Abstract
Osteoarthritis (OA) is a chronic joint disease, which occurs in the elderly. The regulatory mechanisms of circRNAs were involved in the occurrence and development of various diseases. However, the potential regulatory network of circRNA in OA remains further research and clarification. The expression of circ_0114876 was increased in OA tissues and inhibition of circ_0114876 could induce cell viability and suppress inflammation as well as inhibit cell apoptosis in IL-1β induced CHON-001 cells. Circ_0114876 regulated TRAF2 expression via sponging miR-671 in CHON-001 cells. Down-regulated miR-671 expression could reverse the effects of low circ_0114876 expression on cell progression and inflammation in IL-1β induced CHON-001 cells. Overexpression of TRAF2 could weaken the promotion effects of high miR-671 expression on cell progression and inflammation in IL-1β induced CHON-001 cells. Circ_0114876 targeted miR-671 to regulate cell progression and inflammation via modulating TRAF2 expression in IL-1β induced CHON-001 cells, and played an important regulatory mechanism in IL-1β-induced chondrocyte injury, providing a novel diagnostics and therapeutics in OA.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Osteoarthritis (OA) is a common type of joint disease and one of the chronic diseases that are gradually developing and the elderly are the main groups of OA (Brandt 1988; Creamer and Hochberg 1997). At present, the pathogenesis of bones and joints is not clear, and the etiology is very complicated. Therefore, further exploration of the pathogenesis of OA is beneficial to the diagnosis of patients.
Circular RNAs (circRNAs) are a type of non-coding RNA that do not have a 3′ and 5′ UTR terminal tailing structure and looped noncoding RNAs, which directly bind to micro RNA (miRNA) through complementary base pairing, competitively inhibit mRNA expression, and thus exert their biological function (Rong et al. 2017; Chen and Yang 2015; Li et al. 2018, 2019a; Panda 2018). Research has shown that circRNAs could regulate the proliferation, differentiation and apoptosis in many diseases (Xie et al. 2016; Wang and Li 2018; Wei et al. 2019). For example, inhibition of circ_0004491 contributed to cell metastasis in oral squamous cell carcinoma (Li et al. 2019b). Moreover, differentially expressed circRNAs could be used as potential biomarkers to identify OA, for example, circ_0020014 and circ_0332131 (Wang et al. 2019a, b, 2020). The report of Zhou et al. showed that compared with the control group, 119 circRNAs were significantly up-regulated and 136 circRNAs were down-regulated in IL-1β-treated mouse models of OA (Zhou et al. 2018a), indicating that circRNA played an essential role in the pathogenesis and clinical treatment of OA.
More than that, miRNAs also are a type of regulation factors, which are related to the occurrence and development of diseases, such as OA and cancers (Li and Kowdley 2012; Kumar et al. 2013; Chen and Tian 2016; Li et al. 2016). In pancreatic cancer, abnormal miRNA expression could regulate the cell development of cancer cells, suggesting that miRNA could be used as a diagnostic marker and target for pancreatic cancer (Qadir and Faheem 2017). Kopańska et al. 2017 reported that miR-138-5p, miR-146a-5p, miR-335-5p and miR-9-5p were sharply increased in OA patients, implying that these miRNAs were associated with disease prevention and treatment (Kopanska et al. 2017). However, the role of circRNA as miRNA sponges in OA has not been fully elucidated.
To explore the regulatory mechanism of circRNAs in OA, we obtained OA tissues and constructed OA cell model to carry out experiments, and we found that circ_0114876 was upregulated in OA tissues and OA cell model. Furthermore, we also verified that circ_0114876 sponged miR-671-5p (miR-671) to modulate cell progression and inflammation in IL-1β induced CHON-001 cells.
Materials and methods
Patients and tissues
OA tissues were collected from 30 patients who had been diagnosed with OA and normal cartilage tissue was collected from 20 trauma patients at the First Affiliated Hospital of Jinan University. This experiment was approved by the ethics committee of the First Affiliated Hospital of Jinan University. Written informed consent has been obtained from patients. After all the samples were taken out and stored in − 80 ° C refrigerator for the next experiment.
Cell culture, OA model and cell transfection
Human chondrocyte cell line CHON-001 were obtained from American Type Culture Collection (Manassas, VA, USA) and were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen, Carlsbad, CA, USA) with 10% fetal bovine serum (FBS) and streptomycin at 37 °C. Then the IL-1β (10 ng/mL) was applied to simulate CHON-001 cells for 24 h to construct the OA model.
Si-circ_0114876 (si-circ), anti-miR-671, miR-671 mimics (miR-671), oe-TRAF2 and their negative control (si-NC, anti-NC, miR-NC and vector) were obtained from GenomediTech (Shanghai, China). All plasmids and oligonucleotides were transfected into CHON-001 cells using Lipofectamine 2000 (Invitrogen).
Quantitative Real-Time PCR (qRT-PCR) and RNase R treatment
TRIzol Reagen (Invitrogen) was used to separate total RNA in OA tissues and transfected cells. Total RNA was treated with RNase R (Epicentre Biotechnologies, Madison, WI, USA). The TaqMan microRNA assay kits (Applied Biosystems, Foster City, CA, USA) was applied to detect the synthesis of cDNA and qRT-PCR of miR-671 expression. The Transcriptor First Strand cDNA Synthesis Kit (Roche, Basel, Switzerland) was used to synthesize cDNA of circ_0114876 and TNF receptor-associated factor 2 (TRAF2). Then qRT-PCR was performed through using a SYBR Premix ExTaq kit (TaKaRa, Dalian, China) at ABI Prism 7500 (Applied Biosystems). The expression of miR-671 was normalized to U6 and the expression of circ_0114876 and TRAF2 were normalized to GAPDH.
Circ_0114876 Forward: 5′-ACCACTCCAGAAACTTTCCCT-3′; and reverse: 5′-AGCATTGTTGGCACTGACAC-3′;
miR-671 Forward: 5′-ACACTCCAGCTGGGAGGAAGCCCTGGAGGGG-3′, and reverse 5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCTCCAG-3′;
TRAF2 Forward: 5′-CCTACTGCTGAGCTCATTCT-3′; and reverse: 5′- CAATCTTGTCCTGGTCTAGC-3′;
GAPDH Forward: 5′-CATCATCCCTGCCTCTACTGG-3′, and reversed: 5′-GTGGGTGTCGCTGTTGAAGTC-3′;
U6 Forward: 5′-GCTTCGGCAGCACATATACTAAAAT-3′; and reverse: 5′-CGCTTCACGAATTTGCGTGTCAT-3′;
18S Forward: 5′-AACTTAAAGRAATTGACGGA-3′; and reverse: 5′-TCCGTCAATTYCTTTAAGTT-3′;
Actinomycin D assay and cytoplasmic and nuclear extracts
Transfected cells were treated with actinomycin D (2 µg/mL, Sigma Aldrich, St. Louis, MO, USA) at 0 h, 4 h, 8 h, 12 h and 24 h and RNA was extracted and qRT-PCR was used to detect the expression circ_0114876 and GAPDH.
Cytoplasmic and nuclear extracts were applied using PARIS™ kit (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions. U6 and 18S rRNA were employed as control of circ_0114876.
Dual-luciferase reporter assay
The sequences of circ_0114876 or TRAF2 (contained the binding sites of miR-671-5p (miR-671)) and their mutate sequences were amplified and inserted into a Firefly luciferase reporter vector pGL3 (Promega, Madison, WI, USA), named WT-circ_0114876, WT-TRAF2 3′UTR, MUT-circ_0114876 and MUT-TRAF2 3’UTR. Then the WT-circ_0114876, WT-TRAF2 3′UTR, MUT-circ_0114876 and MUT-TRAF2 3′UTR were cotransfected with miR-671 or miR-NC into IL-1β induced CHON-001 cells using Lipofectamine 2000 (Invitrogen). After transfection for 24 h, the luciferase activity was detected with Dual-Luciferase Reporter Assay System (Promega).
Cell apoptosis
Annexin V/PI apoptosis-detection kit (KeyGen BioTech, Shanghai, China) was used to test cell apoptosis. Briefly, cells were seeded into six-well plates and then were added with Annexin V-FITC and PI (10 µl) for 30 min in darkness. The apoptosis rate of transfected cells was detected using flow cytometry (BD Bioscience, Franklin Lake, NJ, USA).
Enzyme-linked immunosorbent assay (ELISA)
The content of IL-6 and IL-8 in each group was measured by Human IL-6 ELISA Kit and Human IL-8 ELISA Kit (Abcam, Cambridge, MA, USA) according to the manufacturer’s instruction. Each experiment was tested in triplicate.
Western blot
Total proteins from cells and tissues were extracted using RIPA buffer. Next, the protein quantification was performed with the BCA Protein Assay Kit (Beyotime, Shanghai, China). The protein was added onto the SDS-PAGE and then transferred onto PVDF membranes (Millipore, Billerica, MA, USA). After blocked with non-fat milk, the membranes were incubated with primary antibodies anti-TRAF2, anti-aggrecan, anti-Matrix Metallopeptidase 13 (MMP13), anti-collagen II and anti-GAPDH (Abcam, 1:1,000) overnight at 4 °C. Next, the membranes were incubated with secondary antibodies horseradish peroxidase (HRP)-conjugated goat-anti-rabbit Immunoglobulin G (IgG) for 1 h. The blot was measured and analyzed with ImageJ software 1.48 (National Institutes of Health, Bethesda, MD, USA).
Cell Counting Kit-8 (CCK-8) assay
Transfected cells were seeded into the 96-well plates at a density of 2000 cells per well. CCK-8 reagent (Beyotime) was used to detect cell proliferation. Briefly, CCK-8 solution (20 µl) was added each well and incubated for 4 h and then the absorbance of samples was detected using a microplate reader (BioRad, Hercules, CA, USA) at a wavelength of 450 nm.
Statistical analysis
These data were performed and analyzed with GraphPad Prism software (La Jolla, CA, USA). Data were repeated in triplicate and were presented as mean ± standard deviation (SD). Student’s t-test and one-way analysis of variance were used to calculate the statistical significance of groups. Pearson’s correlation analysis was applied to determine the relationship among circ_0114876, miR-671 and TRAF2. P < 0.05 was represented as statistically significant.
Results
Circ_0114876 expression was upregulated in OA patients and IL-1β induced CHON-001 cells
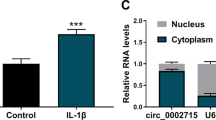
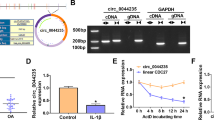
We found that circ_0114876 from the PTPRA gene from the sixth to tenth exon region, the total length of 570nt (Fig. 1a). Also, we found that circ_0114876 significantly higher in OA tissues than in normal tissues (Fig. 1b). Meanwhile, after we treated CHON-001 cells 48 h with IL-1β, the expression of circ_0114876 increased significantly, indicating that IL-1β induced circ_0114876 expression in CHON-001 cells (Fig. 1c). Besides, qRT-PCR detected Actinomycin D treating CHON-001 cells, and the results showed that the expression of circ_0114876 was significantly higher than the expression of GAPDH at 4 h, 8 h, 12 h, 24 h, indicating that the stability of the circ_0114876 was so strong (Fig. 1d). Besides, the results of RNase R enzymeexperiment showed that circ_0114876 resistance to RNase R was beneficial to its stability (Fig. 1e). Cytoplasmic and nuclear extracts experiments determined that circ_0114876 was mainly concentrated in cytoplasm, suggesting that its mechanism of action was carried out in cytoplasm (Fig. 1f). In summary, circ_0114876 might play an important role inIL-1β induced CHON-001 cells and is related to the occurrence of OA.
Circ_0114876 expression was upregulated in OA patients and IL-1β induced CHON-001 cells. a The genomic loci of PTPRA gene and circ_0114876. b qRT-PCR analysis showed that the expression of circ_0114876 was increased in OA tissues. c qRT-PCR analysis showed that the expression of circ_0114876 was induced in chondrocyte treated with IL-1β. d The expression of circ_0114876 was detected in chondrocyte treated with Actinomycin D at 0, 4, 8, 12, 24 h. e qRT-PCR analysis of circ_0114876 after treatment with RNase R in chondrocyte. f Localization of circ_0114876 expression in chondrocytes. *P < 0.05
Circ_0114876 directly interacted with miR-671
Next, we found miR-671 expression was significantly decreased (Fig. 2a). Interestingly, we also found, by using Pearson’s association analysis,that miR-671 expression in OA tissues was negatively correlated with the expression of circ_0114876 (Fig. 2b). Otherwise, IL-1βinhibited the expression of miR-671 in CHON-001 cells (Fig. 2c). Besides, circular RNA Interactome predicted that circ_0114876 had binding sites with miR-671, indicating that miR-671 was a potential target miRNA of circ_0114876 (Fig. 2d). Dual-luciferase reporter assay showed the luciferase activity was significantly decreased when WT-circ_0114876 combined with the miR-671 mimic (miR-671). However, luciferase activity of MUT-circ_0114876 had no effects (Fig. 2e). The results of RIP assay determined that circ_0114876 directly bound the miR-671 in CHON-001 cells (Fig. 2f). Moreover, si-circ transfection decreased circ_0114876 expression and induced miR-671 expression in CHON-001 cells (Fig. 2g, h). Therefore, these data indicated that circ_0114876 could directly target miR-671 in CHON-001 cells.
Circ_0114876 directly interacted with miR-671. a qRT-PCR analysis determined that miR-671 expression was inhibited in OA tissues. b Pearson’s correlation analysis determined the relationship between circ_0114876 and miR-671 in OA tissues. c qRT-PCR analysis showed that the expression of miR-671 was reduced in chondrocyte treated with IL-1β. d Circular RNA Interactome predicted that circ_0114876 had a targeted binding site with miR-671. e Dual-luciferase reporter assay was used to detect the luciferase activities in chondrocytes cotransfected WT-circ_0114876 or MUT-circ_0114876 with miR-NC or miR-671. f RIP assay was applied to determine that circ_0114846 directly bound miR-671 in chondrocytes. g, h qRT-PCR was used to measure the expression of circ_0114876 and miR-671 in si-NC and si-circ groups of chondrocytes. *P < 0.05
Knockdown of circ_0114876 inhibited IL-1β-induced chondrocyte injury via regulating miR-671
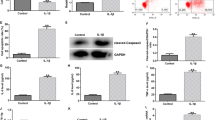
To further study the functions of miR-671 and circ_0114876 in IL-1β-induced chondrocyte injury, we transfected anti-NC and anti-miR-671 to IL-1β induced CHON-001 cells and obtained stablyanti-miR-673 cell lines with low expression of miR-671 (Fig. 3a). The results of CCK-8 assay showed that IL-1β inhibited the proliferation of CHON-001 cells, and inhibition of circ_0114876 increased the cell viability of IL-1β induced CHON-001 cells while reducing the expression of miR-671 could reverse the low expression of circ_0114876 on IL-1β induced CHON-001 cells (Fig. 3b). IL-1β could induce apoptosis of CHON-001 cells by using flow cytometry. Moreover, inhibiting circ_0114876 expression reduced the apoptosis of IL-1β induced CHON-001 cells, which was reversed by inhibition of miR-671 (Fig. 3c, d). ELISA assay detected the levels of inflammatory factors IL-8 and IL-6, and the results showed that IL-1β increased levels of IL-8 and IL-6. Decreasing the expression of miR-671 could reverse the suppression effect of si-circ transfection on IL-8 and IL-6 levels in IL-1β induced CHON-001 cells (Fig. 3e, f). Si-circ transfection induced the protein expression of collagen II and aggrecan in IL-1β induced CHON-001 cells and decreased the protein expression of MMP13, which were impaired through suppression of miR-671 in western blot (Fig. 3g, h). Therefore, circ_0114876 regulated cell progression and inflammation via regulating miR-671 in IL-1β induced CHON-001 cells.
Knockdown of circ_0114876 inhibited IL-1β-induced chondrocyte injury via regulating miR-671. a The expression of miR-672 was detected in chondrocytes transfected with anti-NC and anti-miR-671. Cell viability (b), cell apoptosis (c, d), the level of IL-6 and IL-8 (E and F), and the protein expression of MMP13, collagen II, and aggrecan (g, h) were detected in Control, IL-1β, IL-1β + si-NC, IL-1β + si-circ, IL-1β + si-circ + anti-NC and IL-1β + si-circ + anti-miR-671 groups in chondrocyte. *P < 0.05
TRAF2 was a target mRNA of miR-671
Subsequently, we found that the mRNA and protein of TRAF2 were notably enhanced in OA tissues (Fig. 4a, b). Furthermore, TRAF2 expression was significantly negatively correlated with miR-671 expression in OA tissues (Fig. 4c). In addition, IL-1β significantly induced the mRNA and protein expression of TRAF2 in CHON-001 cells (Fig. 4d, e). We used starBase to predict the target mRNA of TRAF and found that TRAF2 has a targeted binding site to miR-671 (Fig. 4f). And dual-luciferase reporter assay determined that luciferase activity was significantly decreased in CHON-001 cells cotransfected miR-671 with WT-TRAF2 3’UTR, but luciferase activity was no changes in CHON-001 cells cotransfected miR-671 with MUT-TRAF2 3’UTR (Fig. 4g). Furthermore, the mRNA and protein expression of TRAF2 was inhibited by miR-671 transfection but also induced by anti-miR-671 transfection (Fig. 4h, i). Therefore, these results indicated that TRAF2 was a target gene of miR-671 in CHON-001 cells.
TRAF2 was a target mRNA of miR-671. a, b qRT-PCR and western blot analysis showed that the expression of TRAF2 was increased in OA tissues. c Pearson’s correlation analysis determined the relationship between miR-671 and TRAF2 in OA tissues. d, e qRT-PCR and western blot analysis showed that the expression of TRAF2 was induced in chondrocyte treated with IL-1β. f StarBase predicted that miR-671 had a targeted binding site with TRAF2. g Dual-luciferase reporter assay was used to detect the luciferase activities in chondrocytes cotransfected WT-TRAF2 3’UTR or MUT-TRAF2 3’UTR with miR-NC or miR-671. h, i qRT-PCR and western blot were used to measure the mRNA and protein expression of TRAF2 in miR-NC, miR-671, anti-NC and anti-miR-671 of chondrocytes. *P < 0.05
The effect of miR-671 and TRAF2 on IL-1β-induced chondrocyte injury
To verify the mechanism of TRAT2 and miR-671 in OA, we obtained a cell line that overexpressed TRAF2. As shown in Fig. 5a, b, compared with vector group, TART2 mRNA and protein expression in oe-TRAF2 group were significantly increased. More than that, we found that high expression of miR-671 increased cell proliferation, reduced apoptosis and decreased the levels of IL-8 and IL-6 in IL-1β induced CHON-001 cells (Fig. 5c, f). Meanwhile, collagen II and aggrecan protein expression were induced, while MMP13 protein expression was suppressed via overexpression of miR-671 in IL-1β induced CHON-001 cells (Fig. 5g, h). Importantly, promotion of TRAF2 could attenuate the effect of overexpressing miR-671 on cell progression of IL-1β induced CHON-001 cells (Fig. 5c, h). Therefore, miR-671 affected cell progression and inflammation by regulating TRAF2 in IL-1β induced CHON-001 cells.
The effect of miR-671 and TRAF2 on IL-1β-induced chondrocyte injury. a, b The mRNA and protein expression of TRAF2 was detected in chondrocytes transfected with vector and oe-TRAF2. Cell viability (c), cell apoptosis (d), the level of IL-6 and IL-8 (e, f), and the protein expression of MMP13, collagen II, and aggrecan (g, h) were detected in Control, IL-1β, IL-1β + miR-NC, IL-1β + miR-671, IL-1β + miR-671 + vector and IL-1β + miR-671 + oe-TRAF2 groups in chondrocyte. *P < 0.05
Circ_0114876 targeted miR-671 to regulate expression of TRAF2
Finally, we found that si-circ transfection reduced the mRNA and protein expression of TRAF2, which was impaired by decreasing miR-671 expression (Fig. 6a, b). Furthermore, the expression of TRAF2 was significantly positively correlated with the expression of circ_0114786 in OA (Fig. 6c). Thus, circ_0114876 could modulate TRAF2 through sponging miR-671 in OA.
Circ_0114876 targeted miR-671 to regulate expression of TRAF2. a, b The mRNA and protein expression of TRAF2 was detected in chondrocytes transfected with si-NC, si-circ, si-circ + anti-NC and si-circ + anti-miR-671. c Pearson’s correlation analysis determined the relationship between circ_0114876 and TRAF2 in OA tissues. *P < 0.05
Discussion
Increasing evidence suggested that circRNAs played a significant role in disease development (Kong et al. 2016; Lei et al. 2019). Besides, circRNAs were closely related to the physiological and biochemical development of OA, including cell proliferation, apoptosis, invasion, and inflammatory response (Yu and Sun 2018; Zhou et al. 2018a, b, 2019; Shen et al. 2019). For example, the expression of circ_33186 in OA tissue was significantly increased, and the knockdown of circ_33186 promoted the proliferation of IL-1β-treated chondrocytes and inhibited their apoptosis, aggravating OA in mice (Zhou et al. 2019). Moreover, circRNA could affect the occurrence and development of OA by regulating cellular processes such as inflammatory response, basal metabolism, apoptosis and differentiation of cartilage tissue (Liu et al. 2017). RNA-seq sequencing results of Li et al (2019c). showed that circ_0114876 was highly expressed in OA tissues, but its function has not been clear in OA. In this study, we demonstrated the function of circ_0114876 in OA tissues and OA model. Moreover, t the expression of circ_0114876 was significantly increased in OA tissues and IL-1β induced CHON-001 cells. Knockdown of circ_0114876 could enhance cell viability and reduced cell inflammation and apoptosis in IL-1β induced CHON-001 cells. Thus, circ_0114876 was a key factors during the OA progression and affected the cytological physiological responses of OA cells.
It is well known that circRNA could be used as a competitive endogenous RNA in combination with miRNA to regulate the expression of target mRNA (Guan et al. 2019; Kristensen et al. 2019). In this paper, we found and demonstrated that circ_0114876 regulated TRAF2 expression by binding to miR-671. A previous study showed that miR-140-3p, miR-33b-3p and miR-671-3p, acted as biomarkers, were related to metabolic progression of OA. But, miR-671-5p (miR-671) has not been reported in OA progression. Accumulating evidence determined that miR-671 expressed low in gastric cancer, breast cancer, osteosarcoma and OA, and was involved in cell proliferation, apoptosis, cell cycle and radiochemoresistance in cancers (Qiu et al. 2018; Tan et al. 2019; Xin et al. 2019). Our investigation showed that miR-671 was down-regulated in OA tissues and OA cell model and may be a inhibitor in the occurrence of OA. And we found that overexpression of miR-671 promoted cell progression and inhibited inflammation in IL-1β induced CHON-001 cells. Moreover, inhibition of miR-671 could reverse the effects of low circ_0114876 expression on cell progression and inflammation in IL-1β induced CHON-001 cells. These data indicated that circ_0114876 affect OA cell progression and inflammation via sponging miR-671, and that miR-671 play an important role in the treatment of OA as a key biomarker.
TRAF2 is a member of the TNF receptor associated factor protein family and encodes TRAF2 gene at chromosome 9 (Siemienski et al. 1997; Borghi et al. 2016). Various reports have shown that TRAF2 was related to the process of tumor formation and development, which was highly expressed in prostate cancer and breast cancer (Wei et al. 2017; Peramuhendige et al. 2018). Jiang et al. and Zhang et al. reported that TRAF2 expression was increased in OA tissues and IL-1β induced chondrocyte, and affected cell apoptosis, cell proliferation and inflammation in OA (Zhang et al. 2016; Jiang et al. 2018). Interestingly, TRAF2 was highly expressed in OA and IL-1β induced CHON-001 cells consistent with previous study (Zhang et al. 2016; Jiang et al. 2018). More than that, Our research determined that TRAF2 was a target mRNA of miR-671 and overexpression of TRAF2 could weaken the promotion effects of high miR-671 expression on cell progression in IL-1β induced CHON-001 cells. Therefore, circ_0114876 affect TRAF2 to regulate cell progression and inflammation via sponging miR-671, which implying that this mechanism was an indispensable pathway in OA process. However, TARF2 could interact with other factors, including miRNA, and circRNA in OA. Additionally, whether circ_0114876 is related to drug sensitivity of OA is unknown. In this study, we confirmed the role of circ_0114876 in the OA cell process and inflammation, and circ_0114876, as an important biomarker, was related to treatment of OA still need further explored. These explorations were of great significance and can provide basic biological mechanism for the accurate treatment of OA. In summary, this article mainly demonstrated circ_0114876 targeted miR-671 to regulate cell progression and inflammation via modulating TRAF2 expression in IL-1β induced CHON-001 cells, and circ_0114876 promoted IL-1β-induced chondrocyte injury via miR-671/TRAF2 axis, improving the understanding of regulatory mechanism of OA and providing a new therapeutics for treatment of OA.
References
Borghi A, Verstrepen L, Beyaert R (2016) TRAF2 multitasking in TNF receptor-induced signaling to NF-κB, MAP kinases and cell death. Biochem Pharmacol 116:1–10. https://doi.org/10.1016/j.bcp.2016.03.009
Brandt KD (1988) Osteoarthritis. Clin Geriatr Med 4:279–293
Chen H, Tian Y (2016) MiR-15a-5p regulates viability and matrix degradation of human osteoarthritis chondrocytes via targeting VEGFA. Biosci Trends 10:482–488. https://doi.org/10.5582/bst.2016.01187
Chen LL, Yang L (2015) Regulation of circRNA biogenesis. RNA Biol 12:381–388. https://doi.org/10.1080/15476286.2015.1020271
Creamer P, Hochberg MC (1997) Osteoarthritis. Lancet 350:503–508. https://doi.org/10.1016/s0140-6736(97)07226-7
Guan YJ, Ma JY, International WSJCC (2019) Identification of circRNA–miRNA–mRNA regulatory network in gastric cancer by analysis of microarray data. Cancer Cell Int 19:183. https://doi.org/10.1186/s12935-019-0905-z
Jiang J, Zhang J, Wu C, Guo X, Chen C, Bao G et al (2018) Up-regulation of TRAF2 inhibits chondrocytes apoptosis in lumbar facet joint osteoarthritis. Biochem Biophys Res Commun 503:1659–1665. https://doi.org/10.1016/j.bbrc.2018.07.096
Kong DP, Xiao-Yuan ZI, Sun YH, Urology DO, Hospital C (2016) circRNA:research progress and clinical applications. Acad J Second Mil Med Univ. https://doi.org/10.16781/j.0258-879x.2016.03.0330
Kopanska M, Szala D, Czech J, Gablo N, Gargasz K, Trzeciak M et al (2017) MiRNA expression in the cartilage of patients with osteoarthritis. J Orthop Surg Res 12:51. https://doi.org/10.1186/s13018-017-0542-y
Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J (2019) The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet 20:675–691. https://doi.org/10.1038/s41576-019-0158-7
Kumar S, Keerthana R, Pazhanimuthu A, Perumal P (2013) Overexpression of circulating miRNA-21 and miRNA-146a in plasma samples of breast cancer patients. Indian J of Biochem Biophys 50:210–214
Lei B, Xuan XY, Biologicals W-PFJCJo (2019) Progress in research on role of CircRNA in autoimmune diseases. Chin J Biol 32:347–350
Li Y, Kowdley KV (2012) MicroRNAs in common human diseases. Genom Proteom Bioinform 10:246–253. https://doi.org/10.1016/j.gpb.2012.07.005
Li XR, Chu HJ, Lv T, Wang L, Kong SF, Dai SZ (2016) miR-342-3p suppresses proliferation, migration and invasion by targeting FOXM1 in human cervical cancer. FEBS Lett 588:3298–3307. https://doi.org/10.1016/j.febslet.2014.07.020
Li X, Yang L, Chen LL (2018) The biogenesis, functions, and challenges of circular RNAs. Mol Cell 71:428–442. https://doi.org/10.1016/j.molcel.2018.06.034
Li M, Duan L, Li Y, Liu B (2019a) Long noncoding RNA/circular noncoding RNA-miRNA-mRNA axes in cardiovascular diseases. Life Sci 233:116440. https://doi.org/10.1016/j.lfs.2019.04.066
Li X, Zhang H, Wang Y, Sun S, Shen Y, Yang H (2019b) Silencing circular RNA hsa_circ_0004491 promotes metastasis of oral squamous cell carcinoma. Life Sci 239:116883. https://doi.org/10.1016/j.lfs.2019.116883
Li Z, Yuan B, Pei Z, Zhang K, Ding Z, Zhu S et al (2019c) Circ_0136474 and MMP-13 suppressed cell proliferation by competitive binding to miR-127-5p in osteoarthritis. J Cell Mol Med 23:6554–6564. https://doi.org/10.1111/jcmm.14400
Liu Q, Zhang X, Hu X, Yuan L, Cheng J, Jiang Y et al (2017) Emerging roles of circRNA related to the mechanical stress in human cartilage degradation of osteoarthritis. Mol Ther Nucleic Acids 7:223–230. https://doi.org/10.1016/j.omtn.2017.04.004
Panda AC (2018) Circular RNAs act as miRNA sponges. Adv Exp Med Biol 1087:67–79. https://doi.org/10.1007/978-981-13-1426-1_6
Peramuhendige P, Marino S, Bishop RT, de Ridder D, Khogeer A, Baldini I et al (2018) TRAF2 in osteotropic breast cancer cells enhances skeletal tumour growth and promotes osteolysis. Sci Rep 8:39. https://doi.org/10.1038/s41598-017-18327-5
Qadir MI, Faheem A (2017) miRNA: a diagnostic and therapeutic tool for pancreatic cancer. Crit Rev Eukaryot Gene Expr 27:197–204. https://doi.org/10.1615/CritRevEukaryotGeneExpr.2017019494
Qiu T, Wang K, Li X, Jin J (2018) miR-671-5p inhibits gastric cancer cell proliferation and promotes cell apoptosis by targeting URGCP. Exp Ther Med 16:4753–4758. https://doi.org/10.3892/etm.2018.6813
Rong D, Sun H, Li Z, Liu S, Dong C (2017) An emerging function of circRNA-miRNAs-mRNA axis in human diseases. Oncotarget 8:73271–73281. https://doi.org/10.18632/oncotarget.19154
Shen S, Wu Y, Chen J, Xie Z, Huang K, Wang G et al (2019) CircSERPINE2 protects against osteoarthritis by targeting miR-1271 and ETS-related gene. Ann Rheum Dis 78:826–836. https://doi.org/10.1136/annrheumdis-2018-214786
Siemienski K, Peters N, Scheurich P, Wajant H (1997) Organization of the human tumour necrosis factor receptor-associated factor 1 (TRAF1) gene and mapping to chromosome 9q33-34. Gene 195:35–39. https://doi.org/10.1016/s0378-1119(97)00147-9
Tan X, Li Z, Ren S, Rezaei K, Pan Q, Goldstein AT et al (2019) Dynamically decreased miR-671-5p expression is associated with oncogenic transformation and radiochemoresistance in breast cancer. Breast Cancer Res 21:89. https://doi.org/10.1186/s13058-019-1173-5
Wang J, Li H (2018) CircRNA circ_0067934 silencing inhibits the proliferation, migration and invasion of NSCLC cells and correlates with unfavorable prognosis in NSCLC. Eur Rev Med Pharmacol Sci 22:3053–3060. https://doi.org/10.26355/eurrev_201805_15063
Wang T, Wang X, Du Q, Wu N, Liu X, Chen Y et al (2019) The circRNA circP4HB promotes NSCLC aggressiveness and metastasis by sponging miR-133a-5p. Biochem Biophys Res Commun 513:904–911. https://doi.org/10.1016/j.bbrc.2019.04.108
Wang Y, Wu C, Yang Y, Ren Z, Lammi M, Guo X (2019) Preliminary exploration of hsa_circ_0032131 levels in peripheral blood as a potential diagnostic biomarker of osteoarthritis. Genet Test Mol Biomark 23:717–721. https://doi.org/10.1089/gtmb.2019.0036
Wang Y, Wu C, Zhang Y, Yang Y, Ren Z, Lammi M et al (2020) Screening for differentially expressed circRNA between Kashin-Beck disease and osteoarthritis patients based on circRNA chips. Clin Chim Acta 501:92–101. https://doi.org/10.1016/j.cca.2019.10.026
Wei B, Liang J, Hu J, Mi Y, Ruan J, Zhang J et al (2017) TRAF2 is a valuable prognostic biomarker in patients with prostate cancer. Med Sci Monit 23:4192–4204. https://doi.org/10.12659/msm.903500
Wei S, Zheng Y, Jiang Y, Li X, Geng J, Shen Y et al (2019) The circRNA circPTPRA suppresses epithelial-mesenchymal transitioning and metastasis of NSCLC cells by sponging miR-96-5p. EBioMedicine 44:182–193. https://doi.org/10.1016/j.ebiom.2019.05.032
Xie H, Ren X, Xin S, Lan X, Lu G, Lin Y et al (2016) Emerging roles of circRNA_001569 targeting miR-145 in the proliferation and invasion of colorectal cancer. Oncotarget 7:26680–26691. https://doi.org/10.18632/oncotarget.8589
Xin C, Lu S, Li Y, Zhang Y, Tian J, Zhang S et al (2019) miR-671-5p inhibits tumor proliferation by blocking cell cycle in osteosarcoma. DNA Cell Biol 38:996–1004. https://doi.org/10.1089/dna.2019.4870
Yu CX, Sun S (2018) An emerging role for circular RNAs in osteoarthritis. Yonsei Med J 59:349–355. https://doi.org/10.3349/ymj.2018.59.3.349
Zhang G, Sun Y, Wang Y, Liu R, Bao Y, Li Q (2016) MiR-502-5p inhibits IL-1β-induced chondrocyte injury by targeting TRAF2. Cell Immunol 302:50–57. https://doi.org/10.1016/j.cellimm.2016.01.007
Zhou Z, Du D, Chen A, Zhu L (2018a) Circular RNA expression profile of articular chondrocytes in an IL-1β-induced mouse model of osteoarthritis. Gene 644:20–26. https://doi.org/10.1016/j.gene.2017.12.020
Zhou ZB, Du D, Huang GX, Chen A, Zhu L (2018b) Circular RNA Atp9b, a competing endogenous RNA, regulates the progression of osteoarthritis by targeting miR-138-5p. Gene 646:203–209. https://doi.org/10.1016/j.gene.2017.12.064
Zhou ZB, Huang GX, Fu Q, Han B, Lu JJ, Chen AM et al (2019) circRNA.33186 contributes to the pathogenesis of osteoarthritis by sponging miR-127-5p. Mol Ther 27:531–541. https://doi.org/10.1016/j.ymthe.2019.01.006
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, Q., Luo, S., Yang, J. et al. Circ_0114876 promoted IL-1β-induced chondrocyte injury by targeting miR-671/TRAF2 axis. Biotechnol Lett 43, 791–802 (2021). https://doi.org/10.1007/s10529-020-03070-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-020-03070-1