Abstract
Objectives
To alter DNA binding specificity of Vibrio fischeri LuxR and to expand the toolbox for constructing synthetic networks.
Results
A mutation library (about 10,000 individuals) of the DNA binding domain of LuxR were generated. A genetic selection was performed to obtain LuxR mutants that recognize three lux box DNA variants that are not recognized by wild-type LuxR. Six LuxR mutants were identified. The evolved LuxR mutants were further characterized by measuring the transcriptional activities of different combinations of LuxR mutants and lux box variants. Varied transcriptional activities were found in these LuxR–lux box pairs. The background expressions of the evolved LuxR–lux box systems are more tightly regulated than the wild-type LuxR–lux box system.
Conclusion
The LuxR transcriptional system was evolved to recognize three lux box DNAs which are not recognized by wild-type LuxR.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Transcriptional systems functioning in various species and expressing with low background are desirable in many synthetic biology and industrial biotechnology approaches. As an inducible system, the LuxR–lux box and its homologues have been widely used in various organisms, including bacteria (Wei et al. 2004), yeast (Williams et al. 2013) and human cells (Neddermann et al. 2003).
However, as the master regulators of the bacterial quorum sensing system, LuxR family proteins can cross-talk with each other because of their highly conserved structures and biochemical properties. They are normally formed by two conserved domains: chemical inducer-binding domain and DNA binding domain (Koch et al. 2005). Two types of cross-talk, chemical inducer cross-talk and DNA binding cross-talk, become barriers for the wider application in synthetic biology and industrial biotechnology studies (Balagadde et al. 2008; Salis et al. 2009; Wu et al. 2014). Firstly, LuxR homologues can competitively response to chemical inducers (Balagadde et al. 2008). Secondly, 20-bp DNA sequences bound by LuxR family proteins are similar with each other (Salis et al. 2009; Wu et al. 2014).
The chemical inducer-binding specificity of LuxR can be altered. However, little has been done with the DNA binding domain because DNA-binding specificity is difficult to change (Dougherty and Arnold 2009). To expand the toolbox of synthetic biology and biotechnology, different DNA binding patterns of transcriptional factors are significantly desirable. In addition, transcriptional systems with low-background expression are useful to express toxic proteins, or to express fluorescence proteins for bioimaging. Herein, we have obtained LuxR mutants with different transcriptional specificities in order to generate new LuxR–lux box pairs with low-background expression and wide host range. We started with three lux box variants that are not recognized by wild-type LuxR (Antunes et al. 2008). To select the needed LuxR mutants from a mutation library (about 10,000 individuals), a genetic selection system was constructed by fusing the lux box variants to chloramphenicol acetyltransferase (cat) gene. Thus if LuxR mutants could recognize lux box variants, the cat gene will be expressed and those clones will survive from the selection process. Through this selection, six LuxR mutants were identified and we measured the transcriptional activities of different LuxR–lux box combinations to characterize the evolved LuxR–lux box system. We also investigated the background expression of LuxR–lux box pairs by measuring their transcriptional activities under non-induced condition.
Materials and methods
Library construction and genetic selection
Error-prone PCR reactions were performed using EasyTaq DNA polymerase (Transgen, Beijing) (see Collins et al. 2006). The sequence (440–750 bp) of LuxR was amplified. The MegaWHOP (Megaprimer PCR of Whole Plasmid) PCR (95 °C for 5 min, one cycle; 95 °C for 30 s, 68 °C for 6 min, 24 cycles; 68 °C for 10 min, one cycle) was performed in 50 μl using dNTPs mix (0.2 mM), purified error-prone PCR product (250 ng), plasmid DNA template (50 ng) and 1.5 U Phusion polymerase (New England Biolabs). The amplified MegaWHOP product was treated with DpnI (20 U, New England Biolabs) at 37 °C for 1 h and transformed into competent Top ten cells.
During genetic selection, cells were plated onto a selective LB agar plate containing 100 μg ampicillin/ml, 75 μg chloramphenicol/ml and 2 μM decanoyl-dl-homoserine lactone (C10HSL). To estimate the library size, cells were also plated only with 100 μg ampicillin/ml. The survived clones were re-plated on the selective plates to remove any false positives.
Fluorescence measurement
The mediated expression of GFPmut2 in the LuxR mutants was performed as follows: single clones were picked and grown in 2 ml LB medium (100 μg ampicillin/ml) overnight at 37 °C. 10 μl overnight cultures were transferred into fresh 2 ml LB medium (100 μg ampicillin/ml and 2 μM C10HSL) and incubated at 37 °C until the OD600 value reached ~1. 1 ml cultures were centrifuged and washed twice by the same volume of phosphate buffered saline (PBS) at PH 7.2. Cells were resuspended in 1 ml PBS and 200 μl was transferred to each well of a black 96-well microplate with clear bottom. GFPmut2 fluorescence was measured by a microplate reader with excitation at 481 nm excitation and emission at 507 nm. Fluorescence was normalized to the OD. The fluorescence/OD600 value of pBAD/myc-HisA was used as the background fluorescence and this background fluorescence value was subtracted from all fluorescence measurements of LuxR–lux box pairs.
Results and discussion
Genetic selection
Several lux box variants that are not recognized by LuxR have been identified (Antunes et al. 2008). We selected three lux box variants (A, B, C) and confirmed they have significant loss of transcriptional activity mediated by LuxR (about 5–20 % of the transcriptional activity of wild-type LuxR–lux box) (see Fig. 1a). Therefore, we sought to generate new LuxR-like systems by engineering LuxR to recognize three lux box variants.
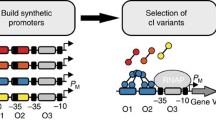
Genetic selection of LuxR mutants recognizing new lux box DNA variants. a Selected lux box variants that have significant loss of transcriptional activity mediated by LuxR. Shown are units of fluorescence due to GFPmut2 production in E. coli containing LuxR (containing mutations: T33A, R67M, S116A and M135I in chemical inducer binding domain and thus binding to C10HSL) and three lux box variants. The data are normalized to optical density at 600 nm and corrected by subtracting background fluorescence from a control strain containing pBAD/myc-HisA (same backbone). Error bars show standard deviations from three independent biological replicates. AU arbitrary unit. b Scheme of genetic selection. During the selection, LuxR is constitutively expressed. Active LuxR mutants are identified by their ability to survive in the presence of chloramphenicol (Cm) due to LuxR-mediated activation of the chloramphenicol acetyl-transferase (cat) gene from the lux box variants
We applied directed evolution approaches to obtain LuxR mutants with various DNA binding patterns and employed a genetic selection to get LuxR mutants that can recognize lux box variants. First, using error-prone PCR and MegaWHOP (Megaprimer PCR of Whole Plasmid), we generated a mutation library (~10,000 individuals) of DNA binding domain (157–250 aa) from a LuxR mutant (four mutations: T33A, R67 M, S116A and M135I in chemical inducer binding domain), which has independent inducer-binding specificity with the wild-type LuxR (Collins et al. 2006). Second, a genetic selection was performed by fusing the chloramphenicol acetyl-transferase (cat) gene to each lux box variant (A, B, C) (Fig. 1b). LuxR mutants that recognize lux box variants will survive in the presence of decanoyl-dl-homoserine lactone (C10HSL) and chloramphenicol.
Characterization of evolved LuxR variants
We characterized six LuxR mutants that finally survived the genetic selection. The transcriptional activities of six LuxR mutants and their corresponding lux box variants were measured by GFPmut2 expression (Fig. 2a). Figure 2b shows the amino acid changes in the LuxR mutants. Notably, D202E in A1, A221 V in C1 and T219A in C3 are located in the Helix–Turn–Helix (HTH) motif, which directly binds to lux box DNA. Interestingly, A1, C1 and C3 also have about 170, 140, and 140 % of transcriptional activities of the wild-type lux box respectively, while LuxR mutants containing mutations outside of the HTH motif do not have such change (Fig. 2c). We also noticed that A1 and C2 share one same mutation, D244H, which is near the area responsible for dimerization of LuxR family proteins (Vannini et al. 2002). It is highly possible that dimerization of LuxR affecting the DNA binding specificity.
Characterization of the evolved LuxR–lux box transcriptional system. a The gfpmut2 transcription mediated by LuxR mutants and corresponding lux box variants. b Amino acid changes of LuxR variants. HTH motif of LuxR directly binds to lux box DNA. A1, C1 and C3 have mutations in HTH motif.c The gfpmut2 transcription mediated by LuxR mutants and wild-type lux box. A1, C1 and C3 with mutations located in HTH motif have higher transcriptional activities towards wild-type lux box than wild-type LuxR. d Varied transcriptional activities of different combinations of LuxR mutants and lux box variants. Shown are units of fluorescence due to GFPmut2 production in E. coli containing LuxR mutants and corresponding lux box variants. Error bars show standard deviations from three independent biological replicates. AU arbitrary unit
To characterize the evolved LuxR transcriptional system, we measured the transcriptional activities of different combinations of LuxR mutants and lux box variants. It is interesting to see that LuxR mutants which were designated to recognize one lux box DNA also have specificities to other two lux box variants (Fig. 2d). For example, transcriptional activities of A1-C and C1–A are similar with those of A1–A and wild-type LuxR–lux box. Also, A1 and C1 have similar DNA specificities to all three lux box variants as indicated by the transcriptional activities. This indicates that mutations in A1 and C1 enhance the binding affinity of LuxR and DNA but do not change the binding specificity to lux box DNA. We also noticed that transcriptional activities of A2–B, C2–B, and C3–B are about 20–35 % of the transcriptional activities of A2-B, C2-B, and C3-B. This indicates that lux box variant B is basically not recognized by A2, C2 and C3 but can be recognized by A1, C1 and B1. In all, transcriptional activities of the evolved LuxR–lux box system vary among different combinations (the transcriptional activities of the evolved LuxR system vary from about 15 to 170 % than the transcriptional activities of wild-type LuxR system). It is applicable in studies which require different transcriptional activities, for example, customized optimization of metabolic pathways (Du et al. 2012).
The tightly regulated expression of evolved LuxR systems
We further measured the transcriptional activities of different combinations of LuxR–lux box under non-induced condition (−C10HSL). The background expressions of evolved LuxR–lux box pairs are from about 20 to 50 % of the background expression of wild-type LuxR–lux pair (Fig. 3). We chose three groups (high, medium and low) of LuxR–lux pairs based on the transcriptional activities under induced condition. Figure 3 presents differences in the GFPmut2 expression under non-induced and induced conditions. Remarkably, the background expressions of the A1–C, C1–A and A1–A, the transcriptional activities of which are about 123, 110, and 100 % of transcriptional activities of wild-type LuxR–lux box pair are about 20–45 %, respectively, of the background expression of the wild-type LuxR–lux box system (Fig. 3). We therefore conclude that the evolved LuxR system is tightly regulated. LuxR mutants-lux box variants can thus be used to manipulate gene expression.
The expression of gfpmut2 transcription mediated by selected LuxR mutants to lux box variants under non-induced condition (−C10HSL) and induced condition (+C10HSL). Three groups of LuxR mutants and lux box variants with different transcriptional activities under induced condition were selected (high, medium and low). GFPmut2 productions under non-induced condition (−C10HSL) and induced condition (+C10HSL) were measured. Error bars show standard deviations from three independent biological replicates. AU arbitrary unit
Conclusion
We have engineered the widely used LuxR system based on directed evolution approaches. The evolved LuxR mutants can recognize promoters that are not recognized by wild-type LuxR. We also found that the transcriptional activities vary among different combinations of LuxR mutants and lux box variants. These LuxR mutants could help to deduce the code of DNA binding specificity of LuxR and are able to decrease the cross-talk between the highly conserved LuxR homologues. The evolved LuxR system is also tightly regulated. The evolved LuxR system is currently available to tune metabolic pathways and construct synthetic networks.
References
Antunes LC, Ferreira RB, Lostroh CP, Greenberg EP (2008) A mutational analysis defines Vibrio fischeri LuxR binding sites. J Bacteriol 190:4392–4397
Balagadde FK et al (2008) A synthetic Escherichia coli predator–prey ecosystem. Mol Syst Biol 4:187
Collins CH, Leadbetter JR, Aronld FH (2006) Dual selection enhances the signaling specificity of a variant of the quorum-sensing transcriptional activator LuxR. Nat Biotechnol 24:708–712
Dougherty MJ, Arnold FH (2009) Directed evolution: new parts and optimized function. Curr Opin Biotechnol 20:486–491
Du J, Yuan Y, Si T, Lian J, Zhao H (2012) Customized optimization of metabolic pathways by combinatorial transcriptional engineering. Nucleic Acids Res 40:e142
Koch B, Liljefors T, Persson T, Nielsen J, Kjelleberg S, Givskov M (2005) The LuxR receptor: the sites of interaction with quorum-sensing signals and inhibitors. Microbiology 151:3589–3602
Neddermann P, Gargioli C, Muraglia E, Sambucini S, Bonelli S, De Francesco R, Cortese R (2003) A novel, inducible, eukaryotic gene expression system based on the quorum-sensing transcription factor TraR. EMBO Rep 4:159–165
Salis H, Tamsir A, Viogt C (2009) Engineering bacterial signals and sensors. Contrib Microbiol 16:194–225
Vannini A, Volpari C, Gargioli C et al (2002) The crystal structure of the quorum sensing protein TraR bound to its autoinducer and target DNA. EMBO J 21:4393–4401
Wei YH, Lai HC, Chen SY, Yeh MS, Chang JS (2004) Biosurfactant production by Serratia marcescens SS-1 and its isogenic strain SMdeltaR defective in SpnR, a quorum-sensing LuxR family protein. Biotechnol Lett 26:799–802
Williams TC, Nielsen LK, Vicker CE (2013) Engineered quorum sensing using pheromone-mediated cell-to-cell communication in Saccharomyces cerevisiae. ACS Synth Biol 2:136–149
Wu F, Menn DJ, Wang X (2014) Quorum-sensing cross-talk-driven synthetic circuits: from unimodality to trimodality. Chem Biol 21:1629–1638
Acknowledgments
This work was supported by National High Technology Research and Development Program of China (2012AA02A704) and the Fundamental Research Funds for the Central Universities (Grant No. WK2070000019).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Lu, Y. Engineering Vibrio fischeri transcriptional activator LuxR for diverse transcriptional activities. Biotechnol Lett 38, 1459–1463 (2016). https://doi.org/10.1007/s10529-016-2134-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-016-2134-z