Abstract
Objectives
This study aimed to discuss the essential amino acid residues and catalytic mechanism of trans-epoxysuccinate hydrolase from Pseudomonas koreensis for the production of meso-tartaric acid.
Results
The optimum conditions of the enzyme were 45 °C and pH 9.0, respectively. It was strongly inhibited by Zn2+, Mn2+ and SDS. Michaelis–Menten enzyme kinetics analysis gave a Km value of 3.50 mM and a kcat of 99.75 s−1, with an exceptional EE value exceeding 99.9%. Multiple sequence alignment and homology modeling revealed that the enzyme belonged to MhpC superfamily and possessed a typical α/β hydrolase folding structure. Site-directed mutagenesis indicated H34, D104, R105, R108, D128, Y147, H149, W150, Y211, and H272 were important catalytic residues. The 18O-labeling study suggested the enzyme acted via two-step catalytic mechanism.
Conclusions
The structure and catalytic mechanism of trans-epoxysuccinate hydrolase were first reported. Ten residues were critical for its catalysis and a two-step mechanism by an Asp-His-Asp catalytic triad was proposed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Epoxysuccinate hydrolases (ESHs) are members of epoxide hydrolases (EHs, EC 3.3.2.3) that catalyze the conversion of epoxysuccinate to tartaric acid (TA). The enzymatic properties of ESHs determine the chiral properties of TA. TA comprises three optically active isomers: L( +)-TA, D(−)-TA and meso-T. The corresponding ESHs for these isomers are named CESH[L], CESH[D], and TESH, respectively (Xuan and Feng 2019).
Meso-TA is a rare enantiomer of TA, is highly sought after as an anti-caking due to its desirable properties such as environmentally safe, low toxicity, and effectiveness in small quantities. Meso-TA holds substantial market potential in chloride production, snow remova, pharmaceuticals, anti-malaria, and new materials. The demand for meso-TA continues to rise (Varga et al. 2013; Dutta and Gellman 2017; Bitew et al. 2019).
Currently, the chemical synthesis method for TA conversion suffers from low efficiency, primarily due to the challenges in separating different TA configurations. Bio-transformation offers several advantages, including mild reaction conditions, excellent chemo-, regio-, and enantio-selectivity, making it a simple and cost-effective approach for TA production (Bao et al. 2020).
The protein structures and catalysis mechanisms of CESH[L] and CESH[D] have been extensively studied. Three genes encoding CESH[L]s from Rhodococcus opacus (RhCESH[L]), Nocardia tartaricans (NoCESH[L]), and Klebsiella sp. BK-58 (KlCESH[L]) have been cloned and sequenced (Pan et al. 2011; Vasu et al. 2012; Cheng et al. 2014). These enzymes belong to the haloacid dehalogenase (HAD)-like superfamily, characterized by the typical Rossmann fold with a flexible cap domain (Novak et al. 2013). Recently, the crystal structures of RhCESH[L] and KlCESH[L] were analyzed, proposing the catalytic triad of RhCESH[L] as D18-H190-E212 (Dong et al. 2024). R55 stabilizes the oxygen of the epoxide and provides a proton to facilitate epoxy ring opening. The amino acids surrounding the carboxyl groups stabilize the substrate, which is securely held within the binding pocket resembling crab claws (Dong et al. 2024). CESH[D] from Bordetella sp. BK-52 belongs to beta-keto acid cleavage enzyme (BKACE) superfamily and possesses a canonical (β/α)8 TIM barrel fold. In this case, a zinc cation interacts with H47-H49-E14, forming a chelate with deep cavity of the barrel center. Combining the study of 18O tracing experiments (Bao et al. 2013), CESH[D] adopts a one-step catalytic mechanism. The crystal structure of CESH[D]-D115A/D-TA demonstrates that catalytic water is activated by D115 and E190, while residue R11 provides a proton to the oxirane oxygen atom (Dong et al. 2018).
However, there have been limited reports on TESH enzyme. In 1955, Martin and Foster initially identified a Flavobacterium sp. capable of converting trans-epoxysuccinate to meso-TA and named this enzyme trans-epoxysuccinate hydrolase (TESH, EC 3.3.2.4) (Martin and Foster 1955; Foster 1960). Subsequently, in 1969, Allen and Jakoby purified TESH from Pseudomonas putida and studied its enzymatic properties (Allen and Jakoby 1969). More recently, we isolated Pseudomonas koreensis BK-9, which exhibited the ability to produce meso-TA, and successfully cloned its TESH gene in our lab (Zhang 2015; Zhang et al. 2016). However, there have been no further reports on the structure and catalytic mechanism of the TESH.
In this study, we extensively characterized the enzymatic properties of TESH from P. koreensis BK-9. We investigated its structure, identified critical amino acid, and proposed its catalytic mechanism using multiple sequence alignment, homologous modeling, molecular docking, site-directed mutagenesis, and isotope labeling. Our finding revealed that ten residues (H34, D104, R105, R108, D128, Y147, H149, W150, Y211, and H272) play a vital role in catalysis, and we proposed a two-step mechanism involving an Asp-His-Asp catalytic triad.
Materials and methods
Strains, plasmids, primers, and culture conditions
P. koreensis BK-9 was isolated by our lab and deposited at the China General Microbiological Culture Collection Center with the identification number CGMCC NO.8395 (Zhang et al. 2016). The BK-9 strain was cultivated following the method described by Bao et al. (2013). The TESH gene of BK-9 (GenBank accession no. WP_151550555) was cloned and expressed in Escherichia coli BL21 (DE3)-pET-15b( +)-TESH by our laboratory (Zhang 2015).
Multiple sequence alignment and homology modeling
The amino acid sequence of TESH from P. koreensis BK-9 was aligned using Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/) to identify conserved sites. The modeled of TESH was generated using Swiss-Model (https://swissmodel.expasy.org/). The docking of the trans-epoxysuccinate with the modeled TESH structure was analyzed by the Discovery Studio 2019 client, and the docking results were visualized by PyMol (Version 2.5).
Construction of site-directed mutagenesis
Based on the reaction mechanism study of the CESH and the alignment results, site-directed mutagenesis was carried out using the PrimeSTAR® Max DNA Polymerase (Takara) with two reverse complement primers. The primers listed in Table S1 were synthesized by Sangon Biotech (Shanghai, China).
Enzyme purification and characterization
Both wild-type and mutant TESHs were expressed in E.coli BL21 (DE3) cells and purified using a His-binding resin column (York Biotech, Shanghai, China). The purification method used for CESH[D] was adapted (Bao et al. 2013), and salt ions were removed via ultrafiltration.
The effect of temperature, pH, metal ions, and surfactants on the activity and stability of TESH were investigated according to the methods used for CESH[L] (Wang et al. 2012). The optimal temperature and pH were determined by varying the assay temperatures (at pH8.0) and pH buffers (at 37 °C) for 30 min. After incubating TESH at different temperatures and pH levels (at room temperature) for 30 min, the enzyme was then adjusted to a standardized condition of 37 °C and pH 8.0 to determine its stability. The relative activity that assayed under standard reaction was taken as 100%. The Km, Vmax and kcat values of the enzymes were determined using Michaelis–Menten plots at increasing trans-epoxysuccinate concentrations ranging from 5 to 170 mM.
Isotope label
The synthesis of TA requires the participation of water molecules, we use H218O to label the reaction system according to the methods used for CESH[D] (Bao et al. 2013). For the single turnover reaction, 200 nmol of TESH and 40 nmol of disodium trans-epoxysuccinate were dissolved in 500 μL of H218O, then incubated the mixture at 30 °C for 12 h. In the case of the multiple turnover experiment, 40 nmol of TESH and 400 nmol disodium trans-epoxysuccinate were mixed in 500 μL of H218O and incubated at 30 °C for 12 h. The reaction was terminated by adding an equal volume of methanol, then measured the molecular mass of the produced tartrate by G6465B Ultivo Triple Quadrupole LC/MS (Agilent, USA) (Bao et al. 2013).
Enzyme assay
TESH activity was assayed at 37 °C for 30 min in 1 mL of 0.2 M disodium trans-epoxysuccinate (20 mM Tris–HCl, pH 8.0). The quantity of meso-TA was determined by high performance liquid chromatography on a chiral column (Chirex 3126 (D)-penicillanmin, 50 × 4.6 mm) at 60 °C with a sample volume of 10 μL. The mobile phase consisted of 88% of 1 mM copper acetate and 50 mM ammonium acetate (pH 4.5) and 12% of isopropanol with a flow rate of 1 mL/min. The detection was done at 280 nm. (Cheng et al. 2014). One unit of TESH activity was defined as the amount of the enzyme that generates 1 μmol of meso-TA per minute. Specific activity was defined as the number of units per milligram of protein. Protein concentration was determined by the Bradford protein assay kit (Shanghai, China).
Results and discussion
Characterization of TESH
The optimum temperature and pH of TESH from P. koreensis BK-9 were determined to be 45 °C and pH 9.0, respectively (Fig. 1A, B). However, when incubated at 45 °C for 30 min, TESH activity significantly decreased, with only 10% of the residual activity remaining at pH levels below 5 or above 10 (Fig. 1A, C), indicating poor temperature and pH stability.
Characterization of TESH from P. koreensis BK-9. Values are mean ± SD calculated from triplicate determinations. The values are shown as relative activities, with the activity under standard condition indicated as 100%. A Effect of temperature on enzyme activity and stability. B Effect of pH on enzyme activity. C Effect of pH on enzyme stability. D Effect of various metal ions and chemicals on enzyme activity. (T test, *P < 0.1, **P < 0.01, ***P < 0.001, ****P < 0.0001)
Statistical analysis indicated that Mg2+, Ca2+, Ba2+ acted as activators can enhance TESH activity (P < 0.01), Zn2+and Mn2+ can inhibited the activity of TESH (P < 0.001). However, adding 10 mM EDTA-Na2 still retained 86.52% activity, varying the concentration of EDTA-Na2 did not cause a significant change in enzyme activity, as indicated in Table S2. These finding suggest that TESH does not rely on metal ions for its function. However, SDS was a strong inhibitor, disrupting the folded protein of the protein and leading to a 95.85% decrease in enzyme activity (Fig. 1D). The kinetic parameters were determined by Michaelis–Menten plotting method (GraphPad Prism 9.5). The values of Km and kcat were calculated to be 3.50 mM and 94.75 s−1, respectively (Fig. S1).
Sequence comparison of TESH and reported EHs
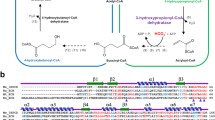
The sequence comparison results indicated that TESH showed no homology to other reported ESHs. While CESH[L]s belong to the HAD like superfamily, CESH[D]s belong to the BKACE superfamily, TESH belongs to MhpC superfamily characterized by the typical α/β hydrolase folding structure. We used Clustal Omega to compare the TESH sequence with 12 structurally characterized proteins from MhpC superfamily. It showed low but considerable sequence identity ranging from 19 to 54%. Several residues were highly conserved, including six nonpolar amino acids (G33, G61, G63, L48, L59, P235), two aspartic acids (D58 and D104), two histidine residues (H103 and H272) and one lysine residue (Y211). These residues includ two motifs and several highly conserved catalytic residues (Fig. 2).
Sequence alignments for TESH from P. koreensis BK-9 and twelve structurally characterized proteins from the MhpC superfamily. The sequences were aligned with Clustal Omega and were shown in order of decreasing sequence identity compared to TESH. Identical amino acids are marked with an asterisk, conserved substitution residues with a colon, semi-conserved substitution residues with a period, and residues mutagenized in this study with a pound sign. α-Helices are underlined, and β-strands are shaded. The sequences are as follows: 3QYJ, putative α/β hydrolase from Nostoc sp. PCC 7120 (Q8Z0Q1); 4NVR, putative acyltransferase from S. enterica (Q8ZRI7); 8SDC, fluroacetate dehalogenase (FAD) DAR3835 from D. aromatica (Q479B8); 1Y37, FAD FA1 from Burkholderia sp.FA1 (Q1JU72); 5SWN, FAD RPA1163 from R. palustris CGA009 (Q6NAM1); 4B9A, putative epoxide hydrolase from P. aeruginosa PA01 (Q9I229); 7AC0, epoxide hydrolase CorEH from Corynebacterium sp. C12 (O52866); 3KD2, CFTR inhibitory factor Cif from P. aeruginosa UCBPP-PA14 (A0A0H2ZD27); 6KXR, putative hydrolase Alp 1U from S. ambofaciens ATCC 23877 (A0A0K2AJY3); 1EHY, Epoxide hydrolase ArEH from A. radiobacter AD1 (O31243); 2CJP, epoxide hydrolase StEH1 from S. tuberosum (Q41415); 5XM6, hydrolase VrEH2 from V. radiata (A0A0R5NGA4) and TESH, trans-epoxysuccinate hydrolase from P. koreensis BK-9 (WP_151550555)
The residue X within the H-G-X-P motif plays a role in stabilizing the negative charge that develops on the carbonyl oxygen of the nucleophilic aspartate. Typically, this residue is an aromatic residue or a charged amino acid (Loo et al. 2006). However, the function of the conserved D-L-R/P-G-X1-G-X2-S/T motif following strand β4 has no been reported yet (Mitusinska et al. 2022). In the case of α/β hydrolase fold epoxide hydrolase, the nucleophile in the catalytic triad is aspartate (Fig. 2, position 1), responsible for attacking the epoxy ring (Loo et al. 2006). The histidine that preceded it is highly conserved but previously neglected. It is electrostatically linked to the general base of potato epoxide hydrolase (StEH1), providing charge balance in its protonated form (Amrein et al. 2015). Y211, a conserved site in EHs, mainly forms hydrogen bonds with oxygen on the epoxide ring and provides a proton to assist ring opening. It usually forms oxygen anion holes with a Tyr-Tyr pair (Yamada et al. 2000) and has also been reported to interact with His and Trp (Bahl and Madden 2012). P235 is located in the loop after strand β7, and there are no reports on this amino acid. As a conserved amino acid in the Asp-His-Asp/Glu catalytic triplet of EHs, histidine (Fig. 2, position 2) is mainly responsible for activating water to form a hydroxyl group, thus hydrolyzing the enzyme–substrate complex. However, the acid in nucleophile-histidine-acid catalytic triad is located either in front of the cap domain (Fig. 2 position 3a) or after the cap domain (Fig. 2 position 3b) (Loo et al. 2006). Based on the results of sequence alignment, it is speculated that TESH adopts the D104-H272-D128 catalytic triad, which is important for the site selection of subsequent molecular docking.
Homology modeling of TESH
We used Swiss-Model to generate the 3D structure of TESH from P. koreensis BK-9, selecting four template structures (AlphaFold, 3QYJ, 4NVR and 8SDC) with the highest amino acid sequence identity to TESH (ranging from 87.29 to 51.69%). The model quality was evaluated by Errat, Verify 3D, Whatcheck, and Procheck (not presented), all four models passed the evaluation, with the model based on 4NVR template had the highest score. The Ramachandran plot (Fig. S2) showed that the percentages of residues falling in disallowed regions, generously allowed regions, favorable regions, and core regions were 0.4, 0.9, 7.5, and 91.2%, respectively. The coverall plot showed that more than 90% of the residues were within the favorable region, indicating a good quality model.
The modeled 3D structure of TESH has the typical eight parallel β-sheets (residues 6–10, 16–22, 27–31, 53–57, 98–103, 121–127, 236–241, and 263–268) surrounded by α-helices and is covered by a cap-domain (Fig. 3A). The structure is similar to that of most α/β hydrolase fold enzyme (Heikinheimo et al. 1999).
Homology modeled structure of TESH based on 4NVR. A, single monomer of TESH highlighting the highly conserved main domain consisting of β-Strands (magenta) surrounded by α-Helices (cyan) and the variable cap domain (orange) typical for members of the α/β hydrolase fold superfamily. The binding pocket is represented by gray. B, substrate and thirteen amino acids that near the binding pocket are shown in green and yellow respectively, hydrogen bonds are dotted in green and salt bonds are dotted in orange
The trans-epoxysuccinate substrate and the four modeled TESH structures were prepared for molecular docking by Discovery Studio. The docking site was defined with D104-D128-H272, and the docking radius was adjusted to 0.5 Å. Thirteen amino acids (H34, D104, R105, R108, D128, I129, Y147, H149, W150, V175, R179, Y211, H272) were found near the binding pocket in four docking results, as labeled on the 4NVR modeled structure by PyMOL (Fig. 3B).
Site-directed mutagenesis
Combing with the study of CESHs, we mutated thirteen amino acid sites to the following rules: H and D were mutated to N; Q to E; R to K; I, V, and W to A; Y to F. The purified protein were shown in Fig. S3. The results of site-directed mutagenesis revealed that six mutants showed relative activities that were only 2% or less of the wild-type enzyme’s activity: R108K, D128N, Y147F, H149N, W150A, and Y211F. Four mutants showed a the lower kcat/Km value than the wild-type: H34N, D104N, R105K, Y211F (Table 1). Although the activity of the H272N mutant did not change significantly, it is possible that there is a backup base in the enzyme (Amrein et al. 2015). Moreover, H272 is highly conserved during evolution. In conclusion, we speculate that the ten residues (H34, D104, R105, R108, D128, Y147, H149, W150, Y211, and H272) played important roles in the catalysis.
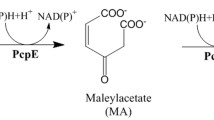
Single and multiple turnover reaction of wild-type TESH in H2 18O
The single and multiple turnover reactions of TESH from P. koreensis BK-9 in H218O showed that most of the tartrate molecules were 16O-labeled in the single turnover reaction (Fig. 4A), while more than 90% tartrate molecules were 18O-labeled in the multiple turnover reaction (Fig. 4B). These results suggested that the catalytic mechanism of the TESH from P. koreensis BK-9 is similar to that of most α/β hydrolases, which involves a two-step catalytic reaction with the forming of an enzyme–substrate ester intermediate.
Ion spray mass spectra of tartrate produced by the TESH from P. koreensis BK-9 in H218O. A, single turnover reaction. B, multiple turnover reaction. The reaction mixtures were analyzed by the system. The step size was 0.1 atomic mass units, and the dwell time was 10 ms per step. The ion spray voltage was set at 4 kV, and the orifice voltage was optimized at 50 V (Bao et al. 2013)
We aligned the ten important catalytic residues of the TESH with the corresponding residues in eight enzymes from MhpC superfamily, which have reported catalytic mechanisms. They respectively are fluoroacetate dehalogenase (FAD) DAR3835 from D. aromatic (Khusnutdinova et al. 2023), FAD RPA1163 from R. palustris (Chan et al. 2011), epoxide hydrolase CorEH from Corynebacterium sp. C12 (Schuiten et al. 2021), bacterial virulence factor EH (Cif) from P. aeruginosa (Bahl et al. 2010), epoxide hydrolase Alp1U from S. ambofaciens (Zhang et al. 2020a), epoxide hydrolase ArEH from A. radiobacter AD1 (Nardini et al. 1999), epoxide hydrolase StEH1 from S. tuberosum (Mowbray et al. 2006), and epoxide hydrolase VrEH2 from V. radiata (Li et al. 2018). We found expect Cif adopted Asp-His-Glu catalytic triad, the MhpC family members shown in the figure all take Asp-His-Asp catalytic triad (Fig. 5A). The docking results revealed that Asp104, Asp128, and His272 were in close proximity, suggesting a potential interaction among them. Additionally, Asp104 was measured to be 2.9 Å away from the carbon atom on the substrate, indicating a favorable position for nucleophilic attack (Fig. 5B). Therefore, we proposed that TESH might adopt an Asp104-His272-Asp128 as catalytic triad.
Structure of TESH and comparison of the active site to other numbers of the MhpC superfamily. A, alignment of the active site of TESH with FAD D3835, FAD RPA1163, CorEH, Cif, Alp1U, ArEH, StEH1, VrEH2 and catalytic residues are highlighted. The catalytic triad Asp-His-Asp/Glu (yellow), epoxide ring-open amino acids (cyan), and substrate/intermediate-stabilizing residues (magenta and orange) are visualized. B, Asp104-His272-Asp128 catalytic triad of TESH. C, the residues help epoxide positioning and ring opening. D, arginine interacting with the carboxyl group in the substrate, orange lines presenting salt bonds. E, the residues that form hydrogen bonds with Asp104
Like most α/β hydrolase fold enzyme, Tyr residue that near the substrate plays an important role in assisting ring opening. Firstly, they have a hydrogen bond with the epoxide oxygen, which can position the substrate in the active site for nucleophilic attack. Then, tyrosine provides a proton to the epoxide oxygen during the ring-opening step (Yamada et al. 2000). While epoxide hydrolases typically employ a Tyr-Tyr or His-Tyr pair to assist in ring-opening (Bahl, Madden 2012), fluoroacetate dehalogenase DAR3835 adopts His152-Trp153-Tyr215 (His155-Trp156-Tyr219 in FAD RPA1163) to stabilize the halogen atom and suggests that the side chain of Tyr150 might also facilitate substrate binding (Khusnutdinova et al. 2023). The docking results (Fig. 5C) showed that Tyr147 and Tyr211 located on different sides of the epoxy ring, with their hydroxyl groups pointed to the oxygen in the ring, which was conducive to providing protons for ring opening. The adjacent chains Tyr147 and His149 interact with two tyrosine residues, forming an oxyanion hole that stabilizes the oxyanion of the alkyl-enzyme intermediate formed by ring opening.
In FADs an Arg-Arg pair serves as the binding site for carboxylic acids on the substrate (Chan et al. 2011; Khusnutdinova et al. 2023). Docking the substrate with the protein revealed that the carboxyl group on the free trans-epoxysuccinic acid had salt bonds with Arg105, Arg108, and Arg179 (Fig. 5D). The results were similar to the research on FAD RPA1163 (Wang et al. 2017; Zhang et al. 2020b), wherein one side of the substrate was secured by Arg111 and Arg114 (corresponding to Arg105 and Arg108 in TESH), while the other side interacted with Trp185 (corresponding to Arg179 in TESH). Mutating Trp185 could obtain a larger binding pocket, thereby improving the conversion ability for bulky substrate. According to the mutation experiment, R179K still maintained 19% enzyme activity relative to wild bacteria (Table 1), so we speculate that Arg179 is not an essential amino acid of TESH. In addition, we found Arg105, Arg108 and His34 formed hydrogen bonds with Asp104 (Fig. 5E). When Asp104 SN2 attack C(S) of the substrate, resulting in a configuration reversal, Arg105, Arg108 and His34 stabilize the negative charge developed on the carbonyl oxygen of the nucleophilic Asp104 (Nardini et al. 1999; Loo et al. 2006; Bahl et al. 2016). On the other side of substrate, Tyr147, His149, Trp150, and Tyr211 stabilize oxygen anion produced by epoxy ring opening, thus facilitating the hydrolysis of the alkyl enzyme intermediate (Fig. 5) (Amrein et al. 2015).
Based on the results and analysis presented, we propose that the TESH adopted a two-step catalytic mechanism involving an Asp104-His272-Asp128 catalytic triad (Fig. 6). The catalytic mechanism of TESH is differs from the reported CESH[L] and CESH[D], as CESH[L] adopts an Asp-His-Glu catalytic triad and its enzyme activity relay on SO42− (Dong et al. 2024). CESH[D] catalyzes through a Zn2+-dependent, one-step mechanism (Dong et al. 2018). Our research on TESH contributes to the understanding of ESHs lineage and provides a theoretical basis for subsequent researches.
Proposed catalytic mechanism for the TESH from P. koreensis BK-9. First, a nucleophilic attack (SN2) occurs on the carbon atom in the oxirane ring of the substrate by Asp104, resulting in the inversion of configuration at C (S) and the formation of an enzyme–substrate intermediate jointed by an ester bond. Then, the ester bond is hydrolyzed by an active water molecule active by His272-Asp128 ion pair, leading to the release of meso-tartaric acid. During the reaction, Tyr147 and Tyr211 provide a proton to stabilize the oxygen anion
Conclusions
The TESH enzyme derived from P. koreensis BK-9 belongs to the MhpC superfamily and possesses a characteristic α/β hydrolase folding structure. Enzymatic experiments revealed that the optimum conditions of the enzyme were at the temperature of 45 °C and a pH of 9.0. Like most EHs, TESH catalysis does not rely on the participation of metal ions. The Michaelis–Menten constant Km was determined to be 3.50 mmol L−1, the constant of catalytic activities was 99.75 s−1, and the enantiomeric purity was higher than 99.9%. Combining site-directed mutagenesis, multiple sequence alignment, and molecular docking results, we propose that the residues H34, D104, R105, R108, D128, Y147, H149, W150, Y211, and H272 are crucial for the catalytic process. Additionally, Isotope labeling study suggested the enzyme acts via an Asp104-His272-Asp128 two-step catalytic mechanism.
References
Allen RH, Jakoby WB (1969) Tartaric acid metabolism. IX. Synthesis with tartrate epoxidase. J Biol Chem 244(8):2078–2084. https://doi.org/10.1016/s0021-9258(18)94369-3
Amrein BA, Bauer P, Duarte F, Carlsson ÅJ, Naworyta A, Mowbray SL, Widersten M, Kamerlin SCL (2015) Expanding the catalytic triad in epoxide hydrolases and related enzymes. ACS Catal 5(10):5702–5713. https://doi.org/10.1021/acscatal.5b01639
Bahl CD, Madden DR (2012) Pseudomonas aeruginosa Cif defines a distinct class of α/β epoxide hydrolases utilizing a His/Tyr ring-opening pair. Protein Pept Lett 19(2):186–193. https://doi.org/10.2174/092986612799080392
Bahl CD, Morisseau C, Bomberger JM, Stanton BA, Hammock BD, O’Toole GA, Madden DR (2010) Crystal structure of the cystic fibrosis transmembrane conductance regulator inhibitory factor Cif reveals novel active-site features of an epoxide hydrolase virulence factor. J Bacteriol 192(7):1785–1795. https://doi.org/10.1128/jb.01348-09
Bahl CD, Hvorecny KL, Morisseau C, Gerber SA, Madden DR (2016) Visualizing the mechanism of epoxide hydrolysis by the bacterial virulence enzyme Cif. Biochemistry 55(5):788–797. https://doi.org/10.1021/acs.biochem.5b01229
Bao W, Pan H, Zhang Z, Cheng Y, Xie Z, Zhang J, Li Y (2013) Analysis of essential amino acid residues for catalytic activity of cis-epoxysuccinate hydrolase from Bordetella sp. BK-52. Appl Microbiol Biot 98:1641–1649. https://doi.org/10.1007/s00253-013-5019-2
Bao W, Liao H, Chen Y, Huang Q, Huang W, Fang R, Liu S (2020) Isolation of a novel strain Aspergillus niger WH-2 for production of L(+)-tartaric acid under acidic condition. Biotechnol Lett 42:605–612. https://doi.org/10.1007/s10529-020-02799-z
Bitew MA, Wawegama NK, Newton HJ, Sansom FM (2019) Meso-tartrate inhibits intracellular replication of Coxiella burnetii, the causative agent of the zoonotic disease Q fever. Pathog Dis 77(8):ftz066. https://doi.org/10.1093/femspd/ftz066
Chan PWY, Yakunin AF, Edwards EA, Pai EF (2011) Mapping the reaction coordinates of enzymatic defluorination. J Am Chem Soc 133(19):7461–7468. https://doi.org/10.1021/ja200277d
Cheng Y, Wang L, Pan H, Bao W, Sun W, Xie Z, Zhang J, Zhao Y (2014) Purification and characterization of a novel cis-epoxysuccinate hydrolase from Klebsiella sp. that produces L(+)-tartaric acid. Biotechnol Lett 36:2325–2330. https://doi.org/10.1007/s10529-014-1614-2
Dong S, Liu X, Cui G, Cui Q, Wang X, Feng Y (2018) Structural insight into the catalytic mechanism of a cis-epoxysuccinate hydrolase producing enantiomerically pure D(-)-tartaric acid. Chem Commun 54(61):8482–8485. https://doi.org/10.1039/c8cc04398a
Dong S, Xuan J, Feng Y, Cui Q (2024) Deciphering the stereo-specific catalytic mechanisms of cis-epoxysuccinate hydrolases producing L(+)-tartaric acid. J Biol Chem 300(2):105635. https://doi.org/10.1016/j.jbc.2024.105635
Dutta S, Gellman AJ (2017) Enantiomer surface chemistry: conglomerate versus racemate formation on surfaces. Chem Soc Rev 46(24):7787–7839. https://doi.org/10.1039/c7cs00555e
Foster JW (1960) Microbiological preparation of meso-tartaric acid. US Patent 2947665
Heikinheimo P, Goldman A, Jeffries C, Ollis DL (1999) Of barn owls and bankers: a lush variety of alpha/beta hydrolases. Structure 7(6):R141–R146. https://doi.org/10.1016/s0969-2126(99)80079-3
Khusnutdinova AN, Batyrova KA, Brown G, Fedorchuk T, Chai YS, Skarina T, Flick R, Petit AP, Savchenko A, Stogios P, Yakunin AF (2023) Structural insights into hydrolytic defluorination of difluoroacetate by microbial fluoroacetate dehalogenases. FEBS J 290(20):4966–4983. https://doi.org/10.1111/febs.16903
Li F, Kong X, Chen Q, Zheng Y, Xu Q, Chen F, Fan L, Lin G, Zhou J, Yu H, Xu J (2018) Regioselectivity engineering of epoxide hydrolase: near-perfect enantioconvergence through a single site mutation. ACS Catal 8:8314–8317. https://doi.org/10.1021/acscatal.8b02622
Loo B, Kingma J, Arand M, Wubbolts MG, Janssen DB (2006) Diversity and biocatalytic potential of epoxide hydrolases identified by genome analysis. Appl Environ Microb 72(4):2905–2917. https://doi.org/10.1128/aem.72.4.2905-2917.2006
Martin WR, Foster JW (1955) Production of trans-epoxysyccinic acid by fungi and its microbiological conversion to meso-tartaric acid. J Bacteriol 70(4):405–414. https://doi.org/10.1128/jb.70.4.405-414.1955
Mitusinska K, Wojsa P, Bzowka M, Raczynska A, Bagrowska W, Samol A, Kapica P, Gora A (2022) Structure-function relationship between soluble epoxide hydrolases structure and their tunnel network. Comput Struct Biotechnol J 20:193–205. https://doi.org/10.1016/j.csbj.2021.10.042
Mowbray SL, Elfstrom LT, Ahlgren KM, Andersson CE, Widersten M (2006) X-ray structure of potato epoxide hydrolase sheds light on substrate specificity in plant enzymes. Protein Sci 15(7):1628–1637. https://doi.org/10.1110/ps.051792106
Nardini M, Ridder IS, Rozeboom HJ, Kalk KH, Rink R, Janssen DB, Dijkstra BW (1999) The X-ray structure of epoxide hydrolase from Agrobacterium radiobacter AD1: an enzyme to detoxify harmful epoxides. J Biol Chem 274(21):14579–14586. https://doi.org/10.1074/jbc.274.21.14579
Novak HR, Sayer C, Isupov MN, Paszkiewicz K, Gotz D, Spragg AM, Littlechild JA (2013) Marine Rhodobacteraceae L-haloacid dehalogenase contains a novel His/Glu dyad that could activate the catalytic water. FEBS J 280(7):1664–1680. https://doi.org/10.1111/febs.12177
Pan H, Xie Z, Bao W, Cheng Y, Zhang J, Li Y (2011) Site-directed mutagenesis of epoxide hydrolase to probe catalytic amino acid residues and reaction mechanism. FEBS Lett 585(15):2545–2550. https://doi.org/10.1016/j.febslet.2011.07.006
Schuiten ED, Badenhorst CPS, Palm GJ, Berndt L, Lammers M, Mican J, Bednar D, Damborsky J, Bornscheuer UT (2021) Promiscuous dehalogenase activity of the epoxide hydrolase CorEH from Corynebacterium sp. C12. ACS Catal 11(10):6113–6120. https://doi.org/10.1021/acscatal.1c00851
Varga G, Docsa T, Gergely P, Juhász L, Somsák L (2013) Synthesis of tartaric acid analogues of FR258900 and their evaluation as glycogen phosphorylase inhibitors. Bioorg Med Chem Lett 23(6):1789–1792. https://doi.org/10.1016/j.bmcl.2013.01.042
Vasu V, Kumaresan J, Babu MG, Meenakshisundaram S (2012) Active site analysis of cis-epoxysuccinate hydrolase from Nocardia tartaricans using homology modeling and site-directed mutagenesis. Appl Microbiol Biot 93:2377–2386. https://doi.org/10.1007/s00253-011-3548-0
Wang Z, Wang Y, Su Z (2012) Purification and characterization of a cis-epoxysuccinic acid hydrolase from Nocardia tartaricans CAS-52, and expression in Escherichia coli. Appl Microbiol Biot 97:2433–2441. https://doi.org/10.1007/s00253-012-4102-4
Wang JB, Ilie A, Yuan S, Reetz MT (2017) Investigating substrate scope and enantioselectivity of a defluorinase by a stereochemical probe. J Am Chem Soc 139(32):11241–11247. https://doi.org/10.1021/jacs.7b06019
Xuan J, Feng Y (2019) Enantiomeric tartaric acid production using cis-Epoxysuccinate hydrolase: history and perspectives. Molecules 24(5):903–914. https://doi.org/10.3390/molecules24050903
Yamada T, Morisseau C, Maxwell JE, Argiriadi MA, Christianson DW, Hammock BD (2000) Biochemical evidence for the involvement of tyrosine in epoxide activation during the catalytic cycle of epoxide hydrolase. J Biol Chem 275(30):23082–23088. https://doi.org/10.1074/jbc.M001464200
Zhang Z (2015) Isolation and heterologous expression of trans-epoxysuccinate hydrolase. Dissertation, Zhejiang University
Zhang J, Xie Z, Zhang Z, Pan H, Bao W (2016) The method for preparing meso-tartaric acid or its salt by Pseudomonas koreensis. Chinese Patent 103923866A
Zhang H, Tian S, Yue Y, Li M, Tong W, Xu G, Chen B, Ma M, Li Y, Wang J (2020a) Semirational design of fluoroacetate dehalogenase RPA1163 for kinetic resolution of α-fluorocarboxylic acids on a gram scale. ACS Catal 10(5):3143–3151. https://doi.org/10.1021/acscatal.9b04804
Zhang L, De BC, Zhang W, Mandi A, Fang Z, Yang C, Zhu Y, Kurtan T, Zhang C (2020b) Mutation of an atypical oxirane oxyanion hole improves regioselectivity of the alpha/beta-fold epoxide hydrolase Alp1U. J Biol Chem 295(50):16987–16997. https://doi.org/10.1074/jbc.RA120.015563
Funding
This study was funded by the Huzhou Scientific and Technological Project (2022GZ56) and the Education of Zhejiang Province of China (Y202248484).
Author information
Authors and Affiliations
Contributions
WNB and HFP contributed to the study conception and design. Material preparation, data collection and analysis were performed by HXL, JFY and RLZ. The first draft of the manuscript was written by HXL and all authors commented on previous version of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
All authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with animals performed by any of the authors.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Liao, H., Pan, H., Yao, J. et al. Essential amino acid residues and catalytic mechanism of trans-epoxysuccinate hydrolase for production of meso-tartaric acid. Biotechnol Lett 46, 739–749 (2024). https://doi.org/10.1007/s10529-024-03490-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-024-03490-3