Abstract
At the core of a biomanufacturing process for recombinant proteins is the production cell line. It influences the productivity and product quality. Its characteristics also dictate process development, as the process is optimized to complement the producing cell to achieve the target productivity and quality. Advances in the past decade, from vector design to cell line screening, have greatly expanded our capability to attain producing cell lines with certain desired traits. Increasing availability of genomic and transcriptomic resources for industrially important cell lines coupled with advances in genome editing technology have opened new avenues for cell line development. These developments are poised to help biosimilar manufacturing, which requires targeting pre-defined product quality attributes, e.g., glycoform, to match the innovator’s range. This review summarizes recent advances and discusses future possibilities in this area.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
For three decades, mammalian cells have been used as the vehicle for the production of therapeutic proteins for the treatment of cancers, rheumatoid arthritis, and many congenic diseases. They have the capability of performing post-translational modifications, including complex tertiary structure formation through multiple disulfide bonds, glycosylation, and phosphorylation, which are important attributes of many therapeutic proteins. Much effort has been devoted to developing alternative host systems, including plants, insect cells, yeasts, and transgenic animals. Nonetheless, the pre-eminent role of mammalian cells remains unsurpassed. During the same period, the productivity of mammalian cell culture processes has steadily increased by nearly two orders of magnitude. Advances in various aspects of process development, such as medium and feed design, vector design and screening strategies for identification of high producing clones, have all contributed to this increase in productivity. This review focuses on advances related to cell line development. A high-producing cell line nowadays can secrete IgG-based protein products at a rate rivalling that of professional secretors in our body (Seth et al. 2007). The method of generating producing cell lines remains largely unchanged over the past three decades. However, the host cells used to derive the producing cell line are often pre-adapted and selected for better traits suitable for industrial manufacturing (Sinacore et al. 1996). The methotrexate (MTX)—dihydrofolate reductase (DHFR) systems and methionine sulfoximine (MSX)—glutamine synthetase (GS) systems have been used to increase the copy numbers of the product genes, consistently yielding very high producing cells. The methods of identifying and isolating higher producing candidate cells have also been refined to increase the chance of isolating them.
In recent years, a torrent of transformative science and technology has been gathering force that can cause changes in the way future cell lines are derived. Genome science and systems analysis have given us new ways of understanding cell physiology, while advances in genome engineering have begun to endow us with the capability of engineering cells with better precision and predictability. This article aims to summarize the new knowledge on cell line development acquired in the past few years and discuss how the insights can be interjected with new tools to forge a new era of cell line development for biopharmaceutical production.
Vectors and modulation of transgene expression
For high productivity, a high transcript level of the transgene is desirable. The expression level of the transgene is affected by the vector and its components as well as the loci of vector integration. Plasmid vectors have been the primary vehicle of delivering the product transgene into the host cells. Most industrial producers have acquired multiple copies of the transgenes through either transfection with a high dose of the plasmid or amplification of the copy number of the transgene after transfection. In addition, viral vectors have also gained interest due to their propensity to integrate into actively transcribed regions of the host cell’s genome, resulting in higher expression levels of the transgene than plasmid transfection (Oberbek et al. 2011).
The transcript level of the transgene is affected by the integration site although, to date, a systematic study on the impact of the integration sites on the transgene expression level is lacking. To minimize the influence of this position effect, cis-acting barrier elements, called insulators that block the propagation of a heterochromatin structure into neighboring euchromatin regions, have been incorporated into the expression vector to isolate the integrated transgene from nearby negative influences in the case where the vector is integrated into a condensed chromosomal region (Ghirlando et al. 2012). The use of such insulating elements significantly increases the number of producing clones and improves the recombinant protein titer (Girod et al. 2005; Boscolo et al. 2012; Hou et al. 2014).
Promoters commonly used in driving the expression of the transgenes are derived from viruses, mice and humans because of their availability. Promoters from Chinese hamster for use in CHO cells have also gained attention. These include a beta-actin promoter (Estes and Zhang 2014), cold-responsive promoters (Thaisuchat et al. 2011; Al-Fageeh and Smales 2013), and time-dynamic promoters (Le et al. 2013). These endogenous promoters offer the possibility of more predictable expression levels, and dynamic control of the transgene expression with various growth stages. In addition, synthetic promoters have been explored to modulate gene expression levels (Hartenbach and Fussenegger 2006; Grabherr et al. 2011; Brown et al. 2014). As the customized combinations of these regulatory motifs do not occur naturally, they tend to be less tissue-specific and species-restricted, and could possibly offer more stable and consistent gene expression than natural ones.
Host cells and engineering host cells
Three decades after the emergence of therapeutic proteins, the list of commonly used host cells for such products remains rather limited but include Chinese hamster ovary (CHO), mouse myeloma NS0 or SP2/0, baby hamster kidney (BHK), and human embryonic kidney (HEK 293) cell lines. The narrow spectra of host cell line choices reflects the mastery in using currently available ones and the reluctance in experimenting new lines out of concerns of the need of seeking regulatory approval for them in spite of differences in proteins expressed from cell lines derived from different species; for example, in their glycosylation capability. Glycans on proteins produced in CHO and NS0 cell lines are somewhat different from each other as well as from those naturally occurring in human proteins (Sheeley et al. 1997; Beck et al. 2008). The small difference in glycans has not been a major concern, as evidenced by the therapeutic use in humans of proteins produced using both the cell types.
Over the years, host cells have been adapted to desired manufacturing culture conditions, including chemically defined medium, suspension growth, and higher mechanical stresses. While the genetic mechanism of such adaptations is not well understood, the fact that many of the acquired traits after adaptation are inheritable, suggests that the root cause of these adapted traits is probably genetic or epigenetic alternations. There have also been efforts to genetically engineer host cells so that the same desirable traits would be carried to new cell lines for different products. With the emergence of biosimilars for which the manufacturers have several years to tailor the candidate producing cell lines to a particular product, it will be tempting to genetically engineer host cell lines for better growth characteristics and product quality such as glycosylation profiles.
For two decades, efforts have been devoted to genetically engineer host cells or produce cells with enhanced growth characteristics or to modulate the glycosylation pattern of the product. Table 1 summarizes reports of cell engineering for cells producing heterologous proteins. Modulating cellular metabolism, especially to reduce lactate production from glucose metabolism has been the goal of many efforts. Another area that has drawn significant effort in engineering cells is tackling the apoptosis pathway by both overexpressing anti-apoptosis genes and suppressing pro-apoptotic genes (reviewed by Krampe and Al-Rubeai 2010). In addition, autophagy is important in the bioprocessing context (reviewed by Kim et al. 2013) and may be an attractive target for cell engineering.
Since most recombinant proteins expressed in animal cells are secreted, there has been keen interest to engineer cells to enhance their secretory pathway. The protein secretion pathway is complex, spanning over many cellular compartments, including ER, Golgi, and involves also vesicle trafficking and post-translational modifications. A cell’s capacity of protein secretion is influenced by its energy metabolism, gene expression and redox balance in addition to the secretory capacity (Seth et al. 2007). Such complexity was further illustrated by the drastically different capabilities of two host cell lines to express two antibody molecules at high levels even though they both expressed another antibody equally well (Hu et al. 2013). It is thus not surprising that reports of genetic manipulation of genes involved in the secretory pathways, such as BiP, PDI, or transcription factor Xbp-1 did not give consistent results Borth et al. (2005), and reviewed by Khan and Schröder (2008).
In comparison, cell engineering targeted at a specific reaction step in the glycosylation pathway has had considerable successes. Cell engineering approaches have been used to make the glycans produced in CHO cells resemble the human glycoform by expressing a heterologous 2,6-sialyl transferase, which is silenced in CHO cells (Lee et al. 1989). The unfucosylated glycan in Asn297 of IgG heavy chain enhances antibody-dependent cellular cytotoxicity (Shields et al. 2002). Knock-out of endogenous fucosyl transferase resulted in the desired alterations in glycan profile (Lee et al. 1989; Yamane-Ohnuki et al. 2004). Using a prokaryotic enzyme to divert the metabolic flux from the synthesis of GDP-l-fucose to GDP-d-rhaborthmnose reduced the GDP-l-fucose substrate pool for the fucosylation reaction and led to reduced fucosylation (von Horsten et al. 2010). However, it is worth noting that glycosylation pathway is very complex. Even though the number of enzymes constituting a pathway is small, their wide range of glycan substrate specificity gives rise to hundreds of possible glycans (Hossler et al. 2006; Spahn and Lewis 2014). To steer the reaction to a particular glycan may not be easily accomplished by tackling a single enzyme alone.
Epigenetics and genome editing on cell line development
Epigenetic regulation
Epigenetic regulations, or the chemical modifications of DNA and histone proteins without DNA sequence alteration, are increasingly being implicated in the development of producing cell lines. Gene silencing has been shown to affect the nutritional requirements of the cells and the transgene expression. A key gene in cholesterol synthesis, Hsd17b7, is silenced (due to methylation in the CpG island upstream of its promoter) in NS0 cells, leading to cholesterol auxotrophy (Seth et al. 2006). Treatment using the demethylation drug 5′-azacytidine or expression of heterologous Hsd17b7 led to the reversal of this phenotype. The methylation of CpGs in the CMV promoter has been linked to decreased transgene expression and productivity (Yang et al. 2010; Kim et al. 2011; Osterlehner et al. 2011). Both 2,6-sialyl transferase and GS are present in the Chinese hamster genome and expressed in liver, but are silenced in CHO cells. Epigenetic intervention may present opportunities in endowing cells with characteristics favorable for industrial production.
Targeted genome editing
Advances in targeted genome editing in the past few years coupled with availability of genome sequencing allows increasing precision in executing desired genetic changes. Efforts will likely be directed towards identifying transgene integration sites that can lead to high expression levels and long-term expression stability. Replacement of the transgene in the genome of the existing high-producing cells with a new product gene may be increasingly used for quick access to a high producer of the new product (Moehle et al. 2007; Nehlsen et al. 2009; Chen et al. 2013). The impact of genome editing need not be limited to the insertion of product genes but can also be used to engineer host cells. The genes modulating the characteristics of producing cells can be targeted to specific sites, possibly under the regulation of a cellular regulatory circuit and be expressed in sync with cellular rhythm at the desired level (Le et al. 2013). Recombinase-Mediated Cassette Exchange (RMCE), one of the earliest targeted gene integration techniques (reviewed by Turan et al. 2013), is based on the replacement of gene cassettes flanked by two recombination target sites. More recently, artificial nucleases like zinc finger nucleases (ZFNs), CRISPR/Cas system and transcription-activator like effector nucleases (TALENs), which combine a DNA sequence-specific binding domain with a non-specific nuclease domain (Fig. 1), have alleviated the requirement of tagging the sites for site specific gene integration (reviewed by Gaj et al. 2013). ZFNs and TALENs employ different DNA recognition elements that also pose different complexity levels in design. ZFNs use a combination of zinc finger domains for DNA specificity, each of ~30 amino acids specifically binds to a 3-bp DNA sequence. The DNA sequence specificity arises from the specific combination of these domains (Maeder et al. 2008). In TALENs, DNA binding domains consist of 33-35 amino acid repeat units, each of which is specific to a single base pair. Several such units are combined to achieve specificity to a target DNA sequence (Zhang et al. 2011).
The more recently developed CRISPR/Cas system, originally identified in bacteria, is a RNA-guided DNA endonuclease (Sander and Joung 2014). Sequence specificity is provided by including a specific guide sequence in the crRNA which binds to the target sequence in the genome and guides the Cas endonuclease to the target. All three systems introduce a double stranded or single stranded break (achieved by the nickase activity instead of the endonuclease activity of the nuclease) in a sequence specific manner, which greatly increases the frequency of homologous recombination at that location and facilitate the integration of the gene of interest. ZFNs have been used to rapidly generate a triple gene knock-out strain of CHO with GS−/−DHFR−/−Fut8−/− (Liu et al. 2010). Recently, the CRISPR/Cas9 technology was used to generate CHO lines with disruptions in COSMC and FUT8. However, off-target gene interruptions caused by the endonucleases poses some concerns for gene therapy applications (Kuscu et al. 2014). Such off-target effects can possibly be reduced by identifying unique target sites in the genome (Cho et al. 2014).
Evaluating and isolating high-producing cells
The increased versatility in performing genetic manipulations has played a major role in the generation of better cell lines in the past two decades. But this increased productivity is also attributed to the expanded capability in identifying and isolating clones that perform well in the manufacturing scales. Importantly, automation of many otherwise labor-intensive procedures in cell line isolation allows more clones to be evaluated and greatly improves the possibility of obtaining a high producer.
Assessing clonal quality and clonal stability
Upon the introduction and amplification of the transgene, single cell cloning is performed, and high producing cells with high productivity and robust growth characteristics are isolated. The selected clones must maintain their productivity and growth characteristics over a large number of generations in the product life cycle. Traditionally, single cell cloning, subsequent cell growth and productivity assessment, and cell stability studies are performed in well plates or flasks. Commercial instrumentations for simultaneous screening of a large number of clones are now standard features in industrial cell line development (Hou et al. 2014).
Microfluidic devices are being increasingly used to isolate single cells at micro- or nano-liter scales (reviewed by Mehling and Tay 2014). At very low cell concentrations used in single cell cloning in 96-well or 384-well plates, cell growth is often limited by the cell density. The small culture volume of the microfluidic device offers the advantage of faster accumulation of autocrine factors to facilitate cell growth during single cell cloning (Hansen et al. 2010). Product concentration assays for assessing productivity and cell isolation for further cultivation in the nano-liter scale microfluidic device are also possible (reviewed by Love et al. 2013). The system still faces some challenges for general users, including water evaporation and medium adsorption to the materials commonly used for fabricating the device (reviewed by Mehling and Tay 2014).
Assessment of process performance and scale-down models
Ideally, the selection of the final production clones should be based on the performance of those clones in culture conditions similar to the manufacturing setting. For cell lines used in a fedbatch manufacturing process, a performance evaluation based on fed-batch mode was reported to yield better outcomes compared to evaluation based on batch mode (Porter et al. 2010a, b). To simulate the manufacturing processes in stirred tanks, commercial miniscale multi-tube/flask systems or miniaturized cell culture systems with various mechanisms of mixing are available to increase the capacity of process performance evaluation (Girard et al. 2001; De Jesus et al. 2004). Some systems allow for control of pH and periodic nutrient feeding (Frison et al. 2002; Chen et al. 2009; Legmann et al. 2009; Hsu et al. 2012; Moses et al. 2012). To alleviate the need of a sensor and a pH control mechanism, a hydrogel based platform of slow release of neutralizing agent in situ for pH maintenance has been used (Pradhan et al. 2012).
To truly simulate manufacturing conditions, the culture system should provide equivalent time profiles of chemical milieu and physical environment (i.e., fluid dynamics and gas–liquid interfacial interactions) in the large scale. Most commercial miniscale systems provide mechanical mixing but do not address fluid dynamic issues. A scale-down model used 2 l reactors to simulate the hydrodynamic stresses at the large scale by oscillations between high and low agitation rates based on agitation power calculations (Sieck et al. 2013). Such studies along with the employment of transcriptome analysis across scales (Jayapal and Goudar 2014), or even extended to metabolomics studies, can provide much insight into the impact of scale up on cellular physiology.
Cell line development for biosimilars
Nearly three decades after the dawn of protein therapeutic biologics, patents for an increasing number of blockbuster biologics such as Remicade (infliximab), Humira (adalimumab) and Epogen (erythropoietin) are coming to the end of their life in various regions. This promises the opening of a large market and much wider availability of biosimilar drugs to patients.
The cell line development strategy for biosimilars differs somewhat from that for innovative biologics. The manufacturers of a biosimilar do not have access to the innovator’s original cell line and bioreactor/purification process. They must employ a different cell line and develop a different process while still “matching” the original protein product in its chemical and physical features, including purity and post-translational modification profile. For the innovator, rapid development and a short timeline to deliver the drug for clinical testing and regulatory approval is critical. But for the biosimilar manufacturers, a span of years is affordable to optimize the cell line and the process. This provides an opportunity to engineer the cell or tweak the process to acquire the desirable productivity and product characteristics, especially the specific glycosylation profiles.
Analytical tools to characterize the product become important in selecting potential cell line candidates and process conditions (Berkowitz et al. 2012). The advances in analytical technology can be a double-edged sword where, on one hand, the reference product can be characterized minutely and, on the other hand, these chemical characterizations can reveal many differences between the biosimilar molecule and the innovator product. Understanding the safety and efficacy impacts of the various attributes, and leveraging this knowledge to target cell line development efforts towards attributes that are critical, continues to challenge process development scientists.
Another challenge during biosimilar development is the choice of host cell line. Some innovator biologics may have been developed using older technologies, expression systems, and especially different cell lines. A range of new expression technologies have emerged since the approval of these early blockbusters. Current production platforms with optimized host cell lines different from innovator cell lines may be able to generate higher titers and improved purity. However, alternate host cell lines may also result in significant differences in the process impurity profiles or in the abundance of product variants.
Impact of genomic technology on cell line development
As industry moves into a post-genomic era and embraces systems biotechnology, we will see more use of genomic and transcriptomic tools in cell line development. The genome sequences of the host cell and the producing cell lines are becoming readily available (Xu et al. 2011; Brinkrolf et al. 2013; Lewis et al. 2013). Targeting the product gene to specific loci that give high transcript level and are less prone to silencing will likely become the norm in cell line construction. Directing genes conferring important trait(s) to specific sites on the genome wherein their expression level and dynamics are desirable may become a reality (Chen et al. 2013). As our understanding of the role of epigenetics on conferring the trait of hyperproductivity improves, targeted epigenetic intervention may also be feasible.
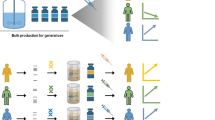
Genomic technology may also facilitate the screening of high producing cells. Through transcriptome and proteome analyses in the past decade, there have been attempts to develop a hyperproductivity gene set, or a collection of genes whose expression is “collectively” altered in high producing cells. Recently, single cell genomic methods for rtPCR and RNA seq based transcriptomic characterization have become available (reviewed by Astley and Al-Rubeai 2008; Eberwine et al. 2013; Nawy 2014). This will enable high throughput transcriptome profiling of producing cells using a very small number of cells. The transcriptome data can allow for faster and more reliable identification of desired clones using the hyperproductivity gene set (reviewed by Vishwanathan et al. 2014). The application of such technologies can help in obtaining next-generation host cells with desired traits. Such Next-Gen host cells can then increase the probability of obtaining hyper producers resulting in faster development of production cell lines (Fig. 2).
Cell line development using Next-Gen host cells. a Development of the Next-Generation host cell Robust host cells with desired characteristics can be obtained by engineering the original host cell line. This robust host cell can then be transfected with a transgene, either with or without targeted integration into desired genomic location, followed by extensive screening for desired traits. This can be expected to result in identification of recombinant clone with optimal traits of growth, quality, secretion and hence productivity. The transgene can then be deleted from this recombinant clone using targeted genome editing to result in a Next-Gen host cell. b Development of the Next-Generation producing cell line The Next-Gen host cell is transfected with the desired new transgene followed by cloning and screening to obtain the final producing cell line. The use of the Next-Gen host cell can potentially result in a high probability of obtaining hyper-producers and a faster cell line development cycle
Single cell based PCR, RNA-seq and genome sequencing may be used to examine the heterogeneity in gene expression or gene/genome sequence in a cell population and may be used to determine the homogeneity of producing cells. Determination of clonality (or the proof that a producing line is indeed originated from a “clone”) is important to manufacturers and regulatory agencies. By surveying “marker” sites on the genome that is unique to each clone, e.g., the transgene integration sites, through single cell PCR, one may be able to determine the clonality of a population of producing cells.
As genomic technology makes inroads into cell line development, the importance of systems biotechnology is also apparent. The availability of transcriptomic and proteomic data has allowed one to have more comprehensive and quantitative understanding of the make-up of metabolic, signalling, secretory, and other pathways. This has allowed for the development of better mathematical models to describe their dynamics. The integration of—omic data with these models may 1 day enable us to choose a production cell line based on the predicted metabolic behavior and product glycosylation pattern.
Concluding remarks
The productivity of protein therapeutics has increased by nearly two orders of magnitude in the past two decades. Efficient development of better cell lines has played a major role in this success. However, the basic method of cell line development remains largely similar over these years. With recent advances in genomics and the development of genome editing tools, we will likely see a transformation of cell line development in the near future. We will likely see increasing efforts on engineering host cells for better growth and metabolic characteristics and other hyperproductivity traits. The deployment of genomic technology and systems approaches to “design” producing cells will likely become the hallmark of the next generation of cell line development.
References
Al-Fageeh MB, Smales CM (2013) Alternative promoters regulate cold inducible RNA-binding (CIRP) gene expression and enhance transgene expression in mammalian cells. Mol Biotechnol 54:238–249
Astley K, Al-Rubeai M (2008) The role of Bcl-2 and its combined effect with p21CIP1 in adaptation of CHO cells to suspension and protein-free culture. Appl Microbiol Biotechnol 78:391–399
Beck A, Wagner-Rousset E, Bussat M-C, Lokteff M, Klinguer-Hamour C, Haeuw J-F, Goetsch L, Wurch T, van Dorsselaer A, Corvaia N (2008) Trends in glycosylation, glycoanalysis and glycoengineering of therapeutic antibodies and Fc-fusion proteins. Curr Pharm Biotechnol 9:482–501
Becker E, Florin L, Pfizenmaier K, Kaufmann H (2008) An XBP-1 dependent bottle-neck in production of IgG subtype antibodies in chemically defined serum-free Chinese hamster ovary (CHO) fed-batch processes. J Biotechnol 135:217–223
Becker E, Florin L, Pfizenmaier K, Kaufmann H (2010) Evaluation of a combinatorial cell engineering approach to overcome apoptotic effects in XBP-1(s) expressing cells. J Biotechnol 146:198–206
Berkowitz SA, Engen JR, Mazzeo JR, Jones GB (2012) Analytical tools for characterizing biopharmaceuticals and the implications for biosimilars. Nat Rev Drug Discov 11:527–540
Borth N, Mattanovich D, Kunert R, Katinger H (2005) Effect of increased expression of protein disulfide isomerase and heavy chain binding protein on antibody secretion in a recombinant CHO cell line. Biotechnol Prog 21:106–111
Boscolo S, Mion F, Licciulli M, Macor P, De Maso L, Brce M, Antoniou MN, Marzari R, Santoro C, Sblattero D (2012) Simple scale-up of recombinant antibody production using an UCOE containing vector. New Biotechnol 29:477–484
Brinkrolf K, Rupp O, Laux H, Kollin F, Ernst W, Linke B, Kofler R, Romand S, Hesse F, Budach WE, Galosy S, Muller D, Noll T, Wienberg J, Jostock T, Leonard M, Grillari J, Tauch A, Goesmann A, Helk B, Mott JE, Puhler A, Borth N (2013) Chinese hamster genome sequenced from sorted chromosomes. Nat Biotechnol 31:694–695
Brown AJ, Sweeney B, Mainwaring DO, James DC (2014) Synthetic promoters for CHO cell engineering. Biotechnol Bioeng 111:1638–1647
Chen KQ, Liu Q, Xie LZ, Sharp PA, Wang DIC (2001) Engineering of a mammalian cell line for reduction of lactate formation and high monoclonal antibody production. Biotechnol Bioeng 72:55–61
Chen A, Chitta R, Chang D, Amanullah A (2009) Twenty-four well plate miniature bioreactor system as a scale-down model for cell culture process development. Biotechnol Bioeng 102:148–160
Chen G, Babb R, Fandl JP (2013) Enhanced expression and stability regions. US Patent 12/793,898
Cho SW, Kim S, Kim Y, Kweon J, Kim HS, Bae S, Kim J-S (2014) Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res 24:132–141
Choi SS, Rhee WJ, Kim EJ, Park TH (2006) Enhancement of recombinant protein production in Chinese hamster ovary cells through anti-apoptosis engineering using 30Kc6 gene. Biotechnol Bioeng 95:459–467
Cost GJ, Freyvert Y, Vafiadis A, Santiago Y, Miller JC, Rebar E, Collingwood TN, Snowden A, Gregory PD (2010) BAK and BAX deletion using zinc-finger nucleases yields apoptosis-resistant CHO cells. Biotechnol Bioeng 105:330–340
De Jesus MJ, Girard P, Bourgeois M, Baumgartner G, Jacko B, Amstutz H, Wurm FM (2004) TubeSpin satellites: a fast track approach for process development with animal cells using shaking technology. Biochem Eng J 17:217–223
Druz A, Son YJ, Betenbaugh M, Shiloach J (2013) Stable inhibition of mmu-miR-466h-5p improves apoptosis resistance and protein production in CHO cells. Metab Eng 16:87–94
Eberwine J, Sul J-Y, Bartfai T, Kim J (2013) The promise of single-cell sequencing. Nat Methods 11:25–27
Estes SD, Zhang W (2014) Beta-actin and RPS21 promoters and uses thereof. US Patent 7423135 B2
Ferrara C, Brunker P, Suter T, Moser S, Puntener U, Umana P (2006) Modulation of therapeutic antibody effector functions by glycosylation engineering: influence of Golgi enzyme localization domain and co-expression of heterologous beta1, 4-N-acetylglucosaminyltransferase III and Golgi alpha-mannosidase II. Biotechnol Bioeng 93:851–861
Figueroa B Jr, Ailor E, Osborne D, Hardwick JM, Reff M, Betenbaugh MJ (2007) Enhanced cell culture performance using inducible anti-apoptotic genes E1B-19K and Aven in the production of a monoclonal antibody with Chinese hamster ovary cells. Biotechnol Bioeng 97:877–892
Frison A, Memmert K, Novartis Pharma A (2002) Fed-batch process development for monoclonal antibody production with Cellferm-pro®. Genet Eng News 22:66–67
Fukuta K, Abe R, Yokomatsu T, Kono N, Asanagi M, Omae F, Minowa MT, Takeuchi M, Makino T (2000) Remodeling of sugar chain structures of human interferon-gamma. Glycobiology 10:421–430
Gaj T, Gersbach CA, Barbas CF 3rd (2013) ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol 31:397–405
Ghirlando R, Files K, Gowher H, Xiao T, Xu Z, Yao H, Felsenfeld G (2012) Chromatin domains, insulators, and the regulation of gene expression. Biochim Biophys Acta 1819:644–651
Girard P, Jordan M, Tsao M, Wurm FM (2001) Small-scale bioreactor system for process development and optimization. Biochem Eng J 7:117–119
Girod PA, Zahn-Zabal M, Mermod N (2005) Use of the chicken lysozyme 5′ matrix attachment region to generate high producer CHO cell lines. Biotechnol Bioeng 91:1–11
Goh JS, Zhang P, Chan KF, Lee MM, Lim SF, Song Z (2010) RCA-I-resistant CHO mutant cells have dysfunctional GnT I and expression of normal GnT I in these mutants enhances sialylation of recombinant erythropoietin. Metab Eng 12:360–368
Grabherr MG, Pontiller J, Mauceli E, Ernst W, Baumann M, Biagi T, Swofford R, Russell P, Zody MC, Di Palma F, Lindblad-Toh K, Grabherr RM (2011) Exploiting nucleotide composition to engineer promoters. PLoS One 6:e20136
Hansen CLG, Lecault V, Piret JM, Singhal A (2010) System and method for microfluidic cell culture. US Patent 13/178,395
Hartenbach S, Fussenegger M (2006) A novel synthetic mammalian promoter derived from an internal ribosome entry site. Biotechnol Bioeng 95:547–559
Hossler P, Goh LT, Lee MM, Hu WS (2006) GlycoVis: visualizing glycan distribution in the protein N-glycosylation pathway in mammalian cells. Biotechnol Bioeng 95:946–960
Hou JJ, Hughes BS, Smede M, Leung KM, Levine K, Rigby S, Gray PP, Munro TP (2014) High-throughput ClonePix FL analysis of mAb-expressing clones using the UCOE expression system. New Biotechnol 31:214–220
Hsu WT, Aulakh RP, Traul DL, Yuk IH (2012) Advanced microscale bioreactor system: a representative scale-down model for bench-top bioreactors. Cytotechnology 64:667–678
Hu Z, Guo D, Yip SS, Zhan D, Misaghi S, Joly JC, Snedecor BR, Shen AY (2013) Chinese hamster ovary (CHO) K1 host cell enables stable cell line development for antibody molecules which are difficult to express in DUXB11-derived dihydrofolate reductase (DHFR) deficient host cell. Biotechnol Prog 29(4):980–985
Hwang SO, Lee GM (2009) Effect of Akt overexpression on programmed cell death in antibody-producing Chinese hamster ovary cells. J Biotechnol 139:89–94
Ifandi V, Al-Rubeai M (2005) Regulation of cell proliferation and apoptosis in CHO-K1 cells by the coexpression of c-Myc and Bcl-2. Biotechnol Prog 21:671–677
Irani N, Wirth M, van Den Heuvel J, Wagner R (1999) Improvement of the primary metabolism of cell cultures by introducing a new cytoplasmic pyruvate carboxylase reaction. Biotechnol Bioeng 66:238–246
Jaluria P, Betenbaugh M, Konstantopoulos K, Shiloach J (2007) Enhancement of cell proliferation in various mammalian cell lines by gene insertion of a cyclin-dependent kinase homolog. BMC Biotechnol 7:71
Jayapal KP, Goudar CT (2014) Transcriptomics as a tool for assessing the scalability of mammalian cell perfusion systems. Adv Biochem Eng Biot 139:227–243
Kanda Y, Imai-Nishiya H, Kuni-Kamochi R, Mori K, Inoue M, Kitajima-Miyama K, Okazaki A, Iida S, Shitara K, Satoh M (2007) Establishment of a GDP-mannose 4,6-dehydratase (GMD) knockout host cell line: a new strategy for generating completely non-fucosylated recombinant therapeutics. J Biotechnol 130:300–310
Khan S, Schröder M (2008) Engineering of chaperone systems and of the unfolded protein response. Cytotechnology 57:207–231
Kim M, O’Callaghan PM, Droms KA, James DC (2011) A mechanistic understanding of production instability in CHO cell lines expressing recombinant monoclonal antibodies. Biotechnol Bioeng 108(10):2434–2446
Kim Y, Baek E, Lee J, Lee G (2013) Autophagy and its implication in Chinese hamster ovary cell culture. Biotechnol Lett 35:1753–1763
Krampe B, Al-Rubeai M (2010) Cell death in mammalian cell culture: molecular mechanisms and cell line engineering strategies. Cytotechnology 62:175–188
Ku SC, Ng DT, Yap MG, Chao SH (2008) Effects of overexpression of X-box binding protein 1 on recombinant protein production in Chinese hamster ovary and NS0 myeloma cells. Biotechnol Bioeng 99:155–164
Kumar N, Gammell P, Clynes M (2007) Proliferation control strategies to improve productivity and survival during CHO based production culture : a summary of recent methods employed and the effects of proliferation control in product secreting CHO cell lines. Cytotechnology 53:33–46
Kuscu C, Arslan S, Singh R, Thorpe J, Adli M (2014) Genome-wide analysis reveals characteristics of off-target sites bound by the Cas9 endonuclease. Nat Biotechnol 32:677–683
Kuystermans D, Al-Rubeai M (2009) cMyc increases cell number through uncoupling of cell division from cell size in CHO cells. BMC Biotechnol 9:76
Le H, Vishwanathan N, Kantardjieff A, Doo I, Srienc M, Zheng X, Somia N, Hu W-S (2013) Dynamic gene expression for metabolic engineering of mammalian cells in culture. Metab Eng 20:212–220
Lee EU, Roth JA, Paulson JC (1989) Alteration of terminal glycosylation sequences on N-linked oligosaccharides of Chinese hamster ovary cells by expression of beta-galatoside alpha 2,6-sialyltransferase. J Biol Chem 264(23):13848–13855
Lee JS, Ha TK, Park JH, Lee GM (2013a) Anti-cell death engineering of CHO cells: co-overexpression of Bcl-2 for apoptosis inhibition, Beclin-1 for autophagy induction. Biotechnol Bioeng 110:2195–2207
Lee KH, Tsutsui T, Honda K, Asano R, Kumagai I, Ohtake H, Omasa T (2013b) Generation of high-producing cell lines by overexpression of cell division cycle 25 homolog A in Chinese hamster ovary cells. J Biosci Bioeng 116:754–760
Legmann R, Schreyer HB, Combs RG, McCormick EL, Russo AP, Rodgers ST (2009) A predictive high-throughput scale-down model of monoclonal antibody production in CHO cells. Biotechnol Bioeng 104:1107–1120
Lewis NE, Liu X, Li Y, Nagarajan H, Yerganian G, O’Brien E, Bordbar A, Roth AM, Rosenbloom J, Bian C, Xie M, Chen W, Li N, Baycin-Hizal D, Latif H, Forster J, Betenbaugh MJ, Famili I, Xu X, Wang J, Palsson BO (2013) Genomic landscapes of Chinese hamster ovary cell lines as revealed by the Cricetulus griseus draft genome. Nat Biotechnol 31:759–765
Liu P-Q, Chan EM, Cost GJ, Zhang L, Wang J, Miller JC, Guschin DY, Reik A, Holmes MC, Mott JE, Collingwood TN, Gregory PD (2010) Generation of a triple-gene knockout mammalian cell line using engineered zinc-finger nucleases. Biotechnol Bioeng 106:97–105
Love KR, Bagh S, Choi J, Love JC (2013) Microtools for single-cell analysis in biopharmaceutical development and manufacturing. Trends Biotechnol 31:280–286
Maeder ML, Thibodeau-Beganny S, Osiak A, Wright DA et al (2008) Rapid “open-source” engineering of customized zinc-finger nucleases for highly efficient gene modification. Mol Cell 31:294–301
Mehling M, Tay S (2014) Microfluidic cell culture. Curr Opin Biotechnol 25:95–102
Moehle EA, Rock JM, Lee YL, Jouvenot Y, DeKelver RC, Gregory PD, Urnov FD, Holmes MC (2007) Targeted gene addition into a specified location in the human genome using designed zinc finger nucleases. Proc Natl Acad Sci U S A 104:3055–3060
Moses S, Manahan M, Ambrogelly A, Ling W (2012) Assessment of AMBR as a model for high-throughput cell culture process development strategy. Adv Biosci Biotechnol 3:918–927
Nawy T (2014) Single-cell sequencing. Nat Methods 11:18
Nehlsen K, Schucht R, da Gama-Norton L, Kromer W, Baer A, Cayli A, Hauser H, Wirth D (2009) Recombinant protein expression by targeting pre-selected chromosomal loci. BMC Biotechnol 9:100
Oberbek A, Matasci M, Hacker DL, Wurm FM (2011) Generation of stable, high-producing CHO cell lines by lentiviral vector-mediated gene transfer in serum-free suspension culture. Biotechnol Bioeng 108:600–610
Ohya T, Hayashi T, Kiyama E, Nishii H, Miki H, Kobayashi K, Honda K, Omasa T, Ohtake H (2008) Improved production of recombinant human antithrombin III in Chinese hamster ovary cells by ATF4 overexpression. Biotechnol Bioeng 100:317–324
Osterlehner A, Simmeth S, Gopfert U (2011) Promoter methylation and transgene copy numbers predict unstable protein production in recombinant Chinese hamster ovary cell lines. Biotechnol Bioeng 108:2670–2681
Paredes C, Prats E, Cairo JJ, Azorin F, Cornudella L, Godia F (1999) Modification of glucose and glutamine metabolism in hybridoma cells through metabolic engineering. Cytotechnology 30:85–93
Porter AJ, Dickson AJ, Racher AJ (2010a) Strategies for selecting recombinant CHO cell lines for cGMP manufacturing: realizing the potential in bioreactors. Biotechnol Prog 26:1446–1454
Porter AJ, Racher AJ, Preziosi R, Dickson AJ (2010b) Strategies for selecting recombinant CHO cell lines for cGMP manufacturing: improving the efficiency of cell line generation. Biotechnol Prog 26:1455–1464
Pradhan K, Pant T, Gadgil M (2012) In situ pH maintenance for mammalian cell cultures in shake flasks and tissue culture flasks. Biotechnol Prog 28:1605–1610
Rahimpour A, Vaziri B, Moazzami R, Nematollahi L, Barkhordari F, Kokabee L, Adeli A, Mahboudi F (2013) Engineering the cellular protein secretory pathway for enhancement of recombinant tissue plasminogen activator expression in Chinese hamster ovary cells: effects of CERT and XBP1s genes. J Microbiol Biotechnol 23:1116–1122
Renner WA, Lee KH, Hatzimanikatis V, Bailey JE, Eppenberger HM (1995) Recombinant cyclin E expression activates proliferation and obviates surface attachment of chinese hamster ovary (CHO) cells in protein-free medium. Biotechnol Bioeng 47:476–482
Sander JD, Joung JK (2014) CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol 32:347–355
Sauerwald TM, Oyler GA, Betenbaugh MJ (2003) Study of caspase inhibitors for limiting death in mammalian cell culture. Biotechnol Bioeng 81:329–340
Seth G, McIvor RS, Hu WS (2006) 17Beta-hydroxysteroid dehydrogenase type 7 (Hsd17b7) reverts cholesterol auxotrophy in NS0 cells. J Biotechnol 121:241–252
Seth G, Charaniya S, Wiaschin KF, Hu WS (2007) In pursuit of a super producer—alternative paths to high producing recombinant mammalian cells. Curr Opin Biotechnol 18:557–564
Sheeley DM, Merrill BM, Taylor LCE (1997) Characterization of monoclonal antibody glycosylation: comparison of expression systems and identification of terminal α-Linked Galactose. Anal Biochem 247:102–110
Shields RL, Lai J, Keck R, O’Connell LY, Hong K, Meng YG, Weikert SH, Presta LG (2002) Lack of fucose on human IgG1 N-linked oligosaccharide improves binding to human Fcgamma RIII and antibody-dependent cellular toxicity. J Biol Chem 277:26733–26740
Sieck JB, Cordes T, Budach WE, Rhiel MH, Suemeghy Z, Leist C, Villiger TK, Morbidelli M, Soos M (2013) Development of a scale-down model of hydrodynamic stress to study the performance of an industrial CHO cell line under simulated production scale bioreactor conditions. J Biotechnol 164:41–49
Sinacore MS, Charlebois TS, Harrison S, Brennan S, Richards T, Hamilton M, Scott S, Brodeur S, Oakes P, Leornard M, Switzer M, Anagnostopoulos A, Foster B, Harris A, Mankowski M, Bond M, Martin S, Adamson SR (1996) CHo DUKX cell lineages preadapted to growth in serum-free suspension culture enable rapid development of cell culture processes for the manufacturing of recombinant proteins. Biotechnol Bioeng 52:518–528
Spahn PN, Lewis NE (2014) Systems glycobiology for glycoengineering. Curr Opin Biotechnol 30C:218–224
Thaisuchat H, Baumann M, Pontiller J, Hesse F, Ernst W (2011) Identification of a novel temperature sensitive promoter in cho cells. BMC Biotechnol 11:51
Tigges M, Fussenegger M (2006) Xbp1-based engineering of secretory capacity enhances the productivity of Chinese hamster ovary cells. Metab Eng 8:264–272
Turan S, Zehe C, Kuehle J, Qiao J, Bode J (2013) Recombinase-mediated cassette exchange (RMCE)—a rapidly-expanding toolbox for targeted genomic modifications. Gene 515:1–27
Vishwanathan N, Le H, Jacob NM, Tsao YS, Ng SW, Loo B, Liu Z, Kantardjieff A, Hu WS (2014) Transcriptome dynamics of transgene amplification in Chinese hamster ovary cells. Biotechnol Bioeng 111:518–528
von Horsten HH, Ogorek C, Blanchard V, Demmler C, Giese C, Winkler K, Kaup M, Berger M, Jordan I, Sandig V (2010) Production of non-fucosylated antibodies by co-expression of heterologous GDP-6-deoxy-d-lyxo-4-hexulose reductase. Glycobiology 20:1607–1618
Weikert S, Papac D, Briggs J, Cowfer D, Tom S, Gawlitzek M, Lofgren J, Mehta S, Chisholm V, Modi N, Eppler S, Carroll K, Chamow S, Peers D, Berman P, Krummen L (1999) Engineering Chinese hamster ovary cells to maximize sialic acid content of recombinant glycoproteins. Nat Biotechnol 17:1116–1121
Wlaschin KF, Hu W-S (2007) Engineering cell metabolism for high-density cell culture via manipulation of sugar transport. J Biotechnol 131:168–176
Wong NS, Yap MG, Wang DI (2006) Enhancing recombinant glycoprotein sialylation through CMP-sialic acid transporter over expression in Chinese hamster ovary cells. Biotechnol Bioeng 93:1005–1016
Xu X, Nagarajan H, Lewis NE, Pan S, Cai Z, Liu X, Chen W, Xie M, Wang W, Hammond S, Andersen MR, Neff N, Passarelli B, Koh W, Fan HC, Wang J, Gui Y, Lee KH, Betenbaugh MJ, Quake SR, Famili I, Palsson BO (2011) The genomic sequence of the Chinese hamster ovary (CHO)-K1 cell line. Nat Biotechnol 29:735–741
Yamane-Ohnuki N, Kinoshita S, Inoue-Urakubo M, Kusunoki M, Iida S, Nakano R, Wakitani M, Niwa R, Sakurada M, Uchida K, Shitara K, Satoh M (2004) Establishment of FUT8 knockout Chinese hamster ovary cells: an ideal host cell line for producing completely defucosylated antibodies with enhanced antibody-dependent cellular cytotoxicity. Biotechnol Bioeng 87:614–622
Yang Y, Mariati J Chusainow, Yap MG (2010) DNA methylation contributes to loss in productivity of monoclonal antibody-producing CHO cell lines. J Biotechnol 147:180–185
Zhang F, Cong L, Lodato S, Kosuri S, Church GM, Arlotta P (2011) Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription. Nat Biotechnol 29:149–153
Zhou M, Crawford Y, Ng D, Tung J, Pynn AF, Meier A, Yuk IH, Vijayasankaran N, Leach K, Joly J, Snedecor B, Shen A (2011) Decreasing lactate level and increasing antibody production in Chinese hamster ovary cells (CHO) by reducing the expression of lactate dehydrogenase and pyruvate dehydrogenase kinases. J Biotechnol 153:27–34
Author information
Authors and Affiliations
Corresponding author
Additional information
Huong Le and Nandita Vishwanathan have contributed equally to this work.
Rights and permissions
About this article
Cite this article
Le, H., Vishwanathan, N., Jacob, N.M. et al. Cell line development for biomanufacturing processes: recent advances and an outlook. Biotechnol Lett 37, 1553–1564 (2015). https://doi.org/10.1007/s10529-015-1843-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-015-1843-z