases (see section below on Data Collection), it allowed us to maximize consistency in sampling individuals across a broad geographic and temporal range.
Sampling locations for this study in Alberta and British Columbia, Canada. Each diamond on the map represents a single sampling locale (N = 17) consisting of a cluster of multiple sites spaced 0.5–35 km apart (mean pairwise distance between sites at the same locale = 4.9 km; total sites = 69), with three to six blue vane traps deployed at each site
Samples were brought back to the University of Calgary (Alberta, Canada) for processing, where female bumble bees were identified to species using the key from Williams et al. (2014) and by referencing samples available in the University of Calgary Invertebrate Collection. Specimens identified as B. l. sylvicola were used in further analyses. We also included individuals identified as B. melanopygus as a closely related, phenotypically similar, and geographically coincident species outgroup.
Data collection
Nuclear DNA
We extracted DNA from thorax muscle tissue of individual female bees using a Qiagen DNEasy Blood and Tissue Kit. We used a slightly modified protocol for extractions that involved freezing tissue in liquid nitrogen and grinding prior to tissue lysis. Extracted DNA was quantified and checked for quality/contamination using a Qubit DNA Broad Range Assay Kit and a NanoDrop Spectrophotometer. 233 individuals were removed from further analysis due to insufficient DNA quantity or quality. Extracted DNA from 566 individual bees (435 B. melanopygus and 131 B. l. sylvicola based on original morphological identification) was sent to the Institute of Integrative Biology and Systems (IBIS—Université Laval, Québec, Canada) for preparation of double-digest restriction associated DNA sequencing (ddRADseq) libraries using PstI and MspI restriction enzymes. Libraries were sequenced at Genome Quebec on a NovaSeq 6000 (350 M reads sequenced using 150 bp paired-end sequencing).
Reads were demultiplexed and barcodes/adapters were removed using STACKS v2.59 (Catchen et al. 2013). We checked read quality using FastQC (Andrews 2010) and used trimmomatic v.039 (Bolger et al. 2014) to trim an additional five bases corresponding to the restriction sequence. Reads were aligned to the B. l. sylvicola genome (assembly ASM1967717v1; Christmas et al. 2021) using the mem function of bwa v0.7.17 (Li and Durbin 2009). Aligned reads were sorted, converted to bam files, and alignment and coverage were checked using samtools v1.13 (Li et al. 2009). Next we used the mpileup functions of bcftools v1.13 with a minimum mapping quality of 30 and a maximum depth of 250, followed by the call function for variant calling (Li 2011).
We used vcftools v0.1.16 to filter variant sites (Supplemental Table S1). First, indel sites were removed to keep only variants corresponding to single nucleotide polymorphisms (SNPs). We removed low confidence SNP calls using a minimum read depth of 8 and a minimum genotype quality of 20. Next, we removed sites with more than 50% of reads missing before further filtering out sites with a minimum mean depth of less than 20 and a maximum mean depth of greater than 158 (corresponding to double the mean site depth). Individuals missing more than 50% of sites were removed before an additional step to remove sites that were missing reads for more than 20% of individuals. We filtered out sites with an observed heterozygosity of greater than 70% to remove loci that were likely paralogous (Taylor et al. 2014). Finally, we removed sites with a minimum allele count of < 3 to ensure that each individual allele was found across at least two individuals and did a final filtering step to remove individuals missing more than 25% of alleles. Having multiple steps to remove both sites and individuals missing data allowed us to iteratively remove the sites and individuals with the greatest amount of missing data. Lastly, we calculated relatedness between individuals using the relatedness2 (Manichaikul et al. 2010) function of vcftools to identify individuals that were likely sisters from the same colony. We used a threshold of 0.20 based on previous literature (Jackson et al. 2018), and kept the individual from each colony that had the lowest proportion of missing alleles.
We also used a second alignment to the more distantly related Bombus terrestris (assembly GCF_000214255.1; Sadd et al. 2015) for our analyses of FST (described below), because it is a chromosome level assembly, and therefore permits plotting the location of SNPs in the genome with greater accuracy. This was also the same assembly used by Christmas et al. (2021) to arrange contigs from the B. l. sylvicola genome into pseudochromosomes, and should allow comparison between our findings and their previously published work. For this second alignment we used similar filtering steps as described above and in Supplemental Table S1, however for FST analyses an additionally filtered dataset was used where SNPs were randomly thinned to a subset of one variant for every 150 bp (corresponding to the sequencing read length) using vcftools v0.1.16.
Mitochondrial DNA
We sent leg samples from 15 individuals that were originally morphologically identified as B. melanopygus and 12 originally identified as B. l. sylvicola to the Canadian Centre for DNA Barcoding (Guelph, ON, Canada) for DNA extraction and sequencing of the 5′ portion of mitochondrial cytochrome c oxidase subunit I (COI-5p)—the region most commonly used for DNA barcoding of insect specimens (Zhou et al. 2019).
We obtained COI-5p sequence data from 176 additional individual samples publicly available in the NCBI online database (Supplemental Table S2). These samples included B. incognitus (N = 5), B. interacti (N = 1), B. johanseni (N = 5), B. lapponicus lapponicus (N = 34), and B. l. sylvicola (N = 45), as well as additional species in the Pyrobombus subgenus used as outgroups (B. bifarius, B. centralis, B. flavifrons, B. frigidus, B. incognitus, B. interacti, B. johanseni, B. melanopygus, B. mixtus, B. sandersoni, B. vancouverensis nearcticus and B. vancouverensis vancouverensis) (Supplemental Table S2). We used muscle v3.8.1551 (Edgar 2004) to align COI-5p sequence reads, followed by trimming using trimAl v1.4 (Capella-Gutiérrez et al. 2009).
Phenotype data
We collected phenotypic data on the colour patterns of a subset of individual bees from each of three genetically distinct populations (44 B. melanopygus, 35 B. l. sylvicola, and 29 cryptic individuals). Because phenotype data was collected following DNA isolation, we were unable to collect colour pattern information for all individuals. In particular, individuals that were smaller and required the full thorax to be sacrificed for DNA extraction could not be included, as well as individuals where multiple DNA extractions were done, requiring all thorax tissue. Spatial locations of individuals used in phenotype data collection and analysis can be found in Supplemental Fig. S1. Colour pattern data was collected by visually categorizing the proportion, in categories spanning 10%, of different colours of setae on the face (between eyes), dorsal head, scutum, inter-alar space, scutellum, lateral thorax and the first five tergum segments (T1-5) based on body components commonly used in bumble bee species identification (Williams et al. 2014). Data on colour pattern was collected by a single technician without knowledge of which genomic category an individual bee had fallen into.
Environmental data
WorldClim bioclimatic data (average climate conditions over the years 1970–2000) were downloaded (Fick and Hijmans 2017) and used along with the raster package in R (Hijmans 2022) to extract bioclimatic variables for each location sampled.
Data analyses
Population differentiation
We used the vcfR package (Knaus and Grünwald 2017) to load thinned SNP data in VCF format into R v4.2.0 (R Core Team 2023) and to convert to a genind object. PCA analysis was done using dudi.pca from the ade4 package (Dray and Dufour 2007). Because this PCA analysis requires a complete dataset, missing values (4.6% of data) were first filled in using the impute function in the LEA package (Frichot and François 2015) after estimating likely ancestral population membership using the snmf function in LEA (based on cross-entropy criterion—in this case the strongest support was for three ancestral populations). To further assess population assignment in the original B. l. sylvicola and B. melanopygus populations we used STRUCTURE software v2.3.4 (Pritchard et al. 2000). Values of K ranging from 1 to 7 were each run five times without informative priors (e.g., location or original group membership). We calculated the rate of change of the likelihood distribution (L′(K)), the second order rate of change (L′′(K)), and the mean second order rate of change for a given K averaged over all runs (ΔK), per Evanno et al. (2005). Lastly, we estimated the genetic distance between individuals based on the proportion of shared alleles (calculated in adegenet (Jombart 2008; Jombart and Ahmed 2011) and subtracted from 1 to convert from similarity to distance). These distances were used in ape (Paradis and Schliep 2019) to generate a neighbour-joining tree with the bionj and ladderize functions.
To place differences between B. l. sylvicola populations in the context of other species differences, we also calculated the genetic distances between mitochondrial DNA sequences of bees from our study and those from other species in the Pyrobombus subgenus available on NCBI. We calculated proportion of shared alleles between all individuals, and Nei’s genetic distance between species using adegenet (Jombart 2008; Jombart and Ahmed 2011). We then created neighbour-joining trees using ape (Paradis and Schliep 2019). We also included a Bayesian approach to assessing relationships between individuals from different taxa using BEAST v1.10.4 (Suchard et al. 2018). For this analysis we assessed 5,000,000 states (burn-in of 500,000) using default settings and priors (including an HKY substitution model), and a Yule Process (Yule 1925; Gernhard 2008) tree prior.
Our final step in assessing population differentiation was to compare the colour phenotype data between individuals. We first checked that each body segment quantified (a) had sufficient variation in color and (b) was not highly correlated with the colour of other segments (Supplemental Fig. S2). We removed the inter-alar space, and the first three tergum segments (T1-3) from further analyses based on lack of variation between individuals. We then used the nnet package (Venables and Ripley 2002) to fit multinomial logistic regression models to test whether phenotypic variables could be used to predict membership in each of the three genomic groups. To check the predictive ability of these data we also fit the model using a training dataset comprising 80% of samples (randomly selected) and used it to predict the remaining 20% of samples. We then estimated the mean predictive accuracy across 1000 iterations of the training model fit using different random samples of data using (a) the group with the maximum probability, regardless of how high the probability was or (b) only predictions with a probability > 90%.
Patterns of population structure and association with climatic variables
To examine the potential for current gene flow we looked at the extent of sympatry between the genetically distinct B. l. sylvicola populations in addition to evidence from STRUCTURE plots. We also used vcftools to calculate FST (Weir and Cockerham 1984) across each of the SNPs in our thinned dataset. We calculated FST values between the two B. l. sylvicola groups as a whole, between the portions of these populations that occurred in sympatry and allopatry, and between northern and southern clusters of individuals within each group (Supplemental Fig. S3). When comparing sympatric and allopatric populations we used a randomly selected subset of 20 individuals from each population (cryptic vs. B. l. sylvicola) and location (sympatric vs. allopatric) to account for differences in sample size that might impact FST. For this comparison we also used the same set of SNPs that were found across both groups (N = 1753).
To test for potential environmental associations for each population we used the WorldClim bioclimatic data. We chose four environmental variables: the precipitation in each of the warmest and coldest quarters, the minimum temperature in the coldest month, and the maximum temperature in the warmest month. Previous studies have shown that both temperature and precipitation may be important for bumble bee gene flow and population distribution (e.g. Jackson et al. 2018). The specific variables were chosen to represent extreme conditions in both temperature and precipitation, while trying to minimize correlations between individual environmental variables and between environmental variables and other geographic variables including latitude, longitude and elevation (Supplemental Figs. S4, S5). We then fit a logistic regression to model the probability that a B. l. sylvicola individual was in the cryptic group based on environmental variables. Sampling year, the time of sampling (whether in the first or second sampling pass), the elevation and the easting and northing coordinates were also included in the model to account for variation that might be attributed to these features. We fit additional models with randomly selected subsets of data to check the predictive power of our model using the same methods described above for our phenotype model. Finally, we fit the same model using only individuals found in the “sympatric zone” (locales where at least one individual from each of the cryptic population and B. l. sylvicola was found) to ensure that trends detected in our broader model were not due strictly to broadly differing environmental conditions between the geographic regions each population was found in.
Results
Sampling and data collection
Following filtering our dataset consisted of 486 individuals (390 B. melanopygus and 96 B. l. sylvicola based on original identification; Supplemental Table S3), and 20,607 SNPs (5176 SNPs in “thinned” dataset aligned to B. terrestris). This included B. melanopygus individuals from all 17 locales sampled and 58 sites. B. l. sylvicola samples from 15 locales and 32 sites were included.
Population differentiation
Nuclear DNA
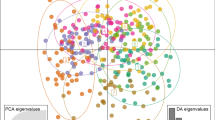
All three analyses methods (PCA, STRUCTURE and neighbour-joining tree) showed strong support for three distinct genomic populations. PCA analysis showed two clusters containing individuals that were originally phenotypically identified as B. melanopygus (N = 299) and B. l. sylvicola (N = 72) differentiated strongly on the first PCA axis (representing 23% of variation). There was a third cluster (N = 114) that contained individuals identified as both B. melanopygus and B. l. sylvicola that was differentiated from the first B. l. sylvicola cluster only on the second PCA axis (representing 8% of variation) (Fig. 2a). One individual appears to have been incorrectly identified as B. melanopygus despite clustering with B. l. sylvicola. There was one additional outlier individual in the PCA plot which did not cluster with any of the other groups, but was located between them all (Fig. 2a).
Plots showing population genomic structure based on SNP data from nuclear DNA. A PCA plot with individuals originally identified as Bombus melanopygus (pink circles) and Bombus lapponicus sylvicola (dark blue circles), with a third cryptic grouping including individuals originally identified as both species in the bottom right. B Structure plot showing individual population assignment. C Neighbour-joining tree using proportion of shared alleles where individual points are assigned colors based on the population assigned in STRUCTURE, corresponding to B. melanopygus (pink), B. l. sylvicola (dark blue), and a third cryptic population (light blue)
We found the strongest support for the STRUCTURE model where K = 4 based on the posterior probability of the data (Pr[X|K]) estimated across all runs (Supplemental Table S4). Summary statistics based on the rate of change in K suggested additional support for K = 2 and K = 3 (Supplemental Fig. S6). In the model using K = 4 as inferred by the posterior probability calculated by STRUCTURE, no individuals were assigned to the fourth population, and only one individual had even partial membership (3.77%) (Fig. 2b). This was the same individual that appeared as an outlier in the PCA plot and in both the STRUCTURE and PCA plot appears to have portions of the genome in common with each of the other three groups (17% cryptic, 48% B. l. sylvicola and 31% B. melanopygus; Fig. 2b). The neighbour-joining tree also shows three clusters of individuals, with the same outlier appearing on a unique branch between B. l. sylvicola and B. melanopygus (Fig. 2c).
Mitochondrial DNA
We found several individuals that had been identified as B. l. sylvicola in NCBI records but that clustered with other species in our individual neighbour joining tree (Supplemental Fig. S7a). These individuals were assumed to be misidentified and were discarded from further analyses, including calculations of genetic distances between species.
Individuals from the cryptic population identified in this study were found on a separate branch from the majority of B. l. sylvicola sampled both from our study and from NCBI (Fig. 3; Supplemental Fig. S7). These cryptic individuals shared a branch with two NCBI individuals that were identified as B. l. sylvicola and that were captured in British Columbia and Alberta, Canada, geographically very close to the cryptic individuals sampled in this study (Fig. 3; Supplemental Fig. S7). Cryptic individuals from our study were also found on a separate branch from B. incognitus and B. l. lapponicus (Fig. 3; Supplemental Fig. S7), although all four of these species were found in a relatively small cluster on the neighbour-joining tree (Fig. 3). The genetic distance between the cryptic population and B. l. sylvicola in this study was 0.027, which is the same as the genetic distance that we calculated between the subspecies B. vancouverensis nearcticus and B. vancouverensis vancouverensis (also 0.027) using similar COI sequence data (Supplemental Table S5).
Neighbour-joining tree based on Nei’s genetic distance between populations and using mitochondrial COI-5p sequence data from bees from this study (colored circles: pink = B. melanopygus, dark blue = B. l. sylvicola, and light blue corresponding to a third cryptic population), and additional samples from data publicly available on NCBI (grey circles; Supplemental Table S2)
Phenotype data
The cryptic population had significantly different proportions of yellow in both the scutum and scutellum when compared to B. melanopygus in our multinomial logistic regression model and was significantly different from both B. melanopygus and the other B. l. sylvicola population in the T4 (Table 1; Fig. 4; Supplemental Fig. S8). While the difference in T5 was not statistically significant, it was notable that the cryptic individuals all had strictly black T5 segments, with no yellow setae visible (Supplemental Fig. S8). Our model had a mean predictive accuracy of 82.5% when the most likely model prediction was used, and an accuracy of 88.9% when only predictions with a probability of > 90% were used (Supplemental Fig. S9).
Patterns of population structure and association with climatic variables
Population structure
The cryptic population occurred parapatrically to the B. l. sylvicola population, with only cryptic individuals found further west into the mountain range and only B. l. sylvicola found in the foothills regions to the east, with a region of overlap in the middle (Fig. 5).
The mean FST value across SNPs (thinned dataset) between the overall B. l. sylvicola and cryptic populations was 0.161 and ranged from − 0.34 to 1. The mean FST when comparing between a subset of individuals from populations occurring in sympatry was 0.230 and was 0.224 for populations occurring in allopatry (the mean FST between all cryptic and B. l. sylvicola individuals in this subset was 0.237). There was no significant difference between mean sympatric and allopatric FST values (Wilcoxon rank sum test, W = 1,508,413, p = 0.35). The mean FST between northern and southern populations of cryptic individuals and B. l. sylvicola was 0.005 and 0.001, respectively.
We found a slight bimodal distribution of FST value frequencies when comparing between the cryptic population and B. l. sylvicola, with a strong peak of values around an FST of 0, and another small peak of values around 1 (Fig. 6a). Higher FST values were distributed in clusters across the genome, most often with one cluster on each chromosome (Fig. 6b; Supplemental Fig. S10). There were two outliers with a very low FST value which were removed from the plot to better visualize the remaining values (Supplemental Fig. S11).
Plots showing Weir and Cockerham FST values calculated for nuclear DNA SNP data. A The distribution of FST values calculated between northern and southern clusters of Bombus lapponicus sylvicola (top) and the cryptic population (middle) (see Supplemental Fig. S3 for map of cluster locations), and between B. l. sylvicola and the cryptic population (bottom). B Manhattan plot of FST values between B. l. sylvicola and cryptic population based on location in genome, with chromosomes differentiated by black and grey colors. Mean FST is shown as a blue dashed line
Association with climate variables
Minimum temperature of the coldest month was the only environmental variable to be a significant component of our logistic regression model predicting the probability that an individual captured was part of the cryptic group (Fig. 7a). Individuals were more likely to be cryptic in areas that had a lower minimum temperature in the coldest month (Fig. 7b). Easting and Northing were also significant variables in the model, but with a small effect (Fig. 7a). Our model had a pseudo-R2 of 0.53, and a mean predictive accuracy of 87.8% (Supplemental Fig. S12).
Results from logistic regression model estimating the probability that an individual sampled belongs to the cryptic population (versus the B. l. sylvicola population) as a function of geographic and environmental variables. A Model estimated coefficients. Reference category for Sampling Time is the earlier sampling period, and for Year is 2017. B Predictive plot showing model estimated probability that an individual belongs to the cryptic population based on the minimum temperature in the coldest month with remaining variables held as mean values (with sampling time = later and year = 2019). Shaded region represents standard error
The logistic regression model that we fit using only data from individuals found in the sympatric geographic area (N = 125) showed similar trends to the model fit using all individuals, but with less power (Supplemental Table S6).
Discussion
We found multiple lines of evidence suggesting that there is cryptic diversity in B. l. sylvicola populations in the Canadian Rocky Mountain region that was previously undetected. We examined both genetic data (nuclear and mitochondrial DNA) and phenotype data (colour patterns) to assess the extent of population differentiation in our sampled groups of B. l. sylvicola and B. melanopygus and found evidence for three distinct populations that could be distinguished both genetically and phenotypically. Furthermore, we explored evidence for evolutionary mechanisms that may have resulted in the observed population divergence and diversity, including gene flow, differential recombination and selection.
Population differentiation
Genetic data
Multiple analyses of two distinct datasets (from both nuclear and mitochondrial DNA) all suggested that the cryptic population described here was distinct from previously identified populations of B. l. sylvicola and B. melanopygus. In all three analyses using nuclear DNA (PCA plot, STRUCTURE, and neighbour-joining tree) we found three groups (Fig. 2). In both the PCA plot and neighbour-joining tree, the third cryptic group appeared more closely related to B. l. sylvicola than to B. melanopygus (Fig. 2), suggesting that this cryptic group falls within the B. l. sylvicola species complex.
We considered the possibility that the cryptic population was due to individuals hybridizing between B. l. sylvicola and B. melanopygus, however there was little evidence for this in any of our analyses. The STRUCTURE plot in particular suggested that the cryptic population had distinct genetic signatures, and no hybrids in the current generation (Fig. 2). The exception to this was a single outlier individual in all three plots, which was estimated to be between the B. l. sylvicola and B. melanopygus clusters and branches in the PCA and neighbour-joining tree, respectively, and may be a hybrid between these two species. In the STRUCTURE plot the same individual had estimated membership in all three clusters, as well as a fourth unique group (Fig. 2). This individual was the only B. l. sylvicola individual from its site included in our analyses, however two other individuals from the same site were sequenced but were excluded in filtering steps due to missing data. Analyses including these additional individuals also resulted in them being grouped in a cluster/branch with the other outlier individual (Supplemental Fig. S13). This may be evidence of a different evolutionary mechanism occurring at that site (perhaps a hybridization event) and could be an interesting avenue for further study.
Mitochondrial DNA also showed individuals from the newly described cryptic population on a separate branch from B. l. sylvicola individuals collected in this study. These cryptic individuals were additionally separate from B. l. sylvicola individuals from NCBI collected in other areas of Canada (Manitoba, Yukon Territories and Newfoundland) and the United States (Colorado and Alaska) (Fig. 3). While the cryptic individuals do share a branch with other samples identified in NCBI as B. l. sylvicola, the samples that it groups with are located in British Columbia and Alberta, in the same region that our sampling occurred (Fig. 3; Supplemental Information Fig. S7). Given this geographic overlap it seems likely that these previously captured individuals also belong to the same cryptic population described here. The geographic distribution of individuals from NCBI clustering with the cryptic population vs. B. l. sylvicola also roughly matches the distribution of individuals from our study in these groups, with the cryptic branch including individuals west of the Rocky Mountains in British Columbia, Washington, and western Alberta and the B. l. sylvicola branch including individuals east into Nunavut, Manitoba, and Colorado.
The cryptic population is also on a separate branch from other recently described species, including B. incognitus (Fig. 3). It is within a relatively small cluster of other populations in the B. lapponicus species complex based on the neighbour-joining tree (Fig. 3), but is part of a distinctive clade in the tree based on Bayesian BEAST analysis (Supplemental Fig. S7b). Given the recent amendment of B. sylvicola to be a subspecies of B. lapponicus (Martinet et al. 2019) and the similar distances between the newly described cryptic population and other populations in the species complex, it seems likely that a subspecies designation would also be most appropriate for this cryptic population, which we provisionally name Bombus lapponicus hibernus. This would also be in line with the similarity in genetic distances between the cryptic population and B. l. sylvicola and the genetic distance between the subspecies B. vancouverensis nearcticus and B. vancouverensis vancouverensis calculated here using NCBI sequence data.
Phenotype data
We found quantifiable differences in color patterns between B. melanopygus, B. l. sylvicola, and the cryptic group B. l. hibernus. In several body segments (i.e. the scutum and T4, which were a key component of original species designations done here), the characteristics of B. l. hibernus were intermediate between and/or overlapping those of B. melanopygus and B. l. sylvicola (Supplemental Information Fig. S8), which may help explain why individuals that were later identified as belonging to the cryptic population were originally sorted into both of these two species groups. This underscores the importance of integrating both phenotype and genetic data into species assessments.
We did not find one single clear phenotypic characteristic that would allow identification of individuals as B. l. hibernus and instead multiple features likely need to be considered together in order to predict which subspecies an individual likely belongs to. For example, an individual is more likely to be identified as B. l. hibernus when the T5 is completely black (Supplemental Fig. S8), and when there is a higher proportion of black setae in both the scutellum and T4 (Fig. 4). Our multinomial logistic regression models suggest that the phenotypic differences between B. l. hibernus and B. melanopygus and B. l. sylvicola can be used in a multinomial regression model to classify population membership with over 80% accuracy.
This finding is in contrast to the assessment of unique phenotypes in B. incognitus compared to B. l. sylvicola, where there were no differences seen in any of the characteristics measured, including color of the head, scutellum, and abdominal segments (Christmas et al. 2021). It should, however, be considered in the context of recent descriptions of B. johanseni, which also has color forms that can resemble both B. l. sylvicola and B. melanopygus while distinct from either (Sheffield et al. 2020). While the focus of this and previous studies has been on morphology, there could also be physiological differences between newly described species/subspecies and existing groups that may be worth further investigation.
Patterns of population structure and association with climatic variables
In addition to quantifying the population divergence between groups, we were interested in examining potential mechanisms driving the differentiation. This differentiation likely occurred over a large timescale, from which we are sampling only one point in time and it is not likely possible to definitively identify the order in which barriers evolved (Nosil 2012). Our intention is to instead provide insight into the factors that currently underlie the structure between populations. To do this we considered three non-mutually exclusive potential processes driving speciation: gene flow, selection, and recombination (Sousa and Hey 2013).
Population structure
Often one of the first considerations in the study of speciation is whether the populations in question occupy the same geographic area: i.e., completely separate (allopatric), overlapping (sympatric) or a combination of the two (parapatric) (Butlin et al. 2008; Wolf and Ellegren 2017). In our study, the cryptic B. l. hibernus and B. l. sylvicola occupied distinct ranges, with a sizable region of overlap between the two groups (Fig. 5). If this current distribution is indicative of the historic range of these populations, it could suggest a process of parapatric divergence. We did not, however, see any evidence of a hybrid zone, indicating that if parapatric divergence did occur here it is now likely in the later stages. It is also possible that the current range does not match the historic range, and that these populations diverged in allopatric conditions followed by one or both expanding into the current overlapping area, i.e., through secondary contact. Our STRUCTURE plot, however, did not show any evidence of contemporary gene flow.
For additional evidence of the extent of gene flow between these populations (both current and historic) we looked at the distribution of FST values. We found that FST values calculated between the cryptic B. l. hibernus and B. l. sylvicola showed a strong peak around an FST of 0 with a smaller peak around an FST of 1 (Fig. 6a). Theory predicts that this pattern can arise where there is speciation in the presence of gene flow, with selection in highly divergent regions balanced in the remainder of the genome by gene flow between populations (Feder et al. 2012; Seehausen et al. 2014). This pattern could also be caused or accentuated by other processes, however, including variation in recombination rates across the genome (e.g. Geraldes et al. 2011; Burri et al. 2015).
The Manhattan plot of FST values across the genome showed regions of high FST (at or approaching 1) on each chromosome plotted (Fig. 6b). This plot resembled the distribution of FST values found by Christmas et al. (2021) between their newly described species B. incognitus and populations of B. l. sylvicola (Supplemental Fig. S10). Using whole-genome sequencing data they were able to estimate the location of centromeres based on the presence of tandem repeats and showed that many of the highly divergent regions found between B. incognitus and B. l. sylvicola corresponded to these centromeric regions (Christmas et al. 2021). We were working with SNP data generated from ddRAD sequencing and had lower resolution to detect cohesive regions of high FST across the genome. However, these regions do seem to correspond to those identified by Christmas et al. (2021) (Supplemental Fig. S10) suggesting that differential rates of recombination, especially around centromeric regions, are a likely factor in the differentiation of the cryptic B. l. hibernus from B. l. sylvicola.
Regions of elevated divergence around centromeres has been predicted to occur when speciation is driven by factors such as Bateson–Dobzhansky–Muller incompatibilities (DMI), which are independent of environmental settings (Seehausen et al. 2014). This pattern can also occur, however, in scenarios of adaptive divergence, where adaptive loci can also accumulate in regions of low recombination (Seehausen et al. 2014). These processes can also act together, and the development of intrinsic barriers such as DMI can both lead to and follow from, extrinsic barriers such as adaptation and selection (Seehausen et al. 2014).
Association with climate variables
Individuals belonging to the cryptic subspecies B. l. hibernus were more likely to be found in locations that reached a lower minimum temperature. Many of these locations were geographically clustered in space, making it difficult to definitively identify this as a key factor in adaptation and speciation. Since we did account for variables such as easting and northing in our model (which improves, conditionally, the independence of these spatially-correlated data in our model) we hypothesize that cold temperature adaptation is a good starting point for future studies into the process of divergence in these populations. The independence of our findings from broad geographic trends was also supported by the model that we fit using only data from sympatric locales where both B. l. sylvicola and B. l. hibernus were found (Supplemental Table S6).
Others (Christmas et al. 2021) have predicted that adaptation to cold temperatures may explain previous examples of speciation in B. l. sylvicola. For example, a period of global cooling followed by global warming (Vimeux et al. 2002; Uemura et al. 2018) occurred prior to the estimated divergence of B. l. sylvicola and its newly described sister species B. incognitus (Christmas et al. 2021). Our findings lend support to this hypothesis and suggest that adaptation to cold temperatures was likely a factor in the divergence of the cryptic B. l. hibernus from B. l. sylvicola as well.
The potential for montane bumble bees to adapt to cold temperatures was also shown in a study of thermal tolerance and gene expression in Bombus vosnesenkii (Pimsler et al. 2020). Queen bees were collected from regions with different temperature regimes and the colonies established from these queens showed critical thermal minima (CTmin) that was associated with the local temperatures in the regions that they came from (Pimsler et al. 2020). Critical thermal maxima, however, did not vary between populations collected in different regions (Pimsler et al. 2020). This further suggests that bumble bees have the potential to be locally adapted to regions based on the coldest temperatures reached there.
Both this and previously published studies (e.g. Ghisbain et al. 2020; Pimsler et al. 2020; Christmas et al. 2021) have found evidence of population and species-level divergence in bumble bees collected from mountain habitats. These mountain regions may harbour other cryptic populations and adaptations that have yet to be discovered. They are also regions that may be especially susceptible to climate change (Guisan et al. 2019). For both reasons, mountain habitats warrant further study to support biodiversity conservation efforts.
Conservation implications
The cryptic genetic and phenotypic diversity found in this study, along with the preliminary evidence for climate specialization in different populations of this species, underscore the importance of considering populations of species independently rather than as one uniform species when planning conservation monitoring and management programs. While genetic diversity may indicate resilience of a population or taxa to changing conditions, it may also indicate specialization of different populations to unique climate niches, especially in heterogenous habitats such as montane regions. These results suggest a need to evaluate whether populations occupying distinct or unique climates would be best considered as separate management units since they could be adapted to, and therefore impacted differently by, changing climates and/or events such as extreme weather that are related to climate change.
Conclusion
We found evidence of a cryptic population of bumble bees in the Canadian Rocky Mountains, likely corresponding to a new subspecies in the B. lapponicus species complex, which we have provisionally named B. l. hibernus. This population is genetically and phenotypically distinguishable from other B. l. sylvicola individuals collected in this study and was found to be parapatric in distribution. It is also genetically distinct from other previously described species in the broader B. lapponicus / B. sylvicola species complex, including B. incognitus (Christmas et al. 2021). Similar to B. incognitus, this new cryptic population was differentiated from an existing B. l. sylvicola population in clustered regions of the genome, likely corresponding to the centromere of each chromosome (Christmas et al. 2021).
While it is difficult to definitively conclude what the catalyst was for the genetic differentiation process between these two groups, they currently appear to occupy areas with different temperature profiles. Specifically, B. l. hibernus is more likely to be found in areas that reach lower minimum temperatures in the coldest quarter, corresponding to previous research predicting (Christmas et al. 2021) and showing (Pimsler et al. 2020) that adaptation to cold temperatures is present in montane bumble bee populations. This evidence of differentiation and of potential adaptation to different temperature regimes is an exciting contribution both for identifying risks to pollinating insects in the face of changing climates and for our understanding of a Holarctic bumble bee species complex that has been the focus of much recent attention.
Data availability
Color phenotype data and genetic data are available on Dryad.
References
Andrews S (2010) FastQC: a quality control tool for high throughput sequence data. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/
Bellard C, Bertelsmeier C, Leadley P, Thuiller W, Courchamp F (2012) Impacts of climate change on the future of biodiversity. Ecol Lett 15:365–377
Bickford D, Lohman DJ, Sodhi NS, Ng PKL, Meier R, Winker K, Ingram KK, Das I (2007) Cryptic species as a window on diversity and conservation. Trends Ecol Evol 22:148–155
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. https://doi.org/10.1093/BIOINFORMATICS/BTU170
Burri R, Nater A, Kawakami T, Mugal CF, Olason PI, Smeds L, Suh A, Dutoit L, Bureš S, Garamszegi LZ et al (2015) Linked selection and recombination rate variation drive the evolution of the genomic landscape of differentiation across the speciation continuum of Ficedula flycatchers. Genome Res 25:1656–1665. https://doi.org/10.1101/GR.196485.115
Butlin RK (2005) Recombination and speciation. Mol Ecol 14:2621–2635. https://doi.org/10.1111/j.1365-294X.2005.02617.x
Butlin RK, Galindo J, Grahame JW (2008) Sympatric, parapatric or allopatric: the most important way to classify speciation? Philos Trans R Soc B Biol Sci 363:2997–3007. https://doi.org/10.1098/rstb.2008.0076
Cameron SA, Hines HM, Williams PH (2007) A comprehensive phylogeny of the bumble bees (Bombus). Biol J Linn Soc 91:161–188. https://doi.org/10.1111/j.1095-8312.2007.00784.x
Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T (2009) trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinforma Appl 25:1972–1973. https://doi.org/10.1093/bioinformatics/btp348
Catchen J, Hohenlohe PA, Bassham S, Amores A, Cresko WA (2013) Stacks: an analysis tool set for population genomics. Mol Ecol 22:3124–3140. https://doi.org/10.1111/MEC.12354
Chapin FSI, Sala OE, Burke IC, Grime P, Hooper DU, Lauenroth WK, Lombard A, Mooney HA, Mosier AR, Naeem S et al (1998) Ecosystem consequences of changing biodiversity: experimental evidence and a research agenda for the future. Bioscience 48:45–52
Chen S, Tang K, Wang X, Li F, Fu C, Liu Y, Faiz AH, Jiang X, Liu S (2022) Multi-locus phylogeny and species delimitations of the striped-back shrew group (Eulipotyphla: Soricidae): implications for cryptic diversity, taxonomy and multiple speciation patterns. Mol Phylogenet Evol 177:107619. https://doi.org/10.1016/J.YMPEV.2022.107619
Christmas MJ, Jones JC, Olsson A, Wallerman O, Bunikis I, Kierczak M, Peona V, Whitley KM, Larva T, Suh A et al (2021) Genetic barriers to historical gene flow between cryptic species of alpine bumblebees revealed by comparative population genomics. Mol Biol Evol 38:3126–3143. https://doi.org/10.1093/molbev/msab086
Clake DJ, Rogers SM, Galpern P (2022) Landscape complementation is a driver of bumble bee (Bombus sp.) abundance in the Canadian rocky mountains. Landsc Ecol 37:713–728. https://doi.org/10.1007/s10980-021-01389-2
Cortés AJ, López-Hernández F (2021) Harnessing crop wild diversity for climate change adaptation. Genes 12:783. https://doi.org/10.3390/genes12050783
Dray S, Dufour AB (2007) The ade4 package: implementing the duality diagram for ecologists. J Stat Softw 22:1–20. https://doi.org/10.18637/jss.v022.i04
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. https://doi.org/10.1093/NAR/GKH340
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620. https://doi.org/10.1111/j.1365-294X.2005.02553.x
Feder JL, Egan SP, Nosil P (2012) The genomics of speciation-with-gene-flow. Trends Genet 28:342–350. https://doi.org/10.1016/J.TIG.2012.03.009
Fick SE, Hijmans RJ (2017) WorldClim 2: new 1-km spatial resolution climate surfaces for global land areas. Int J Climatol 37:4302–4315. https://doi.org/10.1002/joc.5086
Frichot E, François O (2015) LEA: an R package for landscape and ecological association studies. Methods Ecol Evol. https://doi.org/10.1111/2041-210X.12382
Garcia RA, Cabeza M, Rahbek C, Araújo MB (2014) Multiple dimensions of climate change and their implications for biodiversity. Science 344:1247579
Geraldes A, Basset P, Smith KL, Nachman MW (2011) Higher differentiation among subspecies of the house mouse (Mus musculus) in genomic regions with low recombination. Mol Ecol 20:4722–4736. https://doi.org/10.1111/J.1365-294X.2011.05285.X
Gernhard T (2008) The conditioned reconstructed process. J Theor Biol 253:769–778. https://doi.org/10.1016/j.jtbi.2008.04.005
Ghisbain G, Lozier JD, Rahman SR, Ezray BD, Tian L, Ulmer JM, Heraghty SD, Strange JP, Rasmont P, Hines HM (2020) Substantial genetic divergence and lack of recent gene flow support cryptic speciation in a colour polymorphic bumble bee (Bombus bifarius) species complex. Syst Entomol 45:635–652. https://doi.org/10.1111/SYEN.12419
Guisan A, Broennimann O, Buri A, Cianfrani C, D’Amen M, Di Cola V, Fernandes R, Gray SM, Mateo RG, Pinto E et al (2019) Climate change impacts on mountain biodiversity. In: Hannah L, Wilson EO, Lovejoy TE (eds) Biodiversity and climate change transforming the biosphere. Yale University Press, pp 221–236
Hijmans RJ (2022) Raster: geographic data analysis and modeling. R package version 3.5–15. https://CRAN.R-project.org/package=raster
Jackson JM, Pimsler ML, Oyen KJ, Koch-Uhuad JB, Herndon JD, Strange JP, Dillon ME, Lozier JD (2018) Distance, elevation and environment as drivers of diversity and divergence in bumble bees across latitude and altitude. Mol Ecol 27:2926–2942. https://doi.org/10.1111/mec.14735
Johnson KH, Vogt KA, Clark HJ, Schmitz OJ, Vogt DJ (1996) Resilience and stability of ecosystems. Trends Ecol Evol 11:372–377
Jombart T (2008) Adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24:1403–1405. https://doi.org/10.1093/bioinformatics/btn129
Jombart T, Ahmed I (2011) Adegenet 1.3-1: new tools for the analysis of genome-wide SNP data. Bioinformatics 27:3070–3071. https://doi.org/10.1093/bioinformatics/btr521
Knaus BJ, Grünwald NJ (2017) vcfr: a package to manipulate and visualize variant call format data in R. Mol Ecol Resour 17:44–53. https://doi.org/10.1111/1755-0998.12549
Li H (2011) A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27:2987–2993. https://doi.org/10.1093/bioinformatics/btr509
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. https://doi.org/10.1093/BIOINFORMATICS/BTP324
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. https://doi.org/10.1093/bioinformatics/btp352
Loreau M (2000) Biodiversity and ecosystem functioning: recent theoretical advances. Oikos 91:3–17. https://doi.org/10.1034/j.1600-0706.2000.910101.x
Manichaikul A, Mychaleckyj JC, Rich SS, Daly K, Sale M, Chen W (2010) Robust relationship inference in genome-wide association studies. Bioinformatics 26:2867–2873. https://doi.org/10.1093/bioinformatics/btq559
Marske KA, Rahbek C, Nogués-Bravo D (2013) Phylogeography: spanning the ecology-evolution continuum. Ecography 36:1169–1181. https://doi.org/10.1111/j.1600-0587.2013.00244.x
Martinet B, Lecocq T, Brasero N, Gerard M, Urbanova K, Valterova I, Gjershaug JO, Michez D, Rasmont P (2019) Integrative taxonomy of an arctic bumblebee species complex highlights a new cryptic species (Apidae: Bombus). Zool J Linn Soc 187:599–621. https://doi.org/10.1093/zoolinnean/zlz041
Nosil P (2012) The speciation continuum: what factors affect how far speciation proceeds? Ecological speciation. Oxford University Press, pp 192–213
Nosil P, Feder JL (2012) Genomic divergence during speciation: causes and consequences. Philos Trans R Soc B Biol Sci 367:332–342. https://doi.org/10.1098/rstb.2011.0263
Obura DO, Declerck F, Verburg PH, Gupta J, Abrams JF, Bai X, Bunn S, Ebi KL, Gifford L, Gordon C et al (2022) Achieving a nature- and people-positive future. One Earth. https://doi.org/10.1016/j.oneear.2022.11.013
Oliver TH, Isaac NJB, August TA, Woodcock BA, Roy DB, Bullock JM (2015) Declining resilience of ecosystem functions under biodiversity loss. Nat Commun. https://doi.org/10.1038/ncomms10122
Ollerton J (2017) Pollinator diversity: distribution, ecological function and conservation. Annu Rev Ecol Evol Syst 48:353–376. https://doi.org/10.1146/annurev-ecolsys-110316-022919
Owen RE, Whidden TL, Plowright RC (2010) Genetic and morphometric evidence for the conspecific status of the bumble bees, Bombus melanopygus and Bombus edwardsii. J Insect Sci 10:1–18. https://doi.org/10.1673/031.010.10901
Paradis E, Schliep K (2019) ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 35:526–528. https://doi.org/10.1093/bioinformatics/bty633
Pearman PB, D’Amen M, Graham CH, Thuiller W, Zimmermann NE (2010) Within-taxon niche structure: Niche conservatism, divergence and predicted effects of climate change. Ecography 33:990–1003. https://doi.org/10.1111/J.1600-0587.2010.06443.X
Pimsler ML, Oyen KJ, Herndon JD, Jackson JM, Strange P, Dillon ME, Lozier LD (2020) Biogeographic parallels in thermal tolerance and gene expression variation under temperature stress in a widespread bumble bee. Sci Reports 101(10):1–11. https://doi.org/10.1038/S41598-020-73391-8
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959. https://doi.org/10.1093/GENETICS/155.2.945
R Core Team (2023) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/
Riddle BR (2019) Genetic signatures of historical and contemporary responses to climate change. In: Hannah L, Wilson EO, Lovejoy TE (eds) Biodiversity and climate change: transforming the biosphere. Yale University Press, pp 66–76
Sadd BM, Barribeau SM, Bloch G, de Graaf DC, Dearden P, Elsik CG, Gadau J, Grimmelikhuijzen CJP, Hasselmann M, Lozier JD et al (2015) The genomes of two key bumblebee species with primitive eusocial organization. Genome Biol 16:1–31. https://doi.org/10.1186/s13059-015-0623-3
Schluter D, Pennell MW (2017) Speciation gradients and the distribution of biodiversity. Nature 546:48–55. https://doi.org/10.1038/NATURE22897
Schön I, Pieri V, Sherbakov DY, Martens K (2017) Cryptic diversity and speciation in endemic Cytherissa (Ostracoda, Crustacea) from Lake Baikal. Hydrobiologia 800:61–79. https://doi.org/10.1007/S10750-017-3259-3/FIGURES/3
Seehausen O, Butlin RK, Keller I, Wagner CE, Boughman JW, Hohenlohe PA, Peichel CL, Saetre GP et al (2014) Genomics and the origin of species. Nat Rev Genet 15:176–192. https://doi.org/10.1038/nrg3644
Sheffield CS, Oram R, Heron JM (2020) Bombus (Pyrobombus) johanseni Sladen, 1919, a valid North American bumble bee species, with a new synonymy and comparisons to other “red-banded” bumble bee species in North America (Hymenoptera, Apidae, Bombini). Zookeys 2020:59–81. https://doi.org/10.3897/zookeys.984.55816
Sladen FWL (1919) The wasps and bees collected by the Canadian Arctic Expedition, 1913–1918. Report of the Canadian Arctic Expedition 1913–18 3: 25–35
Sousa V, Hey J (2013) Understanding the origin of species with genome-scale data: modelling gene flow. Nat Rev Genet 14:404–414. https://doi.org/10.1038/nrg3446
Suchard MA, Lemey P, Baele G, Ayres DL, Drummond AJ, Rambaut A (2018) Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol 4:vey016. https://doi.org/10.1093/ve/vey016
Taylor SA, Curry RL, White TA, Ferretti V, Lovette I (2014) Spatiotemporally consistent genomic signatures of reproductive isolation in a moving hybrid zone. Evolution 68:3066–3081. https://doi.org/10.1111/EVO.12510
Tilman D, Reich PB, Knops JMH (2006) Biodiversity and ecosystem stability in a decade-long grassland experiment. Nature 441:629–632. https://doi.org/10.1038/nature04742
Trontelj P, Fišer C (2009) Cryptic species diversity should not be trivialised. Syst Biodivers 7:1–3. https://doi.org/10.1017/S1477200008002909
Uemura R, Motoyama H, Masson-Delmotte V, Jouzel J, Kawamura K, Goto-Azuma K, Fujita S, Kuramoto T, Hirabayashi M, Miyake T et al (2018) Asynchrony between Antarctic temperature and CO2 associated with obliquity over the past 720,000 years. Nat Commun 91(9):1–11. https://doi.org/10.1038/s41467-018-03328-3
Ungerer MC, Johnson LC, Herman MA (2008) Ecological genomics: understanding gene and genome function in the natural environment. Heredity 100:178–183
Venables WN, Ripley BD (2002) Modern applied statistics with S, 4th edn. Springer, New York
Vimeux F, Cuffey KM, Jouzel J (2002) New insights into southern Hemisphere temperature changes from Vostok ice cores using deuterium excess correction. Earth Planet Sci Lett 203:829–843. https://doi.org/10.1016/S0012-821X(02)00950-0
Vodă R, Dapporto L, Dincă V, Vila R (2015) Cryptic matters: overlooked species generate most butterfly beta-diversity. Ecography 38:405–409. https://doi.org/10.1111/ECOG.00762
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution (NY) 38:1358–1370. https://doi.org/10.2307/2408641
Williams P, Thorp R, Richardson L, Colla S (2014) Bumble bees of North America. Princeton University Press
Wolf JBW, Ellegren H (2017) Making sense of genomic islands of differentiation in light of speciation. Nat Rev Genet 18:87–100. https://doi.org/10.1038/nrg.2016.133
Yule GU (1925) A mathematicaltheory of evolution, based on the conclusions of Dr. J. C. Willis, F. R. S. Phil Trans R Soc Lond B 213:21–87. https://doi.org/10.1098/rstb.1925.0002
Zhou Z, Guo H, Han L, Chai J, Che X, Shi F (2019) Singleton molecular species delimitation based on COI-5P barcode sequences revealed high cryptic/undescribed diversity for Chinese katydids (Orthoptera: Tettigoniidae). BMC Evol Biol 19:1–19. https://doi.org/10.1186/s12862-019-1404-5
Acknowledgements
The authors would like to thank Hailey Bloom, Jessy Bokvist, David Clake, Brenna Stanford, and Luke Storey for helping to carry out the field work in this study, and Parks Canada for their support in facilitating field sampling. Thanks to Hailey Bloom, Emma Dunlop, and Michael Gavin for assisting with species identification, and to Rebecca Innes for helping to collect phenotypic data. This research occurred on the traditional lands of the Siksikaitsitapi (Blackfoot Confederacy) Siksika, Kainai and Piikani First Nations; the Iyarhe (Stoney) Nakoda First Nation; the Tsuut’ina First Nation, the Métis Nation of Alberta; and the Ktunaxa First Nations.
Funding
This work was funded by Natural Sciences and Engineering Research Council (NSERC) Discovery Grants to Paul Galpern and Sean Rogers, and an Alberta Conservation Association (ACA) Grant in Biodiversity to Danielle Clake. We are grateful to the Digital Research Alliance of Canada for supplying computing resources.
Author information
Authors and Affiliations
Contributions
All authors contributed to research design. DC performed data collection, data analysis and wrote the original draft of the manuscript. PG and SR provided resources, supervision and review and editing of writing.
Corresponding author
Ethics declarations
Competing interests
The authors have no relevant competing interests to disclose.
Additional information
Communicated by Nigel Stork.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Clake, D.J., Rogers, S.M. & Galpern, P. Cryptic genotypic and phenotypic diversity in parapatric bumble bee populations associated with minimum cold temperatures. Biodivers Conserv 33, 485–507 (2024). https://doi.org/10.1007/s10531-023-02753-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10531-023-02753-1