Abstract
Genetic factors are known to play a significant role in the susceptibility of diabetic patients to severe complications such as diabetic nephropathy (DN). This study aimed to evaluate the association between polymorphism of ectonucleotide pyrophosphatase/phosphodiesterase 1 (ENPP1) variants (rs997509, K121Q, rs1799774, and rs7754561) and DN in patients with type 2 diabetes mellitus (T2DM). A total number of 492 patients with T2DM with and without DN were categorized into case and control groups. The extracted DNA samples were genotyped using TaqMan allelic discrimination assay amplified by polymerase chain reaction (PCR). The haplotype analysis among the case and control groups was performed using an expectation-maximization algorithm by the maximum-likelihood method. The analysis of laboratory findings demonstrated significant differences in fasting blood sugar (FBS) and hemoglobin A1c (HbA1c) between the case and control groups (P < 0.05). The results showed that K121Q was significantly related to DN under a recessive model of inheritance (P = 0.006); however, rs1799774 and rs7754561 both were protective for DN under a dominant model of inheritance (P = 0.034 and P = 0.010, respectively) among four studied variants. Two haplotypes, including C-C-delT-G with a frequency < 0.02 and T-A-delT-G with a frequency < 0.01, were associated with the increased risk of DN (P < 0.05). The present study demonstrated that K121Q was associated with the susceptibility of DN; however, rs1799774 and rs7754561 were protecrtive variants for DN in patients with T2DM.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The number of individuals suffering from diabetes mellitus (DM) is increasing at an alarming rate and has been estimated to be doubled by 2030 (Tomino en Gohda 2015). A large number of these patients are from low and middle-income societies (Whiting et al. 2011). In type 2 diabetes mellitus (T2DM), environmental and genetic factors can cause the progressive loss of β-cell volume and/or function that clinically indicates hyperglycemia (Murea et al. 2012). Diabetes leads to different complications such as damaging blood vessels, the heart, kidneys, eyes, and nerves. One of the most common pathological effects of diabetes is chronic kidney disease (CKD) (Stevens en Levin 2013; Alicic et al. 2017).
CKD is more common in some particular groups, including the elderly, youth onset, overweighs and obsesses, and low or middle-income populations. The increasing prevalence of younger patients with T2DM, who expressed an accelerated period of complexity considerably enhance the global burden of CKD (Thomas et al. 2016). One kind of CKD is diabetic nephropathy (DN), which is a long-term kidney disease identified by persistent albuminuria and a progressive failure in renal function and infers a typical pattern of glomerular disease that occurs in 20–50% of those patients with diabetes (Evans et al. 2018; Selby en Taal 2020; Sawaf et al. 2022). The DN associates with hypertension and other cardiovascular morbidity and mortality (Andrésdóttir et al. 2015; Pelle et al. 2022). The pathogenesis of diabetes and following DN is multifactorial, and genetic and epigenetic play an important role in its progress. Genetic background is an unmodifiable risk factor that should be considered a fundamental one (Tziastoudi et al. 2020). A genome-wide association study demonstrated common known loci that express a relation with T2DM. Ectonucleotide pyrophosphatase/phosphodiesterase 1 (ENPP1) is a member of the ENPP family, detected on the long arm of chromosome 6 (6q23.2), and can encode a protein that negatively affects insulin function by blocking the insulin tyrosine-kinase receptor signals (Sortica et al. 2011; Priščáková et al. 2016). Some studies have indicated that patients with T2DM are resistant to either endogenous or exogenous insulin. In coding and noncoding regions, some genetic diversities resulting in enhanced ENPP1 expression in the liver related to resistance to insulin by preventing tyrosine-kinase activity that might play a main role in DN pathological process (Tanyolaç et al. 2009). The K121Q polymorphism of the ENPP1 gene seems to be associate with insulin resistance and DN development (Sortica et al. 2011). The common variant 121Q related to insulin resistance and T2DM has been compared with its 121 K, causing a decrease in receptor autophosphorylation (Frittitta et al. 2001; Cave et al. 2020). A variant in exon 4 of the ENPP1 gene causes the replacement of lysine (K) to glutamine (Q) in codon 121 (K121Q; rs1044498) (Sortica et al. 2011; Bhatti et al. 2018).
The T allele, which is located in the 3’ end of intron 1, is placed in a region, including ENPP1 gene regulatory element that upturns messenger RNA (mRNA) constancy, and is related to ENPP1 overexpression and insulin resistance (Santoro et al. 2009). Two functionally noncoding uncharacterized polymorphisms that enhance the stability of mRNA are rs7754561 diverse in untranslated region 3’ and rs1799774-/T, that have been located in intron 20 of ENPP1 and are related to ENPP1 overexpression and insulin resistance (Huang et al. 2015). The DN is among the leading causes of end-stage renal disease, imposing serious effects on morbidity, mortality, and patient’s life quality. With this background, the present study aimed to cast light on the association of four variants of ENPP1 and their haplotypes related to DN development in patients with T2DM in Guilan province, Iran.
Materials and Methods
Subjects and Study Design
A total number of 492 patients with T2DM were randomly selected and classified into case and control groups (i.e., patients with T2DM with (n=185) and without DN (n=307), respectively). All samples were selected from Guilan province, Iran. For being included, all the individuals with T2DM were identified according to plasma fasting blood sugar (FBS) level > 126 mg/dl or/and hemoglobin A1c (HbA1c) > 6% (Committee 2009; Razi et al. 2018), and also according to applying prescriptions for diabetic patients. The exclusion criteria were included type 1 diabetes, impaired glucose tolerance, metabolic syndrome, and pregnancy diabetes. In addition, patients with positive test results for two of three tests of the microalbuminuric higher than 300 mg of albumin/g of creatinine (Cr) in a six-month period and/or biopsy-proven DN, who were diagnosed by a nephrologist, met the criteria for DN (Caramori et al. 2002). The exclusion criteria of DN were nondiabetic renal diseases, Addison’s disease, and other primary kidney disorders. Serum FBS test, HbA1c level, systolic blood pressure (SBP), and diastolic blood pressure (DBP) were measured. Moreover, the body mass index (BMI) was calculated for all participants. The informed consent of patients was consciously taken to participate in the present investigation. This study was in accordance with the ethical code of human genome/gene research at the Guilan University of Medical Science [IR.GUMS.REC.1394.341].
Genotyping
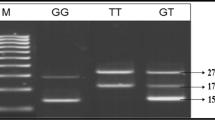
The extracted DNA samples (Keshavarz et al. 2014) that were isolated based on the salting-out method with the standard procedure from whole blood (Miller et al. 1988) were used for the study. Furthermore, sample purifying was confirmed via a NanoDrop spectrophotometer and agarose gel electrophoresis with a sharp single band (Lee et al. 2012; García-Alegría et al. 2020). All four variants of ENPP1 were genotyped on the ABI7300 system (Applied Biosystems, Foster City, CA, US) by TaqMan discrimination assay technology (using the 5’ nuclease assay for amplifying and detecting specific SNP alleles). The TaqMan genotyping reaction was amplified on a real-time polymerase chain reaction (PCR) (denatured at 95 °C for 10 second, 45 cycles for denaturation at 94 °C for 15 second, and annealing for 1 minutes at 60 °C; fluorescence dyes and ABI Prism 7300 HT sequence detector system by Safety Data Sheet software, version 2.1). Genotyping the data was established from over 99% of the genomic DNA samples.
Analysis of Haplotypes
The haplotype analysis of four variants were evaluated for the possible effects of combinations of the variants on DN risk. Significant haplotypes were selected by a frequency higher than 5%. The haplotype analysis among the case and control groups was performed using an expectation-maximization algorithm by the maximum-likelihood method. By comparing the frequency of haplotypes between both groups, permutation p-values were calculated with the basis of 10,000 replications. The data analysis of haplotypes, linkage disequilibrium, and pairwise delta (correlation coefficient) was performed via SNPAlyze software (version 8.1; Dynacom, Japan). Moreover, online analysis with Web-Assotest was performed to find association models (26).
Statistical Analysis
SPSS software (version 24) was used to determine genotype-phenotype and other relations of the studied variants with biochemical and clinical data. In both case and control groups, genotype distribution was performed by Hardy-Weinberg Equilibrium (HWE) (CI: 95%) to determine the association between the variants and DN risk. The data were represented as mean ± standard deviation (SD) and the categorical data were expressed as numbers and percentage. The normality of data was checked using Kolmogorov-Smirnov normality test. Moreover, to compare parametric variants among case and control groups, Chi-square test, Fisher’s exact test, Anova, and independent-sample t-test were performed were performed; and in the case of nonparametric data, Mann Whitney and Kruskal-Wallis were used. All statistical assessments were two-sided and the significant level was considered if P < 0.05.
Results
The participants included 185 patients with T2DM and DN by a mean age of 52.53 ± 9.56 years old that 133 were females and 52 were males. Moreover, 307 patients without DN by a mean age of 51.93 ± 9.10 years old, included 225 females and 82 males. Table 1 shows the clinical characteristics of patients with T2DM in both case and control groups. The results indicated significant differences in FBS and Hb1AC between the two studied groups (P = 0.002 and P < 0.001, respectively). These data demonstrated that the mean levels of FBS and Hb1AC were higher in T2DM patients with DN than those without DN.
Genotype-Phenotype Association of ENPP1 Variants
The genotype and allele frequency of all four investigated variants among 185 patients with T2DM and DN and 307 patients with T2DM and without DN demonstrated that K121Q (P = 0.006), rs1799774 (P = 0.034), and rs7754561 (P = 0.010) were considered significant as a recessive, dominant, and dominant models of inheritance, respectively. The distribution of genotype polymorphisms in the case group was in HWE for rs997509, rs1799774, and rs7754561 (P = 0.975, P = 0.058, and P = 0.985, respectively), but not for K121Q (P = 0.001); and in the control group for rs997509, K121Q, rs1799774, and rs7754561 (P = 0.830, P = 0.130, P = 730, and P = 0.769, respectively). Table 2 shows that the K121Q genotype prevalence among patients with and without DN was significantly different (P = 0.012). This finding showed that K121Q had an association with DN in the recessive inheritance model (P = 0.006). The other significant variant, rs7754561, was different between the two study groups in the allelic level (P = 0.007). Both rs1799774 (P = 0.034; OR 0.66; 95% CI: 0.45–0.97) and rs7754561 (P = 0.010; OR 0.48; 95% CI: 0.28–0.85) played a protective role for DN in patients with T2DM via a dominant model of inheritance.
Haplotype Analysis in Patients with T2DM with and Without DN
All of the above-mentioned variants were examined to find assumed the associated haplotypes. Accordingly, 11 haplotypes with a frequency more than 1% and 5 haplotypes with a frequency more than 5% were obtained. The analysis of haplotypes was performed with haplotype distributions comparing in both case and control groups. None of the haplotypes with a frequency higher than 10% had a significant difference among groups; however, C-C-delT-G and T-A-delT-G were two haplotypes by the allele frequency more than 2% and 1% (P = 0.005 and P = 0.004, respectively), which indicated a weak association of haplotypes in cases and controls (Table 3).
Genotype-Phenotype Sub Analysis
According to Table 4, for other three variants, no significant statistical difference in biochemical and clinical characteristics between the case and control groups in wild type, heterozygote, and homozygote genotypes of patients was reported for K121Q, while in control group of rs1799774 and rs7754561 variants, the mean of FBS and the frequency of gender, respectively, were significantly difference among wild type, heterozygote, and homozygote genotypes (P < 0.05).
Discussion
A dramatically rising global burden of DN that seriously increases the risk of health complications, attracts healthcare system attention to the fundamental reasons for this disease (Zhang et al. 2020). Due to the remarkable effect of genetics on the susceptibility of individuals to various diseases, the study of genes casts light on numerous questions in terms of comorbidities and worsen condition of diseases. The present investigation demonstrated that K121Q had significant associations with DN. Furthermore, rs1799774 and rs7754561 variants were amount of protective variants for DN in T2DM patients. A study has indicated a significant association of KQ polymorphism of the ENPP1 gene with diabetic kidney disease in the Indian population (Chandra et al. 2021), representing a certain effect of genetic factors on DN occurrence.
The significant association of FBS and HbA1c with K121Q through genotype-phenotype analysis revealed that the mean levels of FBS and HbA1c were significantly higher in patients with T2DM with DN than those without DN, which can suggest the association of this variable with DN. A study by Lind et al., reported that the risk of severe complications for patients with diabetes mainly happened at HbA1c levels > 8.6% (Lind et al. 2019). Cave et al. showed the association of the K121Q A allele with a glomerular filtration rate (GFR) decrease in the black population of South Africa (Cave et al. 2020), which indicated that genetic factors play an important role in susceptibility to DN in patients with T2DM. Some studies obtained the important role of genetic variations in DNA methylation and highlighted loci, where methylation and gene-expression changes likely mediate the genotype effect on kidney disease development (Nazir et al. 2014; Sheng et al. 2020). Keen et al. reported the association of rs7754561 and rs1799774 with DN. They also reported that K121Q had no associated with DN (Keene et al. 2008).
An investigation carried out by Gohari-Lasaki et al. suggested that rs997509 and rs7754561 associated with the susceptibility of diabetic retinopathy in patients with T2DM. Moreover, rs7754561 could be a potential genetic indicator for the prognosis and diagnosis of diabetic retinopathy (Gohari-Lasaki et al. 2020). Another study demonstrated that K121Q is significantly expressed in patients with diabetes and bone osteoporosis that resulted in severe complications (Neamati et al. 2017). Ghada et al. reported that ENPP1 K121Q (A/C, rs1044498) variant was associated with DN. They demonstrated that patients, with the minor/risk allele, had significantly higher moderately increased albuminuria/severely increased albuminuria levels and albumin/creatinine ratio compare to those with the wild A allele (Omar et al. 2022).
Moreover, as ENPP1 expressed in many organs including kidney, parathyroid, cartilage, heart, and skeletal muscle, and it is also highly expressed in vascular smooth muscle cells, osteoblasts, and chondrocytes, it has a significant role in regulating calcium and phosphorus and inhibiting soft tissue mineralization that results in many complications in the case of disruption (Cimpean et al. 2017). Increased vascular calcification in patients with end-stage renal disease (ESRD) is one of the complications associated with the ENPP1 K121Q polymorphism (Eller et al. 2008). Wu et al. reported that the levels of serum ENPP1 in non-diabetic ESRD patients were negatively related to the severity of abdominal aortic vascular calcification. They found that among ESRD patients, the severity of vascular calcification correlated with serum ENPP1 value, the severer the vascular calcification, the lower the serum ENPP1 level (Wu et al. 2022). Due to the limited information on the burden of DN, its genetic background and high prevalence in patients with T2DM in Iran (Rabieenia et al. 2020), it is essential that healthcare systems consider effective measures for its diagnosis at the early stage to reduce this complication for better clinical management of patients with diabetic.
Limitation
The limitations of our study were small sample size and diverse ethnicity of population in Guilan province, Iran, which might have resulted in low frequency of homozygotes genotype among the population.
Conclusion
The results of this study revealed that K121Q had an association with DN in patients with T2DM. This study highlighted the role of k121Q variant in susceptibility of patients with T2DM to further severe conditions such as DN.
List of Variables
Ectonucleotide pyrophosphatase/phosphodiesterase 1 (ENPP1); diabetic nephropathy (DN); type 2 diabetes mellitus (T2DM); polymerase chain reaction (PCR); chronic kidney disease (CKD); messenger RNA (mRNA); fasting blood sugar (FBS); Hardy-Weinberg Equilibrium (HWE).
Data Availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Alicic RZ, Rooney MT, Tuttle KR (2017) Diabetic kidney disease: Challenges, Progress, and possibilities. Clin J Am Soc Nephrol 12:2032–2045. https://doi.org/10.2215/CJN.11491116
Andrésdóttir G, Jensen ML, Carstensen B et al (2015) Improved prognosis of diabetic nephropathy in type 1 diabetes. Kidney Int 87:417–426. https://doi.org/10.1038/ki.2014.206
Bhatti GK, Kaur S, Vijayvergiya R et al (2018) ENPP1 K121Q functional variant enhances susceptibility to insulin resistance and dyslipidemia with metabolic syndrome in asian Indians. Dubai Diabetes Endocrinol J 24:8–15. https://doi.org/10.1159/000492478
Caramori ML, Kim Y, Huang C et al (2002) Cellular basis of diabetic nephropathy: 1. Study design and renal structural-functional relationships in patients with long-standing type 1 diabetes. Diabetes 51:506–513. https://doi.org/10.2337/diabetes.51.2.506
Cave EM, Prigge KL, Crowther NJ et al (2020) A polymorphism in the gene encoding the insulin receptor binding protein ENPP-1 is Associated with decreased glomerular filtration rate in an under-investigated indigenous African Population. Kidney Blood Press Res 45:1009–1017. https://doi.org/10.1159/000511213
Chandra S, Singh AK, Singh M et al (2021) Human ENPP1 gene polymorphism in DKD patients: a hospital-based case control study. Int J Diabetes Dev Ctries 41:63–70. https://doi.org/10.1007/s13410-020-00841-4
Cimpean A, Stefan C, Gijsbers R et al (2017) Correction: substrate-specifying determinants of the nucleotide pyrophosphatases/phosphodiesterases NPP1 and NPP2. Biochem J 474:4273–4274
Committee IE (2009) International Expert Committee report on the role of the A1C assay in the diagnosis of diabetes. Diabetes Care 32:1327–1334
Eller P, Hochegger K, Feuchtner GM et al (2008) Impact of ENPP1 genotype on arterial calcification in patients with end-stage renal failure. Nephrol Dial Transplant 23:321–327
Evans K, Pyart R, Steenkamp R et al (2018) UK Renal Registry 20th Annual Report: introduction. Nephron 139(Suppl):1–12
Frittitta L, Ercolino T, Bozzali M et al (2001) A cluster of three single nucleotide polymorphisms in the 3’-Untranslated region of human glycoprotein PC-1 gene stabilizes PC-1 mRNA and is Associated with increased PC-1 protein content and insulin resistance-related abnormalities. Diabetes 50:1952–1955. https://doi.org/10.2337/diabetes.50.8.1952
García-Alegría AM, Anduro-Corona I, Pérez-Martínez CJ et al (2020) Quantification of DNA through the NanoDrop Spectrophotometer: Methodological Validation Using Standard Reference Material and Sprague Dawley Rat and Human DNA. Int J Anal Chem 2020:8896738. https://doi.org/10.1155/2020/8896738
Gohari-Lasaki S, Sharafshah A, Abbaspour S, Keshavarz P (2020) Single locus and haplotype association of ENPP1 gene variants with the development of retinopathy among type 2 diabetic patients. Int Ophthalmol 40:639–647. https://doi.org/10.1007/s10792-019-01224-3
Huang GM, Huang KY, Lee TY, Weng JTY (2015) An interpretable rule-based diagnostic classification of diabetic nephropathy among type 2 diabetes patients. BMC Bioinformatics 16:1–10. https://doi.org/10.1186/1471-2105-16-S1-S5
Keene KL, Mychaleckyj JC, Smith SG et al (2008) Association of the Distal Region of the Ectonucleotide. Diabetes 57:1057–1062. https://doi.org/10.2337/db07-0886.AIM
Keshavarz P, Habibipour R, Ghasemi M et al (2014) Lack of genetic susceptibility of KCNJ11 E23K polymorphism with risk of type 2 diabetes in an iranian population. Endocr Res 39:120–125. https://doi.org/10.3109/07435800.2013.860607
Lee PY, Costumbrado J, Hsu C-Y, Kim YH (2012) Agarose gel electrophoresis for the separation of DNA fragments. J Vis Exp 3923. https://doi.org/10.3791/3923
Lind M, Pivodic A, Svensson AM et al (2019) HbA 1c level as a risk factor for retinopathy and nephropathy in children and adults with type 1 diabetes: swedish population based cohort stuady. BMJ 366. https://doi.org/10.1136/bmj.l4894
Miller SA, Dykes DD, Polesky HF (1988) A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 16:1215. https://doi.org/10.1093/nar/16.3.1215
Murea M, Ma L, Freedman BI (2012) Genetic and environmental factors associated with type 2 diabetes and diabetic vascular complications. Rev Diabet Stud 9:6–22. https://doi.org/10.1900/RDS.2012.9.6
Nazir N, Siddiqui K, Al-Qasim S, Al-Naqeb D (2014) Meta-analysis of diabetic nephropathy associated genetic variants in inflammation and angiogenesis involved in different biochemical pathways. BMC Med Genet 15:1–14. https://doi.org/10.1186/s12881-014-0103-8
Neamati N, Hosseini SR, Hajiahmadi M et al (2017) The ENPP1 K121Q polymorphism modulates developing of bone disorders in type 2 diabetes: a cross sectional study. Gene 637:100–107. https://doi.org/10.1016/j.gene.2017.09.042
Omar GA, Khalid MM, Ghanem AI (2022) Ectonucleotide pyrophosphatase/phosphodiesterase 1 (K121Q rs1044498) polymorphism is associated with diabetic nephropathy but not obesity among type-2 diabetes mellitus egyptian patients. J Med Sci Res 5:72
Pelle MC, Provenzano M, Busutti M et al (2022) Up-Date on diabetic nephropathy. Life 12:1202
Priščáková P, Minárik G, Repiská V (2016) Candidate gene studies of diabetic retinopathy in human. Mol Biol Rep 43:1327–1345. https://doi.org/10.1007/s11033-016-4075-y
Rabieenia E, Jalali R, Mohammadi M (2020) Prevalence of nephropathy in patients with type 2 diabetes in Iran: a systematic review and meta-analysis based on geographic information system (GIS). Diabetes Metab Syndr 14:1543–1550. https://doi.org/10.1016/j.dsx.2020.08.007
Razi F, Khashayar P, Ghodssi-Ghassemabadi R et al (2018) Optimal HbA1c cut-off point for diagnosis of type 2 diabetes in iranian adults. Can J Diabetes 42. https://doi.org/10.1016/j.jcjd.2018.03.005
Santoro N, Cirillo G, Lepore MG et al (2009) Effect of the rs997509 polymorphism on the association between ectonucleotide pyrophosphatase phosphodiesterase 1 and metabolic syndrome and impaired glucose tolerance in childhood obesity. J Clin Endocrinol Metab 94:300–305. https://doi.org/10.1210/jc.2008-1659
Sawaf H, Thomas G, Taliercio JJ et al (2022) Therapeutic advances in diabetic nephropathy. J Clin Med 11:378
Selby NM, Taal MW (2020) An updated overview of diabetic nephropathy: diagnosis, prognosis, treatment goals and latest guidelines. Diabetes Obes Metab 22:3–15. https://doi.org/10.1111/dom.14007
Sheng X, Qiu C, Liu H et al (2020) Systematic integrated analysis of genetic and epigenetic variation in diabetic kidney disease. Proc Natl Acad Sci U S A 117:29013–29024. https://doi.org/10.1073/pnas.2005905117
Sortica DA, Crispim D, Zaffari GP et al (2011) The role of ecto-nucleotide pyrophosphatase/phosphodiesterase 1 in diabetic nephropathy. Arq Bras Endocrinol Metabol 55:677–685. https://doi.org/10.1590/s0004-27302011000900002
Stevens PE, Levin A (2013) Evaluation and management of chronic kidney disease: Synopsis of the kidney disease: improving global outcomes 2012 clinical practice guideline. Ann Intern Med 158:825–830. https://doi.org/10.7326/0003-4819-158-11-201306040-00007
Tanyolaç S, Bremer AA, Hodoglugil U et al (2009) Genetic variants of the ENPP1/PC-1 gene are associated with hypertriglyceridemia in male subjects. Metab Syndr Relat Disord 7:543–548. https://doi.org/10.1089/met.2009.0027
Thomas MC, Cooper ME, Zimmet P (2016) Changing epidemiology of type 2 diabetes mellitus and associated chronic kidney disease. Nat Rev Nephrol 12:73–81. https://doi.org/10.1038/nrneph.2015.173
Tomino Y, Gohda T (2015) The Prevalence and Management of Diabetic Nephropathy in Asia. Kidney Dis 1:52–60. https://doi.org/10.1159/000381757
Tziastoudi M, Stefanidis I, Zintzaras E (2020) The genetic map of diabetic nephropathy: evidence from a systematic review and meta-analysis of genetic association studies. Clin Kidney J 13:768–781. https://doi.org/10.1093/ckj/sfaa077
Whiting DR, Guariguata L, Weil C, Shaw J (2011) IDF Diabetes Atlas: global estimates of the prevalence of diabetes for 2011 and 2030. Diabetes Res Clin Pract 94:311–321. https://doi.org/10.1016/j.diabres.2011.10.029
Wu X, Di F, Shen S et al (2022) Levels of serum ecto-nucleotide pyrophosphatase/phosphodiesterase 1 (ENPP1) predicts severity of abdominal aortic calcification in end-stage renal disease patients receiving regular dialysis. Hemodial Int 26:23–29. https://doi.org/10.1111/hdi.12969
Zhang X-X, Kong J, Yun K (2020) Prevalence of diabetic nephropathy among patients with type 2 diabetes mellitus in China: a meta-analysis of observational studies. J Diabetes Res 2020:Web assotest.
Acknowledgements
We thankfully express our thanks to all those blood donors of the survey subjects for their contribution to the DNA samples.
Funding
No funding.
Author information
Authors and Affiliations
Contributions
[Parvaneh Keshavarz] and [Niloofar Faraji] contributed to the study design, Material preparation, and performing molecular technics. Data collection and statistical analysis were performed by [Saima Abbaspour], and [Farzam Ajamian]. The whole draft was written by [Niloofar Faraji] and [Parvaneh Keshavarz] and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors have no relevant financial or non-financial interests to disclose.
Ethical Approval
All procedures performed in studies involving human participants were in accordance with the ethical code of human genome/gene research at the Guilan University of Medical Science [IR.GUMS.REC.1394.341].
Consent to participate
Informed consent was obtained from all individual participants included in the study.
Consent for publication
The authors affirm that human research participants provided informed consent for publication of the study.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Faraji, N., Abbaspour, S., Ajamian, F. et al. Role of ENPP1 Gene Variants in the Susceptibility to Diabetic Nephropathy in Patients with type 2 Diabetes Mellitus. Biochem Genet 61, 2710–2723 (2023). https://doi.org/10.1007/s10528-023-10402-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10528-023-10402-z