Abstract
Objectives
To find a simple enzymatic strategy for the efficient synthesis of the expensive 5′-hydroxyomeprazole sulfide, a recently identified minor human metabolite, from omeprazole sulfide, which is an inexpensive substrate.
Results
The practical synthetic strategy for the 5′-OH omeprazole sulfide was accomplished with a set of highly active CYP102A1 mutants, which were obtained by blue colony screening from CYP102A1 libraries with a high conversion yield. The mutant and even the wild-type enzyme of CYP102A1 catalyzed the high regioselective (98 %) C-H hydroxylation of omeprazole sulfide to 5′-OH omeprazole sulfide with a high conversion yield (85–90 %).
Conclusions
A highly efficient synthesis of 5′-OH omeprazole sulfide was developed using CYP102A1 from Bacillus megaterium as a biocatalyst.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
During the process of drug development, all significant metabolites should be characterized to evaluate drug efficacy and toxicity (Obach 2013). Although some drug metabolites of concern are prepared by chemical methods, other metabolites can be prepared using cytochrome P450 enzymes, including human P450s (Yun et al. 2006). Some bacterial P450s, particularly CYP102A1 from Bacillus megaterium, are competitive biocatalysts for metabolite production due to their high activities and stabilities (Yun et al. 2007; Whitehouse et al. 2012).
Omeprazole, a proton pump inhibitor, has been widely used as an acid inhibitor to treat gastric acid hypersecretion disorders (Wedemeyer and Blume 2014; Saccar 2009). Omeprazole is primarily metabolized in human livers by CYP2C19 and CYP3A4. While CYP2C19 favors the C-5′ hydroxylation of R-omeprazole, CYP3A4 mainly mediates sulfoxidation of the S-form (Li et al. 2005). Some other minor metabolites have also been identified (Supplementary Fig. 1). A set of CYP102A1 mutants can catalyze the regioselective C-5′ hydroxylation of the S- and R-omeprazole (Ryu et al. 2014; Butler et al. 2013, 2014). Omeprazole sulfide is a major human metabolite of omeprazole (Rezk et al. 2006) but its metabolic fate has not yet been reported. A minor metabolite of omeprazole (5′-OH omeprazole sulfide) was identified in human urine (Nevado et al. 2014). However, a systematic approach has not been performed to study the drug efficacy and toxicity of omeprazole metabolites, such as 5′-OH omeprazole and 5′-OH omeprazole sulfide.
The aim of this study was to find a simple enzymatic strategy for the efficient synthesis of the expensive 5′-OH omeprazole sulfide from omeprazole sulfide, which is an inexpensive substrate. Here we found that CYP102A1 can catalyze the regioselective C-H hydroxylation of omeprazole sulfide to 5′-OH omeprazole sulfide with a high yield (Fig. 1).
Materials and methods
Materials
Omeprazole (racemic mixture), 5′-hydroxyomeprazole (5′-OH omeprazole), 5′-hydroxyomeprazole sulfide (5′-OH omeprazole sulfide) and NADPH were purchased from Sigma-Aldrich. Omeprazole sulfide was obtained from Santa Cruz Biotechnology (Dallas, Texas). All other chemicals and supplies used were from standard sources.
Hydroxylation of omeprazole sulfide
The initial investigations to determine the catalytic activity of the CYP102A1 mutants in the hydroxylation of omeprazole sulfide were performed using HPLC (see Ryu et al. 2014). The reconstituted enzyme system contained 0.20 μM CYP102A1, an NADPH regeneration system (10 mM glucose 6-phosphate, 0.50 mM NADP+, and 1 IU yeast glucose-6-phosphate dehydrogenase/ml), and 1 mM omeprazole sulfide in 0.25 ml of potassium phosphate buffer (0.1 M, pH 7.4). Reaction mixtures were incubated for 10 min at 37 °C. Mutated amino acid residues of the CYP102A1 mutants used in this study are shown at the Supplementary Table 1.
The steady-state kinetics (k cat and K M) of the WT and mutant enzymes were determined in 0.25 ml potassium phosphate buffer (0.1 M, pH 7.4). The reaction mixtures contained 0.04 μM CYP102A1, an NADPH regeneration system, and omeprazole sulfide (5–1000 μM). The samples were incubated at 37 °C for 5 min. A stock solution of omeprazole sulfide (100 mM) was prepared in dimethyl sulfoxide and diluted in the enzymatic reactions to a final organic solvent concentration of <1 % (v/v). Product formation was analyzed by HPLC and quantified by comparing their concentrations to those of standard compounds. The samples (20 μl) were injected onto a Gemini C18 column (4.6 × 150 mm, 5 μm; Phenomenex, Torrance, CA) with an acetonitrile/5 mM potassium phosphate buffer (pH 7.3) (40:60, v/v) as mobile phase. Eluates were detected at 302 nm. The kinetic parameters (k cat and K M) were calculated using Michaelis–Menten nonlinear regression analysis with GraphPad Prism (GraphPad Software, San Diego, CA).
To determine the total turnover numbers (TTNs) (mol product/mol catalyst) of each CYP102A1 mutant, the reaction mixture contained 0.20 μM mutant enzyme, an NADPH-generating system, and 10 mM omeprazole sulfide in 0.25 ml potassium phosphate buffer (0.10 M, pH 7.4). The reaction was initiated by the addition of the NADPH-generating system and incubated at 37 °C for 3 h. The formation rate of the 5′-OH omeprazole sulfide was determined by HPLC as described above.
NADPH oxidation
The reaction was performed in a spectrophotometric cuvette maintained at 25 °C. The reaction mixture contained 0.1 μM CYP102A1 and 1 mM omeprazole sulfide in 1 ml of potassium phosphate buffer (0.1 M, pH 7.4). The reactions were initiated by the addition of 10 μl 10 mM NADPH (final concentration, 100 μM), and the decrease in A 340 was monitored for 1 min. The rates of NADPH oxidation were calculated using ε340 = 6.22 M−1c cm−1 for NADPH.
Thermal stabilities of the CYP102A1 mutants
To estimate the enzyme thermal stabilities of the CYP102A1 mutants, WT CYP102A1 and its mutants (#10, #375, #387, and #389) were incubated between 30 and 70 °C for 5 min followed by cooling to 4 °C. After centrifugation, the protein solutions were mixed with sodium hydrosulfite in a microtiter plate and incubated for 2 min with carbon monoxide in a sealed chamber. The enzyme solution contained 1 μM purified CYP102A1 in 100 mM potassium phosphate (pH 7.4). The temperature for 50 % inactivation (T 50) of the entire enzyme was determined from the differences in the CO-binding difference spectra before and after heat treatment. The thermal stability (T 50 , °C) was estimated using GraphPad Prism software (GraphPad Software, San Diego, CA).
Results and discussion
Hydroxylation of omeprazole sulfide by CYP102A1
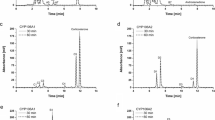
To determine whether CYP102A1 can hydroxylate omeprazole sulfide, the rates at which the wild-type (WT) CYP102A1, chimera M16V2, and its 50 mutants formed the omeprazole sulfide metabolites were examined in a preliminary test. The WT enzyme, M16V2, and the mutants produced one major metabolite (5′-OH omeprazole sulfide) (Fig. 2). Only eleven of the mutants showed higher activity than M16V2, which was used as a template to produce the CYP102A1 libraries. Mutants #328, #375, #387, and #389 exhibited 2.9- to 4.7-fold increases in omeprazole sulfide C-5′ hydroxylation activity compared with M16V2. The omeprazole sulfide C-5′ hydroxylation activity of mutant #387 was 13-fold higher than that of WT. Interestingly, mutant #10 also showed high catalytic activity that was comparable to that of #387. Although the activity of mutant #10 was comparable to that of #387, these two mutants have quite different mutations. Mutant #10 has mutations in the substrate channel and active site (van Vugt-Lussenburg et al. 2006). However, mutant #387 includes three mutations (F11L/Q110P/R190Q) outside the active site and substrate channel and an additional 20 mutations in the reductase domain. It showed a 4.7-fold increase in omeprazole sulfide C-5′ hydroxylation activity compared with M16V2. Finally, mutant #387 has activity comparable to that of mutant #10. The identities of the major metabolites and substrate were confirmed by comparing the HPLC (Fig. 3), LC-MS (Fig. 4), and NMR results (Fig. 5; Supplementary Fig. 3; Supplementary Table 2) to those of standard compounds. Although two minor metabolites were observed from the HPLC trace and their retention times matched 5′-OH omeprazole and omeprazole, their chemical structures were not identified because of their low abundance (Fig. 3c).
Formation rate of the omeprazole sulfide metabolite catalyzed by the CYP102A1 mutants. The formation rates of the products were determined by HPLC. The values are presented as the mean ± S.E. of duplicate measurements. The reactions contained P450 (0.2 μM), an NADPH regeneration system (10 mM glucose 6-phosphate, 0.5 mM NADP+, and 1 IU yeast glucose-6-phosphate dehydrogenase/ml) and omeprazole sulfide (1 mM) in 0.25 ml potassium phosphate buffer (0.1 M, pH 7.4). The reaction mixtures were incubated at 37 °C for 10 min
HPLC chromatograms of the omeprazole sulfide metabolites produced by WT CYP102A1 (b) and mutants (c). The peaks were identified by comparing their retention times with those of the following standards (a): authentic 5′-OH omeprazole (t R = 2.23 min), 5′-OH omeprazole sulfide (t R = 3.33 min), omeprazole (t R = 3.79 min) and omeprazole sulfide (t R = 8.87 min). The reactions contained P450 (0.2 μM), an NADPH regeneration system and omeprazole sulfide (1 mM) in a final volume of 0.25 ml of potassium phosphate buffer (0.1 M, pH 7.4). The reaction mixtures were incubated at 37 °C for 10 min
LC-MS analyses of the product derived from the hydroxylation of omeprazole sulfide by the CYP102A1 mutant. Extracted ion chromatograms were contracted from the incubation of omeprazole sulfide with CYP102A1 mutant #10 in the absence (a) and presence (b) of NADPH and the peaks of omeprazole sulfide and 5′-OH omeprazole sulfide are shown at 7.3 and 6.1 min, respectively. The MS spectra demonstrate that the protonated molecular ions of for omeprazole sulfide (c) and 5′-OH omeprazole sulfide (d) were 346 and 330, respectively
NMR analysis of the product derived from the hydroxylation of omeprazole sulfide by the CYP102A1 mutant. The chemical structure (a), key ROE results (b), and 1H NMR spectrum (c) of 5′-OH omeprazole sulfide are shown. In the inset of (c), the expanded spectrum at 3.81 ppm is shown. Two singlet peaks from 4′-OCH3 and 5-OCH3 protons can be clearly observed
Catalytic properties of omeprazole sulfide C-5′ hydroxylation by CYP102A1
Mutants #10, #375, #387, and #389, that exhibited high product formation rates, were selected for additional kinetic analyses and a binding titration study. Table 1 shows the steady-state kinetics, rates of NADPH oxidation and product formation, and coupling efficiency during omeprazole sulfide hydroxylation by the WT enzyme and the selected mutants. The three selected mutants exhibited 4- to 61-fold increases in catalytic efficiency (k cat/K M) compared with WT, which was primarily due to the significantly increased k cat values (21- to 104-fold), with little change in K M (Table 1). Mutant #375 showed the highest catalytic efficiency among the tested enzymes. Compared with the WT enzyme, all three mutants exhibited increases (3.2- to 14-fold) in NADPH oxidation and product formation (12- to 39-fold) in the presence of the substrate. All mutants exhibited an improved coupling efficiency ranging from 39 to 82 % in the presence of omeprazole sulfide compared with that of the WT enzyme (16 %). The mutants also exhibited significantly higher (7130–17,500) TTNs for the formation of the 5′-OH product from omeprazole sulfide than that of the WT (890) enzyme (Table 1).
Selective conversion of omeprazole sulfide to 5′-OH omeprazole sulfide by CYP102A1
When the mutants were incubated with 1 mM omeprazole sulfide for 10 min, mutants #10, #375, #387 and #389 exhibited ideal combinations of high conversion percentages (65–89 %) and high selectivities (95–98 %) for the production of 5′-OH omeprazole sulfide (Fig. 3c; Table 2). This result indicates that 5′-OH omeprazole sulfide can be effectively obtained from omeprazole sulfide with a high conversion rate and high selectivity.
The WT enzyme showed very low omeprazole 5′-hydroxylation activities toward S-omeprazole (0.16 min−1) and R-omeprazole (0.37 min−1), which were only 0.46 and 1.9 % of those of the highly active mutant #10, respectively (Ryu et al. 2014). However, a simple change from a sulfoxide to a sulfide in omeprazole caused a significant increase in the k cat value for the 5′-hydroxylation activity to 7.7 min−1 (Table 1).
According to the S-omeprazole-bound crystal structure of the heme domain of the CYP102A1 mutant (F87V/A82F), omeprazole sulfide was bound to the active site instead of omeprazole (Butler et al. 2014). Based on the structure of the heme domain bound to omeprazole sulfide, we gained insights into the possible binding features, along with the distance (4.1 Å) between the C-5′ of the substrate and the heme iron (Butler et al. 2014). This result suggests that C-5′ hydroxylation is more likely to occur rather than sulfur oxygenation. Mutant #10 is a well-known, highly active mutant toward several human substrates and has mutations in the substrate channel and active site (van Vugt-Lussenburg et al. 2006). However, the other highly active mutants #375, #387, and #389 have mutations outside of the active site and substrate channel. The mutations appear to have different effects on the conformation of the active site upon binding to the substrates (Supplementary Fig. 4).
Thermal stabilities of CYP102A1 mutants
To estimate the stabilities of the mutants, we determined the thermal stabilities (T 50, °C) of the mutants and compared them to that of WT (Table 3). T 50 values of the mutants varied from 46.5 to 52.2 °C. All mutants showed a decrease of the T 50 value by 3.9–9.6 °C when compared to that of WT (56.1 ± 1.9 °C). Although the four mutants selected among the tested mutants had very high k cat values and TTNs toward omeprazole sulfide, they showed apparently decreased thermal stabilities. The mutations appear to increase the catalytic activities and productivities but reduce the thermal stabilities of the mutant. Further experiments are required to increase the thermal stabilities of highly active mutants for biotechnological applications.
Conclusion
Omeprazole sulfide is an efficient and inexpensive substrate for preparing 5′-OH omeprazole sulfide, which is a human metabolite of omeprazole but is very expensive to purchase. Here we report an efficient one-step synthesis of 5′-OH omeprazole sulfide from omeprazole sulfide using CYP102A1. The proposed catalytic mechanism proceeds via a single step of regioselective 5′-hydroxylation of omeprazole sulfide by CYP102A1 mutants. The CYP102A1 mutants can catalyze the highly regioselective hydroxylation of omeprazole sulfide to produce 5′-OH omeprazole sulfide with a high conversion yield. This strategy, which uses regioselective C-H hydroxylation, should be applicable to the production of metabolites for other proton pump inhibitor drugs with similar chemical structures. In addition, the hydroxylated metabolites can be used as “drug leads” to avoid interindividual variations in drug metabolizing enzymes and drug–drug interactions with or without further modification of the hydroxylated group.
References
Butler CF, Peet C, Mason AE, Voice MW, Leys D, Munro AW (2013) Key mutations alter the cytochrome P450 BM3 conformational landscape and remove inherent substrate bias. J Biol Chem 288:25387–25399
Butler CF, Peet C, McLean KJ, Baynham MT, Blankley RT, Fisher K, Rigby SE, Leys D, Voice MW, Munro AW (2014) Human P450-like oxidation of diverse proton pump inhibitor drug by ‘gatekeeper’ mutants of flavocytochrome P450 BM3. Biochem J 460:247–259
Li XQ, Weidolf L, Simonsson R, Andersson TB (2005) Enantiomer/enantiomer interactions between the S- and R-isomers of omeprazole in human cytochrome P450 enzymes: major role of CYP2C19 and CYP3A4. J Pharmacol Exp Ther 315:777–787
Nevado JJ, Peñalvo GC, Dorado RM, Robledo VR (2014) Simultaneous determination of omeprazole and their main metabolites in human urine samples by capillary electrophoresis using electrospray ionization-mass spectrometry detection. J Pharm Biomed Anal 92:211–219
Obach RS (2013) Pharmacologically active drug metabolites: impact on drug discovery and pharmacotherapy. Pharmacol Rev 65:578–640
Rezk NL, Brown KC, Kashuba AD (2006) A simple and sensitive bioanalytical assay for simultaneous determination of omeprazole and its three major metabolites in human blood plasma using RP-HPLC after a simple liquid–liquid extraction procedure. J Chromatogr B 844:314–321
Ryu SH, Park BY, Kim SY, Park SH, Jung HJ, Park M, Park KD, Ahn T, Kang HS, Yun CH (2014) Regioselective hydroxylation of omeprazole enantiomers by bacterial CYP102A1 mutants. Drug Metab Dispos 42:1493–1497
Saccar CL (2009) The pharmacology of esomeprazole and its role in gastric acid related diseases. Expert Opin Drug Metab Toxicol 5:1113–1124
van Vugt-Lussenburg BM, Damsten MC, Maasdijk DM, Vermeulen NP, Commandeur JN (2006) Heterotropic and homotropic cooperativity by a drug-metabolising mutant of cytochrome P450 BM3. Biochem Biophys Res Commun 346:810–818
Wedemeyer RS, Blume H (2014) Pharmacokinetic drug interaction profiles of proton pump inhibitors: an update. Drug Saf 10:187–194
Whitehouse CJ, Bell SG, Wong LL (2012) P450 BM3 (CYP102A1): connecting the dots. Chem Soc Rev 41:1218–1260
Yun CH, Yim SK, Kim DH, Ahn T (2006) Functional expression of human cytochrome P450 enzymes in Escherichia coli. Curr Drug Metab 7:411–429
Yun CH, Kim KH, Kim DH, Jung HC, Pan JG (2007) The bacterial P450 BM3: a prototype for a biocatalyst with human P450 activities. Trends Biotechnol 25:289–298
Acknowledgments
This research was supported by the National Research Foundation of Korea (Grant NRF-2016R1A2B4006978) and by the Next-Generation BioGreen 21 program (SSAC Grant# PJ011058), Rural Development Administration, Republic of Korea.
Supporting information
Supplementary Methods-Generation of the CYP102A1 libraries.
Supplementary Methods-Screening of the CYP102A1 libraries.
Supplementary Methods-Liquid chromatography-mass spectrometry analysis.
Supplementary Methods-Identification of the omeprazole sulfide metabolite by NMR spectroscopy.
Supplementary Table 1. Mutated amino acid residues of the CYP102A1 mutants used in this study.
Supplementary Table 2. 1H chemical shifts for 5'-OH omeprazole sulfide.
Supplementary Figure 1. Metabolic pathway of omeprazole in humans.
Supplementary Figure 2. Screening of CYP102A1 mutant libraries.
Supplementary Figure 3. Close look at the 3.81 ppm and NOE results of 5′-OH omeprazole sulfide.
Supplementary Figure 4. The mutation sites for CYP102A1 M16V2 and its mutants.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Jang, HH., Ryu, SH., Le, TK. et al. Regioselective C-H hydroxylation of omeprazole sulfide by Bacillus megaterium CYP102A1 to produce a human metabolite. Biotechnol Lett 39, 105–112 (2017). https://doi.org/10.1007/s10529-016-2211-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-016-2211-3