Abstract
Objectives
To characterize the ent-copalyl diphosphate (ent-CPP) synthase involved in the biosynthetic pathway of andrographolides in a medicinal plant, Andrographis paniculata.
Results
The ent-CPP synthase (ent-CPS) gene was cloned from A. paniculata and its encoded ApCPS was demonstrated to react with (E,E,E)-geranylgeranyl diphosphate to form ent-CPP through recombinant expression in Escherichia coli. Site-directed mutagenesis of the Asp to Ala in the conserved DXDD motif of ApCPS resulted in loss of function. One Arg is located in the conserved position close to DXDD motif indicating the involvement of ApCPS in specialized metabolism. In addition, RT-PCR analysis revealed that ApCPS was expressed in all tissues of A. paniculata at all growth stages, which is consistent with andrographolides accumulating in these organs. Methyl jasmonate induced ApCPS gene expression, matching inducible accumulation of andrographolides in vivo.
Conclusions
ApCPS is the first ent-CPS characterized in A. paniculata and is suggested to be involved in biosynthesis of andrographolides that have high pharmaceutical values.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Labdane-related diterpenoids, with over 7000 members, exhibit broad activities and are involved in many biological aspects, including plant growth and development or plant pathogen defense mediated by the phytohormones gibberellic acids and phytoalexins (Peters 2010). All labdane-related diterpenoids have the core structure of a bicyclic labdane, which is derived from copalyl diphosphate (CPP). CPP is cyclized from geranylgeranyl diphosphate (GGPP) through CPP synthase (CPS). Three types of CPP with different stereochemical configurations, ent-, syn- and normal, are produced by three differential CPSs respectively and are involved in biosynthesis of various labdane-related diterpenoids (Fig. 1a; Prisic et al. 2004; Xu et al. 2004; Gao et al. 2009). ent-CPP, formed by ent-CPS, is the direct precursor of gibberellic acid biosynthesis and also is involved in the metabolism of many natural products including phytoalexins in rice and maize (Bensen et al. 1995; Prisic et al. 2004; Harris et al. 2005) and the sweetener steviol glycoside in Stevia rebaudiana (Richman et al. 1999).
Andrographis paniculata is a traditional medicinal plant and is used to treat infections and inflammation in China and South Asia. Its major bioactive constituents are andrographolides (Fig. 1b; Pholphana et al. 2013). Andrographolides are labdane-related diterpenoids and exhibit various bioactivities including anti-cancer (Luo et al. 2014), anti-virus (Chen et al. 2009), antimicrobial and anti-inflammatory activities (Chua 2014), suggesting potential pharmaceutical values. However, the biosynthetic pathway of andrographolides has not been elucidated. Based on their chemical structure, andrographolides biosynthesis should be initiated from the cyclization of GGPP to form ent-CPP catalyzed by ent-CPS. One CPS gene (ApCPS, GenBank: JN216843) has been cloned previously from A. paniculata but not yet characterized biochemically. Here we characterized the function of CPS with its recombinant expression in E. coli. The CPP product was further identified as ent-CPP through co-expression of ApCPS and rice kaurene synthase, enabling the formation of ent-kaurene. Gene expression analysis of ApCPS showed that it ubiquitously expressed in whole plant and was induced by methyl jasmonate, consistent with andrographolides accumulation and indicating participation in its biosynthesis.
Materials and methods
Material
Andrographis paniculata was grown in the greenhouse at 28 °C, 14 h light/10 h dark. All chemical reagents used are analytic grade unless described specifically.
Gene cloning and recombinant constructs
The coding sequence of ApCPS was amplified from leaf cDNA of 5 weeks old A. paniculata using primers of ApCPS-FL-F and ApCPS-FL-R (Supplementary Table 1). The resulted fragment was ligated into pGM-T vector (Tiangen, Beijing) for sequencing verification, and subsequently subcloned into pGEX vector using BamHI and XhoI to be pGEX/ApCPS for recombinant expression. The putative mature protein sequence was constructed by deleting the first 27 amino acids plastid transit peptide at the N-terminus using PCR, which was also inserted into pGEX vector to be pGEX/ApCPS d27 for expression in E. coli.
Recombinant expression
The full length and putative mature ApCPS were recombinant expressed in E. coli C41 (DE3) strain (Lucigen) with co-expression of GGPP synthase (GGPPs) from giant fir constructed as pGG (Cyr et al. 2007). pGG derived plasmid, pGG/An2 harboring maize ent-CPP synthase An2 was used as positive control for ent-CPP production (Harris et al. 2005). pGG and pGG/An2 were kindly provided by Prof. Reuben Peters at Iowa State University. The enzymatic product was twice extracted with equal volumes of hexane and concentrated by rotary evaporation. The residue was resuspended in 200 μl hexane for GC–MS analysis. To identify the stereochemistry of ApCPS enzymatic product, rice kaurene synthase, OsKS with ent-CPP specific reactivity was incorporated into the above expression system (Xu et al. 2007). Wheat terpene synthase, TaKSL1, with syn- and normal CPP reactivity was also co-expressed with ApCPS as above (Zhou et al. 2012). The procedure of product extraction and analysis is same as described above.
GC–MS analysis
GC–MS analysis was performed on Agilent 6890-5973 instrument with quadruple mass spectrometer and HP5 column. Samples in hexane (1 μl) were injected in GC–MS in splitless mode with the program: 70 °C for 2 min, raising to 280 °C at 10 °C /min, and holding for 2 min. After an acquisition delay of 10 min, MS data was collected from m/z of 50 to 400.
Site-directed mutagenesis
Site-directed mutagenesis of ApCPS was carried out at the conserved DXDD motif in pGEX/ApCPS d27 using the Quickchange kit (Strategene). The first Asp (D388) at DXDD motif was mutated to Ala using primers of D388A-F and R (codon is underlined, Supplementary Table 1). The resulted construct pGEX/ApCPS d27-D388A was sequenced to verify the successful mutation and subsequently tested for the enzymatic activity as above.
Gene expression analysis
Constitutive gene expression of ApCPS in different tissues was analyzed using semi-quantitative RT-PCR. Roots, leaves and stems were collected from 5 week old A. paniculata plants. Flowers and siliques were sampled at flowering stage. Radicals and hypocotyls were collected from germinating seeds. All tissues were ground to a fine powder in liquid N2 for RNA extraction with Trizol. cDNA was synthesized using the M-MLV reverse transcriptase kit (Takara) following the manufacture protocol. RT-PCR was performed for ApCPS with primers of CPS-F and CPS-R. ACTIN of A. paniculata (GenBank: JX444056) was used as internal control with primers of actin-F and actin-R. Gene expression of ApCPS with methyl jasmonate (MeJA) treatment was analyzed by quantitative RT-PCR analysis (qRT-PCR). 5 week old A. paniculata plants were treated with 25 μM MeJA for 0, 12, 24 and 48 h for RNA extraction and cDNA synthesis as above. qRT-PCR was performed on a Bio-Rad CFX96 instrument using the SsoFast Eva Green Supermix (Bio-Rad) with ACTIN as the reference gene. Primers are listed in Supplementary Table 1.
TLC analysis of andrographolides
A. paniculata tissues (~500 mg each) were ground in liquid N2, stirred with 10 ml methanol for 1 h, filtered and the extract concentrated by rotary evaporation. The residue was dissolved in 0.5 ml methanol for analysis by TLC. (see Fig. 4 below). Andrographolides standards were purchased from Sigma.
Bioinformatics analysis
Gene and amino acid sequence alignment were performed using CLC sequence viewer 7.0 (CLC bio). All primers were designed using Oligo 7 and Primer 3 (bioinfo.ut.ee/primer3-0.4.0/), and the resulted amplicon was verified with sequencing. The plastid localization of ApCPS was predicted online at TargetP and ChloroP (www.cbs.dtu.dk/services/TargetP).
Results and discussion
Recombinant expression and production identification
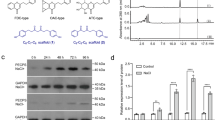
The coding sequence of ApCPS was cloned, and the full length and transit peptide truncated variants were co-expressed with GGPPs in E. coli as before (Cyr et al. 2007). CPP is usually depyrophosphorylated to copalol by the endogenous phosphatase in E. coli and could be extracted with organic solvent for GC–MS analysis (Wu et al. 2012). GC–MS analysis of fermentation products revealed CPP production from GGPP by full length ApCPS and truncated variant ApCPS d27 (Fig. 2a, b), with identical retention time and mass spectra of ent-CPP standard.
GC–MS analysis of ApCPS enzymatic product. a Extracted ion chromatograms (275 m/z) of GC–MS analysis for co-expression products of ApCPS and its truncated variant ApCPS d27 (putative mature ApCPS with truncation of 27 aa plastid transit peptide) with GGPPs in E. coli. ent-copalol produced by An2 (maize ent-CPP synthase) was used as the authentic standard. b Mass spectra of copalol produced by ApCPS (Peak 1, RT = 19.16 min) and ApCPS d27 (Peak 2, RT = 19.16 min) in comparison to that of ent-copalol (RT = 19.16 min). c Total ion chromatograms of GC–MS analysis for co-expression products of ApCPS d27 and its mutant ApCPS d27-D388A with OsKS in E. coli. ent-kaurene (Peak 3) was detected in co-expression products of ApCPS d27 but not for ApCPS d27-D388A mutant. The authentic standard of ent-kaurene produced by co-expression of OsKS and An2 was used for comparison. GC–MS analysis of co-expression product of ApCPS d27 with TaKSL1 (wheat terpene synthase with reactivity of syn- and normal CPP) also was shown; however, no terpene was detected. d Mass spectra of co-expression product of ApCPS d27 with OsKS (Peak 3, RT = 17.79 min) and the authentic standard of ent-kaurene (RT = 17.79 min)
Stereochemistry analysis of ApCPS enzymatic product
There are three types of CPP with different stereochemistry, among which ent-CPP and normal CPP are hard to separate and distinguish by GC–MS analysis (Wu et al. 2012). To investigate the stereochemistry of CPP produced by ApCPS, OsKS with ent-CPP specific reactivity was co-expressed with ApCPS d27 and GGPPs (Xu et al. 2007). ent-Kaurene was clearly detected from the co-expression extract, which demonstrated ent-CPP production by ApCPS (Fig. 2c, d). To test whether minor syn- and/or nomal CPP was produced by ApCPS, TaKSL1 with reactivity of syn- and normal CPP (Zhou et al. 2012), was co-expressed with ApCPS d27; however, no terpene product was detected (Fig. 2c), indicating no syn- and/or normal CPP was produced by ApCPS.
Conserved domain identification of ApCPS
The DXDD conserved motif is found in all CPSs and is required for catalysis (Prisic et al. 2007; Zi et al. 2014). It was also observed in ApCPS (Fig. 3). To verify the catalytic function of this motif in ApCPS, site-directed mutagenesis was employed to change the first Asp of DXDD motif to Ala (D388A), which resulted in loss of CPS function (Fig. 2c). This result demonstrated that ApCPS catalyzed the ent-CPP formation depending on DXDD motif as other CPSs (Prisic et al. 2007; Zi et al. 2014).
Conserved DXDD motif and Arg of ApCPS with alignment with characterized CPSs. DXDD motif is labeled with solid line, and the conserved Arg or His is marked with asterisk. The enzymes are separated according to physiological function by a line. The CPS in the upper section were characterized to be involved in specialized/secondary metabolism (2°) including ApCPS identified in this study, AgAS (U50768, Abies grandis), SmCPS (EU003997, Salvia miltiorrhiza), OsCPS2 (AY602991) and OsCPS4 (AY530101) from rice (Oryza sativa). The enzymes in the lower section were identified as CPSs in gibberellic acid metabolism: LsCPS (AB031204, Lactuca sativa), CmCPS (AB109763, Cucurbita maxima), PsCPS (U63652, Pisum sativum), SrCPS (AF034545, Stevia rebaudiana), OsCPS1 (NM_001053085, Oryza sativa), and AtCPS (U11034, Arabidopsis thaliana)
One conserved amino acid, His versus Arg, close to DXDD motif was identified and related to CPS roles in primary or specialized metabolism respectively (Prisic and Peters, 2007; Mann et al. 2010). ApCPS has an Arg at this position (Fig. 3), suggesting putative involvement in specialized metabolism (i.e. andrographolides biosynthesis).
Gene expression of ApCPS
ent-CPP formation catalyzed by ent-CPS is the key step for gibberellic acid metabolism, as well as for andrographolides biosynthesis. To identify ApCPS gene function in vivo, semi-quatitative RT-PCR was adopted to analyze gene expression of ApCPS in different A. paniculata tissues. Figure 4a shows that ApCPS was expressed in all tested tissues including stems, leaves, flowers, siliques, and rapid growing tissues, such as hypocotyls and radicles, which corresponds to the accumulation of andrographolides in vegetative and reproductive organs (Fig. 4b). A trace amount of andrographolides accumulated in roots and radicals (Fig. 4b), which is consistent with low gene expression of ApCPS in these tissues (Fig. 4a). These results are also consistent with ubiquitous distribution of andrographolides at all growth stages (Pholphana et al. 2013). Gibberellic acid metabolism is regulated in planta temporally and spatially (Sun and Kamiya 1994), and active gibberellic acids as well as their biosynthetic genes are usually located in rapidly growing tissues. Although ApCPS was expressed in young tissues where gibberellic acid is synthesized, andrographolides also accumulated in these tissues (Fig. 4a, b). Thus, ubiquitous expression of ApCPS in A. paniculata tissues does not match the limited distribution of gibberellic acids (Sun and Kamiya 1994).
Gene expression of ApCPS and andrographolides accumulation in tissues of A. paniculata. a RT-PCR analysis of ApCPS gene expression in different tissues. R root, S stem, L leaf, F flower, Si silique, Ra radicle, H hypocotyl. ACTIN is the endogenous control. b TLC analysis of andrographolides accumulation in the same tissues as above. 5 ul andrographolides extract in methanol for each sample was loaded on the silica gel TLC plate. The developing solvent was chloroform/ethyl acetate/methanol (4:3:0.4, by vol.). andrographolides were stained with pink using the chromogenic agent [2 M potassium hydroxide/ 2 % (v/v) 3, 5-dinitrobenzoic acid in ethanol, 1:1, v/v]. Standards (Sd) were labeled as 1 (14-deoxy-11, 12-didehydroandrographolide) and 2 (andrographolide). c qRT-PCR analysis of ApCPS gene expression with MeJA treatment. The relative expression levels of ApCPS were normalized to the reference gene and determined by the \( \Delta^{{\Delta C_{\text{t}} }} \)-method. All qRT-PCR analysis was replicated in trice. The error bars indicate the standard deviation
CPS usually exhibits inducible gene expression in response to elicitation, such as by MeJA treatment (Prisic et al. 2004; Xu et al. 2004; Zi et al. 2014). qRT-PCR analysis showed that MeJA induced ApCPS gene expression strongly within 12 h (Fig. 4c), consistent with inducible accumulation of andrographolides by MeJA (Sharma et al. 2015).
Taken together, constitutive and inducible gene expression of ApCPS matched the corresponding accumulation of andrographolides in vivo, indicating involvement in andrographolides biosynthesis.
Conclusions
Characterization of an a ent-CPS from A. paniculata is reported. It catalyzed CPP formation from GGPP. The CPP produced by ApCPS was identified as ent-CPP through co-expression of ApCPS and OsKS, enabling ent-kaurene formation. Site-directed mutagenesis of the conserved Asp in the DXDD motif resulted in loss of function for ApCPS. The specific Arg near to DXDD motif of CPS that is involved in specialized metabolism was also detected in ApCPS. In addition, ubiquitous gene expression of ApCPS matches andrographolides accumulation in all tissues of A. paniculata. Importantly, MeJA induced ApCPS gene expression, which corresponds to inducible accumulation of andrographolides by MeJA. These results suggest that ApCPS is involved in andrographolides biosynthesis and this is the first report of ent-CPS characterization in A. paniculata, a medicinal plant containing andrographolides with high pharmaceutical values.
References
Bensen RJ, Johal GS, Crane VC, Tossberg JT, Schnable PS, Meeley RB, Briggs SP (1995) Cloning and characterization of the maize An1 gene. Plant Cell 7:75–84
Chen JX, Xue HJ, Ye WC, Fang BH, Liu YH, Yuan SH, Yu P, Wang YQ (2009) Activity of andrographolide and its derivatives against influenza virus in vivo and in vitro. Biol Pharm Bull 32:1385–1391
Chua LS (2014) Review on liver inflammation and antiinflammatory activity of Andrographis paniculata for hepatoprotection. Phytother Res 28:1589–1598
Cyr A, Wilderman PR, Determan M, Peters RJ (2007) A modular approach for facile biosynthesis of labdane-related diterpenes. J Amer Chem Soc 129:6684–6685
Gao W, Hillwig ML, Huang L, Cui G, Wang X, Kong J, Yang B, Peters RJ (2009) A functional genomics approach to tanshinone biosynthesis provides stereochemical insights. Org Lett 11:5170–5173
Harris LJ, Saparno A, Johnston A, Prisic S, Xu M, Allard S, Kathiresan A, Ouellet T, Peters RJ (2005) The maize An2 gene is induced by Fusarium attack and encodes an ent-copalyl diphosphate synthase. Plant Mol Biol 59:881–894
Luo X, Luo W, Lin C, Zhang L, Li Y (2014) Andrographolide inhibits proliferation of human lung cancer cells and the related mechanisms. Intern J Clin Exper Med 7:4220–4225
Mann FM, Prisic S, Davenport EK, Determan MK, Coates RM, Peters RJ (2010) A single residue switch for Mg(2+)-dependent inhibition characterizes plant class II diterpene cyclases from primary and secondary metabolism. J Biol Chem 285:20558–20563
Peters RJ (2010) Two rings in them all: the labdane-related diterpenoids. Nat Prod Rep 27:1521–1530
Pholphana N, Rangkadilok N, Saehun J, Ritruechai S, Satayavivad J (2013) Changes in the contents of four active diterpenoids at different growth stages in Andrographis paniculata (Burm.f.) Nees (Chuanxinlian). Chin Med 8:2
Prisic S, Peters RJ (2007) Synergistic substrate inhibition of ent-copalyl diphosphate synthase: a potential feed-forward inhibition mechanism limiting gibberellin metabolism. Plant Physiol 144:445–454
Prisic S, Xu M, Wilderman PR, Peters RJ (2004) Rice contains two disparate ent-copalyl diphosphate synthases with distinct metabolic functions. Plant Physiol 136:4228–4236
Prisic S, Xu J, Coates RM, Peters RJ (2007) Probing the role of the DXDD motif in class II diterpene cyclases. ChemBioChem 8:869–874
Richman AS, Gijzen M, Starratt AN, Yang Z, Brandle JE (1999) Diterpene synthesis in Stevia rebaudiana: recruitment and up-regulation of key enzymes from the gibberellin biosynthetic pathway. Plant J 19:411–421
Sharma SN, Jha Z, Sinha RK, Geda AK (2015) Jasmonate-induced biosynthesis of andrographolide in Andrographis paniculata. Physiol Plant 153:221–229
Sun TP, Kamiya Y (1994) The Arabidopsis gibberellic acid1 locus encodes the cyclase ent-kaurene synthetase A of gibberellin biosynthesis. Plant Cell 6:1509–1518
Wu Y, Zhou K, Toyomasu T, Sugawara C, Oku M, Abe S, Usui M, Mitsuhashi W, Chono M, Chandler PM, Peters RJ (2012) Functional characterization of wheat copalyl diphosphate synthases sheds light on the early evolution of labdane-related diterpenoid metabolism in the cereals. Phytochemistry 84:40–46
Xu M, Hillwig ML, Prisic S, Coates RM, Peters RJ (2004) Functional identification of rice syn-copalyl diphosphate synthase and its role in initiating biosynthesis of diterpenoid phytoalexin/allelopathic natural products. Plant J 39:309–318
Xu M, Wilderman PR, Morrone D, Xu J, Roy A, Margis-Pinheiro M, Upadhyaya NM, Coates RM, Peters RJ (2007) Functional characterization of the rice kaurene synthase-like gene family. Phytochemistry 68:312–326
Zhou K, Xu M, Tiernan M, Xie Q, Toyomasu T, Sugawara C, Oku M, Usui M, Mitsuhashi W, Chono M, Chandler PM, Peters RJ (2012) Functional characterization of wheat ent-kaurene(-like) synthases indicates continuing evolution of labdane-related diterpenoid metabolism in the cereals. Phytochemistry 84:47–55
Zi J, Mafu S, Peters RJ (2014) To gibberellins and beyond! Surveying the evolution of (di)terpenoid metabolism. Ann Rev Plant Biol 65:259–286
Acknowledgments
This work was supported by fund for distinguished young scientist of Sichuan Province (2014JQ0038) and start-up fund from Sichuan Agricultural University to Q.W.
Supporting information
Supplementary Table 1: Primers used.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Shen, Q., Li, L., Jiang, Y. et al. Functional characterization of ent-copalyl diphosphate synthase from Andrographis paniculata with putative involvement in andrographolides biosynthesis. Biotechnol Lett 38, 131–137 (2016). https://doi.org/10.1007/s10529-015-1961-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-015-1961-7