Abstract
This study aimed to investigate more detailed functions of circ_0005276 in prostate cancer (PCa) and provide a novel mechanism for circ_0005276 action. The expression of circ_0005276, microRNA-128-3p (miR-128-3p) and DEP domain containing 1B (DEPDC1B) was detected by quantitative real-time PCR. In functional assays, cell proliferation was determined by CCK-8 assay and EdU assay. Cell migration and invasion were determined by transwell assay. The ability of angiogenesis was determined by tube formation assay. Cell apoptosis was determined by flow cytometry assay. The potential binding relationship between miR-128-3p and circ_0005276 or DEPDC1B was ascertained by dual-luciferase reporter assay and RIP assay. Mouse models were used to verify the role of circ_0005276 in vivo. The upregulation of circ_0005276 was determined in PCa tissues and cells. Circ_0005276 knockdown inhibited proliferation, migration, invasion and angiogenesis in PCa cells, and circ_0005276 knockdown also blocks tumor growth in vivo. Mechanism analysis discovered that miR-128-3p was a target of circ_0005276, and miR-128-3p inhibition recovered circ_0005276 knockdown-inhibited proliferation, migration, invasion and angiogenesis. In addition, DEPDC1B was a target of miR-128-3p, and miR-128-3p restoration-inhibited proliferation, migration, invasion and angiogenesis were rescued by DEPDC1B overexpression. Circ_0005276 might promote the development of PCa by activating the expression of DEPDC1B via targeting miR-128-3p.
Graphical Abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Prostate cancer (PCa) represents the most prevalent type of cancer among men, with approximate 1,414,259 new cases and 375,304 new deaths worldwide in 2020 (Sung et al. 2021). The 5-year survival is high (98.9%) for patients with non-metastatic PCa, while the 5 years are low (only 20.2%) for patients with metastatic PCa (Siegel et al. 2018). PCa is mostly indolent at first and can later develop into aggressive disease (Attard et al. 2016). Therefore, an in-depth understanding of PCa pathogenesis is necessary to improve the early diagnosis of PCa.
Published studies suggest that circular RNAs (circRNAs) are vital regulators in the development of human diseases, including tumorigenesis (Kristensen et al. 2018). CircRNA is a type of non-coding RNA (ncRNA) that undergoes back-splicing from precursor mRNA (Hua et al. 2019). Recent studies have revealed the widespread and valued functions of circRNA molecules in PCa. For example, circRNA-102004 was highly expressed in PCa tissues, and it functioned as an oncogene by promoting PCa cell migration, invasion, and proliferation (Si-Tu et al. 2019). Circ_0005276, deriving from X-linked inhibitor of apoptosis protein (XIAP), was reported to contribute to PCa progression (Feng et al. 2019). Nonetheless, the role of circ_0005276 in PCa is not completely understood.
CircRNA may function as miRNA sponge to sequester miRNA expression (Witkos et al. 2018). Therefore, the study of circRNA-targeted miRNAs is constructive to explore the functional mechanisms of circRNAs. The advance of bioinformatics makes it easier to predict circRNA-targeted miRNAs (Jiang and Ye 2019). To clarify the mechanism of circ_0005276 action in PCa, we characterized miRNAs targeted by circ_0005276 and obtained miR-128-3p. MiR-128-3p, as a tumor suppressor in PCa, has been previously reported (Khan et al. 2010). Here, we further demonstrated the interplays between circ_0005276 and miR-128-3p in PCa (Table 1).
DEP domain containing 1B (DEPDC1B) was widely reported to be an oncogene in various cancers, including PCa (Bai et al. 2017). In this study, we showed that miR-128-3p depleted DEPDC1B expression by binding to DEPDC1B 3’ untranslated region (3’ UTR), suggesting that DEPDC1B might be involved in PCa progression via miR-128-3p-mediated manner. However, the crosstalk between miR-128-3p and DEPDC1B has not been clarified.
Herein, we investigated the function of circ_0005276 in PCa cells in vitro and in nude mice in vivo. Importantly, we determined the crosstalk between miR-128-3p and circ_0005276 or DEPDC1B, thus providing a new mechanism regarding circ_0005276 function in PCa.
Materials and Methods
Clinical Samples
PCa tumor tissues (n = 30) and matched normal tissues (n = 30) were obtained from Huangshi Central Hospital, Affiliated Hospital of Hubei Polytechnic University. The written informed consent was obtained from each patient. For specimen storage, these tissues frozen by liquid nitrogen were preserved at − 80 °C condition. This study obtained the permission of the Ethics Committee of Huangshi Central Hospital, Affiliated Hospital of Hubei Polytechnic University.
Cell Lines
PCa cells (LNCaP and DU145) were bought from Procell (Wuhan, China) and cultured in RPMI1640 medium (Procell) added with 10% FBS (Procell). Human normal prostatic epithelial cells (RWPE-1; Procell) were used as non-cancer control and cultured in Keratinocyte serum-free medium (K-SFM; Procell) supplemented with 0.05-mg/mL bovine pituitary extract (BPE; Procell) and 5-ng/mL epidermal growth factor (EGF; Procell).
Quantitative Real-time PCR (qPCR)
Total RNA was conventionally isolated using Trizol reagent (Invitrogen, Carlsbad, CA, USA) and checked by Nanodrop 2000 (Thermo Fisher Scientific, Waltham, MA, USA). The assay of cDNA synthesis was carried out using TruScript First-Strand cDNA Synthesis Kit (Norgenbiotek, Ontario, Canada) or microScript microRNA cDNA Synthesis Kit (Norgenbiotek). SYBR GreenER™ qPCR SuperMix Universal (Invitrogen) was then used for qPCR amplification. GAPDH or U6 was used as the internal reference, using the 2−ΔΔCt method for relative expression calculation. Primer sequences are shown here: circ_0005276, F 5’-ACTAGAAGAATTGGTGAAGGTGA-3’, and R 5′-TAGCATGTTGTTCCCAAGGG-3′; XIAP, F 5′-ACTTCGGGTTTCACGACTCC-3′, and R 5′-CCGAGCCCCAATCTGGAAAT-3′; miR-128-3p, F 5′-GGTCACAGTGAACCGGTC-3′, and R 5′-GTGCAGGGTCCGAGGT-3′; DEPDC1B, F 5′-ATAGAGGAGCGTGTGGCTCA-3′, and R 5′-TGACGGCAAAATGATGGAGC-3′; GAPDH, F 5′-CATGGGTGTGAACCATGAGAAGTA-3′, and R 5′-CAGTAGAGGCAGGGATGATGTTCT-3′; U6, F 5′-TGCGGGTGCTCGCTTCGGCAGC-3′, and R 5′-CCAGTGCAGGGTCCGAGGT-3′.
RNase R and Actinomycin D Treatment
For circ_0005276 stability and circularity analysis, total RNA was administered with RNase R (3 U/μg; Epicenter Technologies, Madison, WI, USA). Actinomycin D (2 μg/mL; Sigma-Aldrich) was supplemented into culture medium, and the treated cells were collected for RNA isolation. After treatment with RNase R or actinomycin D, qPCR was performed to determine the expression of circ_0005276 and its linear transcript.
Oligo (dT)18 Primer Amplification
To detect the circular structure of circ_0005276, oligo (dT)18 primers and random primers purchased from Thermo Fisher Scientific were used to synthesize cDNA.
Cell Transfection
Circ_0005276 small interference RNA (si-circ_0005276) and siRNA negative control (si-NC) were customized by Geneseed (Guangzhou, China). MiR-128-3p mimic, miR-128-3p inhibitor and their matched negative control (miRNA NC and inhibitor NC), fusion overexpression vector pcDNA-DEPDC1B (pc-DEPDC1B), and black negative control (pc-NC) were purchased from Ribobio (Guangzhou, China). LNCaP and DU145 cells were subjected to transfections using Lipofectamine 3000 reagent (Invitrogen).
Cell Counting Kit-8 (CCK-8) Assay
LNCaP and DU145 cells were seeded into 96-well plates (2000 cell/well). After incubation for 24 h, cells were treated with CCK-8 reagent (10 μL/well; Dojindo, Kumamoto, Japan) for another 2 h. Cell viability was examined using a microplate absorbance reader (Bio-Rad, Hercules, CA, USA).
5-Ethynyl-2’-deoxyuridine (EdU) assay
EdU assay was carried out using EdU Imaging Kit (Life technologies, Gaithersburg, MD, USA). Cells were plated into 96-well plates at 60–70% confluence and labeled with 20-μM EdU solution for 2 h. Cells were then fixed using 3.7% paraformaldehyde in PBS for 20 min in the dark and washed using PBS. Cells were stained with anti-EdU working solution, and cell nuclei were labeled by 4’, 6-diamidino-2-phenylindole (DAPI). The number of EdU-positive cells was observed using a fluorescent microscope (Olympus, Tokyo, Japan).
Transwell Assay
Cells suspended into serum-free culture medium were added into the upper chambers (Corning Incorporated, Corning, NY, USA) coated with or without Matrigel (BD Biosciences; San Jose, CA, USA) for cell invasion or cell migration, respectively. Culture medium added with 20% FBS was filled into the bottom of chambers. After incubation for 24 h, cells that migrated or invaded into the lower surface of chambers were immobilized with paraformaldehyde and stained with crystal violet (Invitrogen). The number of migrated or invaded cells was counted by light microscopy (Olympus) in five random fields per replicate.
Tube Formation Assay
The conditioned mediums of LNCaP and DU145 cells were collected at 48-h incubation in serum-free culture medium. 50-μL ice-cold Matrigel (BD Biosciences) was plated into 96-well plates for 30 min. Human umbilical vein endothelial cells (HUVECs; 4 × 104) in 100-μL conditioned medium were pipetted into the well and cultured for 5 h at 37 °C. Tube formation in each well was observed by a light microscope (Olympus).
Flow Cytometry Assay
To detect cell apoptosis, cells after treatment were examined using Cell Apoptosis Kit with Annexin V FITC and PI, for flow cytometry (Invitrogen). This assay was conducted in line with the guideline. BD FACSVerse™ Flow Cytometer (BD Biosciences) was used for the identification of apoptotic cells.
Dual-Luciferase Reporter Assay
The binding site between miR-128-3p and circ_0005276 or DEPDC1B 3’UTR was provided by circBANK (http://www.circbank.cn/) or targetscan (http://www.targetscan.org/vert_72/). The mutant-type (MUT) and wild-type (WT) sequence of circ_0005276 were inserted into pmirGLO vector (Promega, Madison, WI, USA) to obtain reporter plasmids (WT-circ_0005276 and MUT-circ_0005276). Luciferase reporter plasmids (WT-DEPDC1B-3’UTR and MUT-DEPDC1B-3’UTR) were also constructed. For dual-luciferase reporter assay, LNCaP and DU145 cells were cotransfected with miR-128-3p mimic (or miRNA NC) and WT-circ_0005276 (or MUT-circ_0005276) and incubated for 48 h. Dual-Luciferase® Reporter Assay System (Promega) was employed to analyze luciferase activity.
RNA Immunoprecipitation (RIP) Assay
Imprint RIP Kit (Sigma-Aldrich, St. Louis, MO, USA) was used for RIP assay. Ago2 antibody (Sigma-Aldrich), a key component of the miRNA-containing RISC complex, was used to enrich miR-128-3p RISC, with mouse IgG (Sigma-Aldrich) as a negative control. The enrichment of miR-128-3p and circ_0005276 in the precipitates was ascertained by qPCR.
Western Blot
Total protein was obtained using ReadyPrep™ Protein Extraction Kit (Bio-Rad) and examined using Bio-Rad protein assay reagent (Bio-Rad). Protein samples were then loaded on 10% SDS-PAGE, transferred to PVDF membranes (Bio-Rad), and blocked in 5% skim milk. Membranes were then incubated with the specific primary antibodies, including anti-Bax (PA5-11,378; Invitrogen), anti-Bcl-2 (PA5-27,094; Invitrogen), anti-DEPDC1B (PA5-67,248; Invitrogen), anti-PI3K (MA5-15,030; Invitrogen), anti-Phospho-PI3K (PA5-104,853; Invitrogen), anti-AKT (MA5-15,591; Invitrogen), anti-Phospho-AKT (44-602G; Invitrogen), and anti-GAPDH (PA1-16,777; Invitrogen). Membranes were then reacted with a secondary horseradish peroxidase-conjugated secondary antibody (31,460; Invitrogen). The protein blots were viewed using enhanced chemiluminescence (Bio-Rad).
Mouse Models
To construct mouse models, balb/c mice (male, 6-week-old) were bought from Vital River (Beijing, China) and then regularly housed. Mice were randomly assigned into 2 groups (n = 6 per group) and administered with the injection of DU145 cells infecting with lentivirus-integrated sh-circ_0005276 (Genepharma) or sh-NC. During tumor growth, tumor volume (length × width2 × 0.5) was weekly recorded. We excised tumor tissues after 4 weeks and measured tumor weight. Animal study was approved by the Animal Care and Use Committee of Huangshi Central Hospital, Affiliated Hospital of Hubei Polytechnic University.
Statistical Analysis
Each assay was performed 3 times to obtain the experimental data. The data were then processed and analyzed using GraphPad Prism 7 (GraphPad Inc., La Jolla, CA, USA). Multiple comparisons in various groups were performed using Student’s t test or analysis of variance (ANOVA, followed by Tukey’s multiple comparisons test). Pearson’s correlation analysis was used to analyze the correlation between two sets. The data were eventually shown as mean ± standard deviation (SD). P-value less than 0.05 was considered to be significant in statistics.
Results
Circ_0005276 was Highly Expressed in PCa Tumor Tissues and Cells
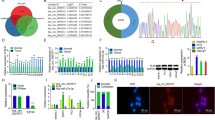
We identified the expression and characteristics of circ_0005276 in PCa. Circ_0005276 was notably upregulated in PCa tumor tissues (Fig. 1A), as well as in PCa cell lines (LNCaP and DU145) compared to non-cancer cell line (RWPE-1) (Fig. 1B). Circ_0005276, compared to XIAP, was resistant to RNase R digestion and Actinomycin D treatment (Fig. 1C and D). Besides, compared to random primers, circ_0005276 could not be effectively detected and amplified by oligo (dT)18 primers (Fig. 1E). As shown in Fig. 1F, circ_0005276 was back-spliced from the exon5, exon6, and exon7 of XIAP mRNA, with 222 bp in length (Fig. 1F). These results demonstrated that circ_0005276 was upregulated in PCa, and circ_0005276 was a circular molecule with high stability.
The upregulation of circ_0005276 was determined in PCa tissues and cells. A The expression of circ_0005276 in tumor tissues and normal tissues was detected by qPCR. B The expression of circ_0005276 in LNCaP, DU145, and RWPE-1 cells was detected by qPCR. C and D The stability of circ_0005276 was ascertained using RNase R and Actinomycin D. E The circularity of circ_0005276 was determined using oligo(dT)18 primers and random primers. F The formation of circ_0005276. *P < 0.05
Circ_0005276 Knockdown Inhibited PCa Cell Malignant Phenotypes
We investigated the functions of circ_0005276 in vitro by loss function assays, and the results are shown in supplementary file, Fig. S1. Circ_0005276 abundance was significantly decreased in LNCaP and DU145 cells transfected with si-circ_0005276 relative to si-NC (Fig. S1A). In terms of function, circ_0005276 knockdown notably reduced cell viability, proliferation, migration, invasion, and angiogenesis in LNCaP and DU145 cells by multiple tests (Fig. S1B–F). Moreover, circ_0005276 knockdown notably increased cell apoptotic ability (Fig. S1G). Bax level was reinforced, while Bcl-2 expression was decreased in LNCaP and DU145 cells after circ_0005276 knockdown (Fig. S1H and S1I), suggesting that circ_0005276 knockdown promoted cell apoptosis. These results revealed that circ_0005276 knockdown blocked PCa development in vitro.
MiR-128-3p was a Target of circ_0005276
We next investigated circ_0005276-targeted miRNAs. The binging site between circ_0005276 and miR-128-3p was predicted by circBANK (Fig. 2A). The level of miR-128-3p was remarkably enhanced in LNCaP and DU145 cells transfected with miR-128-3p mimic (Fig. 2B). MiR-128-3p mimic and WT-circ_0005276 cotransfection remarkably reduced luciferase activity in LNCaP and DU145 cells (Fig. 2C and D). In RIP assay, the expression of circ_0005276 and miR-128-3p was abundantly detected in Ago2 antibody-coupled compounds (Fig. 2E and F). The expression of miR-128-3p was downregulated in PCa tumor tissues and cell lines (Fig. 2G and 2I), and miR-128-3p expression in PCa samples was inversely correlated with circ_0005276 expression (Fig. 2H). The expression of miR-128-3p was significantly weakened in LNCaP and DU145 cells with miR-128-3p inhibitor transfection (Fig. 2J). The expression of miR-128-3p was notably increased in LNCaP and DU145 cells after si-circ_0005276 transfection but considerably repressed by si-circ_0005276 + miR-128-3p inhibitor transfection (Fig. 2K). These results suggested that miR-128-3p was a target of circ_0005276.
MiR-128-3p was targeted by circ_0005276. A The putative binding between circ_0005276 and miR-128-3p was obtained from circBANK. B The efficiency of miR-128-3p mimic was ascertained by qPCR. C and D The relationship between circ_0005276 and miR-128-3p was ensured by dual-luciferase reporter analysis. E and F Their binding was validated by RIP assay. G MiR-128-3p expression in tumor samples and normal samples was ascertained by qPCR. H The correlation between circ_0005276 expression and miR-128-3p expression in PCa. I The expression of miR-128-3p in LNCaP, DU145, and RWPE-1 cells was detected by qPCR. J The efficiency of miR-128-3p inhibitor. K The expression of miR-128-3p in LNCaP and DU145 cells introduced with si-circ_0005276 or si-circ_0005276 + miR-128-3p inhibitor was detected by qPCR. *P < 0.05
MiR-128-3p Inhibition Reversed the Effects of circ_0005276 Knockdown
Rescue experiments were performed to determine whether circ_0005276 regulated PTC progression by targeting miR-128-3p. The results showed that the viability, proliferation, migration, invasion, and angiogenesis of PTC cells were weakened by si-circ_0005276 but partly enhanced by si-circ_0005276 + miR-128-3p inhibitor in LNCaP and DU145 cells (Fig. 3A–E; Fig. S2A-S2D). In addition, circ_0005276 knockdown-enhanced cell apoptotic rate was largely reduced by the transfection of miR-128-3p inhibitor (Fig. 3F). The expression of Bax elevated in LNCaP and DU145 cells with si-circ_0005276 transfection was partially repressed by si-circ_0005276 + miR-128-3p inhibitor, while the expression of Bcl-2 depleted in cells containing si-circ_0005276 was partially restored in cells containing si-circ_0005276 + miR-128-3p inhibitor (Fig. 3G and H). These results demonstrated that circ_0005276 regulated PCa development by targeting miR-128-3p.
Circ_0005276 knockdown-inhibited proliferation, migration, invasion, and angiogenesis were recovered by miR-128-3p inhibition. The following functional assays were performed in LNCaP and DU145 cells transfected with si-circ_0005276, si-NC, si-circ_0005276 + miR-128-3p inhibitor or si-circ_0005276 + inhibitor NC. A and B Cell proliferation was investigated by CCK-8 assay and EdU assay. C and D Cell migration and cell invasion by transwell assay. E The ability of angiogenesis by tube formation assay. (F) Cell apoptosis was investigated by flow cytometry assay. G and H The protein levels of Bax and Bcl-2. *P < 0.05
DEPDC1B was Targeted by miR-128-3p
We next investigated the target genes of miR-128-3p. The binding site between miR-128-3p and WT-DEPDC1B 3’UTR was predicted by bioinformatics tool targetscan (Fig. 4A). In dual-luciferase reporter assay, miR-128-3p mimic and WT-DEPDC1B-3’UTR transfection markedly diminished luciferase activity in LNCaP and DU145 cells (Fig. 4B and C). DEPDC1B was strikingly overexpressed in PCa tumor tissues by qPCR and western blot assays (Fig. 4D and F), and DEPDC1B expression was negatively associated with miR-128-3p expression (Fig. 4E). As expected, the expression of DEPDC1B was enhanced in PCa cell lines (LNCaP and DU145) compared with that in non-cancer cells (RWPE-1) by western blot assay (Fig. 4G). DEPDC1B expression was strikingly increased in LNCaP and DU145 cells transfected with pc-DEPDC1B relative to pc-NC (Fig. 4H). DEPDC1B expression was strikingly declined in LNCaP and DU145 cells containing miR-128-3p mimic but largely recovered in cells containing miR-128-3p mimic + pc-DEPDC1B (Fig. 4I). These results suggested that DEPDC1B was a target of miR-128-3p.
DEPDC1B was targeted by miR-128-3p. A The binding between miR-128-3p and DEPDC1B was obtained by targetscan. B and C The relationship between miR-128-3p and DEPDC1B was verified by dual-luciferase reporter analysis. D and F The expression of DEPDC1B in tumor samples and normal samples was ascertained by qPCR and western blot. E The correlation between miR-128-3p expression and DEPDC1B expression in PCa. G The protein level of DEPDC1B in LNCaP, DU145, and RWPE-1 cells. (H) The overexpression efficiency of pc-DEPDC1B. I The expression of DEPDC1B protein in LNCaP and DU145 cells with miR-128-3p mimic, miRNA NC, miR-128-3p mimic + pc-DEPDC1B, or miR-128-3p mimic + pc-NC. *P < 0.05
MiR-128-3p Inhibited the Progression of PCa In Vitro by Suppressing DEPDC1B
In terms of function, cell viability, proliferation, migration, invasion, and angiogenesis were prominently depleted in LNCaP and DU145 cells with miR-128-3p mimic transfection compared to miRNA NC, while these malignant cell behaviors were partly recovered in LNCaP and DU145 cells transfected with miR-128-3p mimic + pc-DEPDC1B compared to miR-128-3p mimic + pc-NC (Fig. 5A–E; Fig. S3A–D). In addition, miR-128-3p restoration-induced cell apoptosis was largely suppressed by DEPDC1B overexpression (Fig. 5F). Moreover, Bax expression elevated in LNCaP and DU145 cells introduced with miR-128-3p mimic was partially repressed in cells introduced with miR-128-3p mimic + pc-DEPDC1B, while the expression of Bcl-2 was opposite to Bax expression (Fig. 5G and H). These results indicated that miR-128-3p restoration inhibited PCa development in vitro by sequestering DEPDC1B.
MiR-128-3p enrichment-inhibited proliferation, migration, invasion, and angiogenesis were recovered and were restored by DEPDC1B overexpression. The following assays were performed in LNCaP and DU145 cells with miR-128-3p mimic, miRNA NC, miR-128-3p mimic + pc-DEPDC1B, or miR-128-3p mimic + pc-NC transfection. A and B Cell proliferation was examined using CCK-8 assay and EdU assay. C and D Cell migration and cell invasion were examined using transwell assay. E The ability of angiogenesis was examined by tube formation assay. F Cell apoptosis was examined by flow cytometry assay. G and H The protein levels of Bax and Bcl-2 were measured by western blot. *P < 0.05
Circ_0005276 Positively Regulated DEPDC1B Expression by Targeting miR-128-3p
Interestingly, we discovered that DEPDC1B expression was strikingly decreased in LNCaP and DU145 cells by si-circ_0005276 transfection relative to si-NC, while DEPDC1B expression was largely recovered in LNCaP and DU145 cells after si-circ_0005276 + miR-128-3p inhibitor transfection relative to si-circ_0005276 + inhibitor NC (Fig. 6A). Moreover, DEPDC1B expression in PCa samples was positively correlated with circ_0005276 expression (Fig. 6B). These results hinted that circ_0005276 functioned as miR-128-3p sponge to enrich the expression of DEPDC1B.
The expression of DEPDC1B was inhibited by si-circ_0005276 but recovered by miR-128-3p inhibitor in LNCaP and DU145 cells. A The expression of DEPDC1B protein in LNCaP and DU145 cells introduced with si-circ_0005276, si-NC, si-circ_0005276 + miR-128-3p inhibitor, or si-circ_0005276 + inhibitor NC. B The correlation between circ_0005276 expression and DEPDC1B expression in PCa. *P < 0.05
Circ_0005276 Knockdown Inhibited the Activity of the PI3K/AKT Signaling
The activation of the PI3K/AKT pathway was closely associated with cancer development. Interestingly, we found that the phosphorylation levels of PI3K and AKT were obviously elevated by DEPDC1B overexpression in LNCaP and DU145 cells (Fig. 7A). In addition, the phosphorylation levels of PI3K and AKT were strikingly decreased in LNCaP and DU145 cells by si-circ_0005276 relative to si-NC, while the phosphorylation levels of PI3K and AKT were partly restored in cells with si-circ_0005276 + miR-128-3p inhibitor cotransfection compared to si-circ_0005276 + inhibitor NC (Fig. 7B). The data suggested that circ_0005276 knockdown inhibited the activity of the PI3K/AKT pathway by enriching miR-128-3p.
The PI3K/AKT signaling pathway was inhibited by circ_0005276 knockdown. A The levels of p-PI3K and p-AKT in LNCaP and DU145 cells overexpressing DEPDC1B. B The protein levels of p-PI3K and p-AKT in LNCaP and DU145 cells containing si-circ_0005276, si-NC, si-circ_0005276 + miR-128-3p inhibitor, or si-circ_0005276 + inhibitor NC. *P < 0.05
Circ_0005276 Knockdown Suppressed Solid Tumor Growth In Vivo
Animal study was conducted to determine the role of circ_0005276 in vivo. The results showed that circ_0005276 downregulation significantly suppressed tumor growth, resulting in poor tumor volume and tumor weight (Fig. 8A and B). Moreover, in sh-circ_0005276-introduced tumor samples, the expression of circ_0005276 and DEPDC1B was strikingly declined, while the expression of miR-128-3p was strikingly reinforced (Fig. 8C–E), suggesting that circ_0005276 knockdown inhibited the expression of DEPDC1B by increasing miR-128-3p, thus suppressing tumor growth.
Circ_0005276 knockdown inhibited solid tumor growth. A and B Tumor volume was weekly measured, and tumor tissues were excised after 4 weeks to measure tumor weight. C The expression of circ_0005276 in tumor tissues. D The expression of miR-128-3p in tumor tissues. E The expression of DEPDC1B protein in tumor tissues. *P < 0.05
Discussion
Our present study discovered the upregulation of circ_0005276 in PCa. Circ_0005276 downregulation significantly inhibited PCa growth and development. Mechanism analyses disclosed that circ_0005276 indirectly upregulated the expression of DEPDC1B by targeting miR-128-3p. Moreover, we found that circ_0005276 governed the miR-128-3p/DEPDC1B axis to regulate the activity of the PI3K/AKT signaling, thus contributing to the development of PCa.
Accumulating studies illustrate that circRNAs can act as either oncogenic drivers or tumor suppressors in cancer development (Chen and Huang 2018). For instance, circ_0016068 was strongly expressed in PCa, and circ_0016068 promoted PCa cell growth and invasion (Li et al. 2020). Circ_0001427 harbored lower expression in PCa tissues, and circ_0001427 upregulation blocked PCa cell proliferation and invasion (Wu et al. 2019). As for circ_0005276, it was declaimed to be highly regulated in PCa, and the knockdown of circ_0005276 restricted PCa cell proliferation, migration, and invasion (Feng et al. 2019). Consistent with these data, we noticed that circ_00052765 knockdown substantially sequestered the ability of angiogenesis and promoted cell apoptosis. Circ_0005276 was also reported to be upregulated in epithelial ovarian cancer tissues, and circ_0005276 knockdown largely attenuated cancer cell migration (Liu et al. 2020). These data stated that circ_0005276 might be an oncogenic driver in various cancers. Furthermore, we found that circ_0005276 knockdown reduced the phosphorylation levels of PI3K and AKT, suggesting that circ_0005276 knockdown inactivated the PI3K/AKT signaling. Accumulating studies unveiled that the activation of PI3K/AKT signaling was linked to the progression of multiple cancers (Chen et al. 2016; Mirza-Aghazadeh-Attari et al. 2020), and targeting PI3K/AKT signaling pathway might be an effective strategy for cancer therapy (Mirza-Aghazadeh-Attari et al. 2020). Herein, we found that circ_0005276 deregulation regulated the activity of the PI3K/AKT signaling pathway, thus affecting PCa progression.
Circ_0005276 functioned as miR-128-3p sponge and suppressed miR-128-3p expression. Previous studies clarified that miR-128-3p expression was markedly declined in invasive PCa cell lines relative to benign prostate epithelial cells, and the deficiency of miR-128-3p promoted PCa cell invasion (Khan et al. 2010). Besides, miR-128-3p restoration strengthened PCa cell chemosensitivity (Sun et al. 2015). In addition, miR-128-3p was deemed to be targeted by TUG1, and miR-128-3p inhibition promoted PCa cell proliferation and invasion that were suppressed by TUG1 knockdown (Hao et al. 2020). In agreement with these findings, we also verified the tumor-suppressive role of miR-128-3p in PCa and discovered that miR-128-3p absence recovered PCa cell proliferation, migration, invasion, and angiogenesis that were blocked by circ_0005276 knockdown, suggesting that circ_0005276 promoted PCa malignant characteristics by targeting miR-128-3p.
DEPDC1B was screened as a target of miR-128-3p in our study. The upregulation of DEPDC1B was identified in PCa tissues, and elevated DEPDC1B expression predicted the advanced stage of PCa, lymph node metastasis, and short survival time (Bai et al. 2017). Besides, DEPDC1B was closely linked to the metastasis status of PCa, and DEPDC1B promoted epithelial–mesenchymal transition (EMT) to induce PCa cell migration and invasion (Chen et al. 2020). Considering the importance of DEPDC1B in PCa development, we further noticed that miR-128-3p restoration-inhibited PCa cell proliferation, migration, invasion, and angiogenesis were largely restored by DEPDC1B overexpression, indicating that miR-128-3p inhibited the expression and function of DEPDC1B. Besides, DEPDC1B overexpression markedly enhanced the phosphorylation levels of PI3K and AKT, hinting its high expression activated the PI3K/AKT signaling pathway. More importantly, we identified that DEPDC1B level was notably sequestered by circ_0005276 knockdown but recovered by miR-128-3p inhibition, hinting that circ_0005276 acted as miR-128-3p sponges to regulate the expression of DEPDC1B.
Conclusion
Circ_0005276 was an oncogenic driver in PCa, and circ_0005276 promoted PCa development by promoting PCa cell proliferation, migration, invasion, and angiogenesis. Our study provided convincing evidence for the first time that circ_0005276 aggravated the progression of PCa by modulating the miR-128-3p/DEPDC1B pathway at least in part. We supposed that circ_0005276 was a key regulator that contributed to the diagnosis and treatment of PCa.
Data Availability
The analyzed datasets generated during the present study are available from the corresponding author on reasonable request.
References
Attard G et al (2016) Prostate cancer. Lancet 387(10013):70–82
Bai S et al (2017) High levels of DEPDC1B predict shorter biochemical recurrence-free survival of patients with prostate cancer. Oncol Lett 14(6):6801–6808
Chen B, Huang S (2018) Circular RNA: An emerging non-coding RNA as a regulator and biomarker in cancer. Cancer Lett 418:41–50
Chen H et al (2016) The PI3K/AKT pathway in the pathogenesis of prostate cancer. Front Biosci (landmark Ed) 21:1084–1091
Chen Y et al (2020) Mitophagy impairment is involved in sevoflurane-induced cognitive dysfunction in aged rats. Aging (albany NY) 12(17):17235–17256
Feng Y et al (2019) Circular RNA circ0005276 promotes the proliferation and migration of prostate cancer cells by interacting with FUS to transcriptionally activate XIAP. Cell Death Dis 10(11):792
Hao SD et al (2020) Long non-coding TUG1 accelerates prostate cancer progression through regulating miR-128-3p/YES1 axis. Eur Rev Med Pharmacol Sci 24(2):619–632
Hua JT, Chen S, He HH (2019) Landscape of noncoding RNA in prostate cancer. Trends Genet 35(11):840–851
Jiang WD, Ye ZH (2019) Integrated analysis of a competing endogenous RNA network in renal cell carcinoma using bioinformatics tools. Biosci Rep. https://doi.org/10.1042/BSR20190996
Khan AP et al (2010) Quantitative proteomic profiling of prostate cancer reveals a role for miR-128 in prostate cancer. Mol Cell Proteomics 9(2):298–312
Kristensen LS et al (2018) Circular RNAs in cancer: opportunities and challenges in the field. Oncogene 37(5):555–565
Li Q et al (2020) Circular RNA circ-0016068 promotes the growth, migration, and invasion of prostate cancer cells by regulating the miR-330-3p/BMI-1 axis as a competing endogenous RNA. Front Cell Dev Biol 8:827
Liu ZH et al (2020) Circ_0005276 aggravates the development of epithelial ovarian cancer by targeting ADAM9. Eur Rev Med Pharmacol Sci 24(20):10375–10382
Mirza-Aghazadeh-Attari M et al (2020) Targeting PI3K/Akt/mTOR signaling pathway by polyphenols: Implication for cancer therapy. Life Sci 255:117481
Siegel RL, Miller KD, Jemal A (2018) Cancer statistics, 2018. CA Cancer J Clin 68(1):7–30
Si-Tu J et al (2019) Upregulated circular RNA circ-102004 that promotes cell proliferation in prostate cancer. Int J Biol Macromol 122:1235–1243
Sun X et al (2015) miR-128 modulates chemosensitivity and invasion of prostate cancer cells through targeting ZEB1. Jpn J Clin Oncol 45(5):474–482
Sung H et al (2021) Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 71(3):209–249
Witkos TM et al (2018) A potential role of extended simple sequence repeats in competing endogenous RNA crosstalk. RNA Biol 15(11):1399–1409
Wu G et al (2019) Preclinical study using circular RNA 17 and micro RNA 181c–5p to suppress the enzalutamide-resistant prostate cancer progression. Cell Death Dis 10(2):37
Acknowledgements
Not applicable.
Funding
No funding was received.
Author information
Authors and Affiliations
Contributions
WL and WW made substantial contribution to conception and design, acquisition of the data, or analysis and interpretation of the data; take part in drafting the article or revising it critically for important intellectual content; gave final approval of the revision to be published; and agree to be accountable for all aspect of the work.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethical Approval
The present study was approved by the ethical review committee of Huangshi Central Hospital, Affiliated Hospital of Hubei Polytechnic University with approval No. 20210318.
Consent to Participate
Written informed consent was obtained from all enrolled patients.
Consent for Publication
Patients agree to participate in this work.
Informed consent
The research has been carried out in accordance with the World Medical Association Declaration of Helsinki and that all subjects provided written informed consent.
Research Involving Human and Animal Studies
Animal studies were performed in compliance with the ARRIVE guidelines and the Basel Declaration. All animals received humane care according to the National Institutes of Health (USA) guidelines.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
10528_2022_10328_MOESM1_ESM.tif
Supplementary file1 Circ_0005276 knockdown inhibited proliferation, migration, invasion, and angiogenesis in PCa cells. (A) The efficiency of si-circ_0005276 was determined by qPCR. (B) The effect of circ_0005276 knockdown on cell proliferation was assessed by CCK-8 assay. (C) The effect of circ_0005276 knockdown on cell proliferation was assessed by EdU assay. (D and E) The effect of circ_0005276 knockdown on cell migratory and invasive capacity was assessed by transwell assay. (F) The ability of angiogenesis caused by circ_0005276 knockdown was determined by tube formation assay. (G) The effect of circ_0005276 absence on cell apoptosis was determined by flow cytometry. (H and I) The expression of Bax and Bcl-2 affected by circ_0005276 knockdown was determined by western blot. *P<0.05 (TIF 667 KB)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Li, W., Wu, W. Circ_0005276 Promotes Prostate Cancer Progression Through the Crosstalk of miR-128-3p/DEPDC1B Axis. Biochem Genet 61, 1987–2003 (2023). https://doi.org/10.1007/s10528-022-10328-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10528-022-10328-y