Abstract
Cervical cancer (CC) is the fourth most common cancer in women, and circular RNAs (circRNAs) have been shown to regulate CC development. However, the role of circ_0006646 in CC progression is still unclear. The levels of circ_0006646, miR-758-3p, and ribonucleotide reductase regulatory subunit M2 (RRM2) were evaluated by quantitative real-time PCR. Cell proliferation was tested by cell counting kit 8 and 5-ethynyl-2′-deoxyuridine assays. Flow cytometry was used to test cell apoptosis. Migration and invasion were estimated by transwell assay. Western blot assay was performed to examine protein expression. Dual-luciferase reporter assay, RIP assay, and RNA pull down assay were used to analyze the connection between miR-758-3p and circ_0006646 or RRM2. Tumor growth was detected by in vivo experiments. Exosomes were isolated form CC patients and healthy controls. Circ_0006646 expression was elevated in CC cells, and its knockdown suppressed CC cell growth, migration, and invasion. MiR-758-3p was sponged by circ_0006646, and RRM2 was targeted by miR-758-3p. In addition, the effects of circ_0006646 depletion on CC cell progression were overturned by miR-758-3p inhibitor, and either RRM2 overexpression reversed those effects of miR-758-3p overexpression on CC cell progression. Circ_0006646 was highly expressed in the exosomes of CC patients. Circ_0006646 expedited CC cell growth and metastasis by regulating miR-758-3p/RRM2 axis, and exosomal circ_0006646 might be a potential diagnostic indicator of CC.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cervical cancer (CC) occurs in the cervix and is a fourth universal malignant cancer of the female (Revathidevi et al. 2021). The prognosis and 5-year survival rate of CC patients are not ideal, and thus CC has become one of the main cancers that kill women (Chen et al. 2020; Biewenga et al. 2011). Recent studies have found that targeted therapy is very important in the treatment and prognosis of cancers (Pérez-Herrero and Fernández-Medarde 2015).
Circular RNAs (circRNAs) play a vital part in promoting or inhibiting cancers development (Chen et al. 2018). Circ_0000515 was significantly aggrandized in CC, and high circ_0000515 expression suggested poor prognosis in patients with CC (Tang et al. 2019). Circ_0019435 was highly expressed in CC, and its knockdown blocked the growth and epithelial-mesenchymal transition (EMT) of CC cells (Wang and Zhuo 2021). As an oncogene, circ_0007534 was significantly fortified in CC, and the invasion and development of CC cells were restricted by circ_0007534 silencing (Sun et al. 2021). CircBank (http://www.circbank.cn/infoCirc.html?id=hsa_circPTK2_018) has shown that circ_0006646 is located in chr8: 141,889,569–141,900,868 of protein tyrosine kinase 2 (PTK2), with a length of 394 bp. Li et al. found that circ_0006646 might be a potential biomarker for CC (Li et al. 2020). But the function of circ_0006646 in CC is still indistinct.
As non-coding RNAs, the sponge role of circRNAs and microRNAs (miRNAs) in cancer has been extensively studied (Tang et al. 2021). MiRNAs have been confirmed to serve as tumor promotor or suppresser to regulate the development of cancer (Barbato et al. 2017; Pardini et al. 2018). MiRNAs play an irreplaceable role in the treatment, prognosis, and diagnosis of CC (Shen et al. 2020). MiR-489 was a tumor-restrained miRNA in CC, and its overexpression inhibited the evolution of CC through the PI3K/AKT/P53 pathway (Juan et al. 2018). In addition, circ_0000228 sponged miR-195-5p and thus accelerated the development of CC (Liu et al. 2021). MiR-758-3p has often been studied for its function as an inhibitor of cancer (Chen et al. 2019). And previous studies indicated that miR-758-3p was associated with the treatment and prognosis of CC (Ding et al. 2021; Xie et al. 2021). However, the regulatory relationship between miR-758-3p and circ_0006646 in CC is still not understood.
Ribonucleotide reductase regulatory subunit M2 (RRM2) is one of two distinct subunits encoding ribonucleotide reductase (Nordlund and Reichard 2006). Studies had demonstrated that RRM2 was upregulated in lung adenocarcinoma (LUAD), and low RRM2 expression in LUAD patients hinted better survival and prognosis (Ma et al. 2020; Jin et al. 2020). In liver cancer, RRM2 was robustly intensified in liver cancer, and serum RRM2 was useful as a biomarker to improve diagnostic efficiency of liver cancer (Yang et al. 2020). In glioma, RRM2 was triggered in glioma tissues and cells, and RRM2 confined the growth by ERK1/2 and AKT pathway (Sun et al. 2019). Wang et al. noticed that RRM2 was obviously augmented in CC, and its overexpression implied poor overall survival of CC patients (Wang et al. 2020). But, the relationships among RRM2, circ_0006646, and miR-758-3p in CC have not been reported.
The function and mechanism of circ_0006646 in CC were inquired for the first time in our research through function recovery experiments and in vivo assay, providing a new promising biomarker for the treatment and diagnosis of CC.
Materials and Methods
Cell Culture
Human CC cell lines (SiHa, CaSki, C33A, and HeLa) and human cervical epithelial cell line Ect1/E6E7 were received from American Type Culture Collection (ATCC, Manassas, VA, USA), and hatched in EMEM (Merck, Darmstadt, Germany) with 10% FBS (Merck).
Quantitative Real-Time PCR (qRT-PCR)
TRI reagent from Merck was for isolating RNA. The concentration of RNA was measured by ultraviolet photometer (UNICO, Shanghai, China). SYBR Green Quantitative RT-QPCR Kit (Merck) was employed to detect circ_0006646, miR-758-3p, and RRM2 expression. The data of qRT-PCR were examined by 2−ΔΔCT method. β-actin or U6 was the internal reference. Primers in Table 1 were obtained from TaKaRa (Tokyo, Japan).
Cell Localization Experiment
Cytoplasmic & Nuclear RNA Purification Kit (100) (Norgen Biotek Corp, Ontario, Canada) was hired to detect the location of circ_0006646. 1 × 107 SiHa and C33A cells were gathered utilizing Trypsin (Merck), and lysed in a 50 µM DTT 1 × cytoplasmic lysis buffer for 15 min, and the lysate was centrifuged at maximum speed for 3 min to collect cytoplasm; the cells were re-suspended with nuclear extraction buffer. And the nuclear suspension was stirred in a rotator at 4 °C for 1 h, and then centrifuged at 4 °C for 5 min at 16,000 × g to collect the supernatant, which was the nuclear extraction solution. RNA was extracted from cytoplasmic and nuclear extracts, and the contents of circ_0006646, GAPDH, and U6 were detected by qRT-PCR to verify the location of circ_0006646. And GAPDH and U6 were cytoplasm and nucleus controls, respectively.
Cell Transfection
Silencing expression vectors of circ_0006646 (small interfering (si)-circ_0006646#1, si-circ_0006646#2 and short hairpin (sh)-circ_0006646), miR-758-3p inhibitor (anti-miR-758-3p), miR-758-3p and RRM2 overexpression vectors (miR-758-3p and pcNDA-RRM2) and their respective negative controls were established by TaKaRa. The transfection was carried out with the guidance of X-Tremegene 360 transfection reagent (Roche, Basel, Switzerland).
Cell Counting Kit 8 (CCK8) Assay
100 µl SiHa or C33A cell suspensions were seeded in 96-well plates for 24 h and then transfected with different diagrams. The old cell culture medium was sucked away at 0, 24, 48, and 72 h after transfection, and the new culture medium with 10 μl CCK8 solution (Merck) was added and trained at 37 °C for 4 h. The OD 450 nm value was determined with Accuris™ SmartReader™ 96 microplate reader (Merck).
5-Ethynyl-2′-Deoxyuridine (EdU) Assay.
5-Ethynyl-2′-Deoxyuridine (EdU) Assay
BeyoClick™ EdU Cell Proliferation Kit with Alexa Fluor 488 (Beyotime, Shanghai, China) was hired for EdU assay. Transfected SiHa and C33A cells were seeded in the 6-well plates and co-cultured with the same volume of 20 µM EdU working solution for 2 h, and hatched with 1 ml of 4% paraformaldehyde (Beyotime) and permeable solution for 15 min, respectively. Then SiHa and C33A cells were trained with 0.5 ml click reaction solution for half an hour away from light and dyed with 1 ml of 10 µg/ml DAPI staining solution (Beyotime) for 3 min for nuclear staining. And then Fluorescence detection was performed.
Cell Apoptosis Assay
AnnexinV-FITC/PI Apoptosis Detection Kit (Solarbio, Beijing, China) was employed for cell apoptosis detection. 5 × 105 SiHa or C33A cells were suspended with phosphate-buffered saline PBS (Merck) and centrifuged at 4 °C with 1000 rpm for 10 min. The cells were mixed with 200 µl binding buffer, 10 µl AnnexinV-FITC and 10 µl PI and fostered at 24 °C for 15 min under dark conditions, and then 300 µl binding buffer was added, and cell apoptosis rate was evaluated by flow cytometry within 1 h.
Western Blot
The protein expression of RRM2, apoptosis suppressor B-cell lymphona-2 (Bcl-2), apoptosis promoter Bcl-2-Associated X (Bax), stromal cell markers N-cadherin and Vimentin, epithelial cell marker E-cadherin, and exosome surface markers CD83, TSG101, and calnexin were estimated by western blot assay. Shortly, 250 µl RIPA Lysis Buffer (Servicebio, Wuhan, China), 25 µl PMSF (100 mM) (Servicebio), and 25 µl protease inhibitor (Servicebio) were used to lyse cells for 3 min, and then cells were harvested into the new EP tube with a cell scraper and centrifuged, the supernatant was collected to form the total protein solution. BCA Protein quantitative detection Kit (Servicebio) was used for detecting protein concentration.
20 µg protein was shifted to gel by SDS-PAGE Gel Preparation Kit (Servicebio) and then to polyvinylidene fluoride with pore size of 0.45 µM (Servicebio) by transfer electrophoresis tank (Servicebio). Then 5% Bovine Serum Albumin (BSA) (Servicebio) was used for sealing membrane and the membrane was hatched with primary antibodies against N-cadherin (1:500, GB111009, Servicebio), Vimentin (1:500, GB111308, Servicebio), E-cadherin (1:1000, GB11082, Servicebio), RRM2 (1:1000, ab172476, Abcam, Cambridge, MA, USA), beta actin (β-actin) (1:1000, ab8227, Abcam), CD83 (1:1000, ab244204, Abcam), TSG101 (1:1000, ab125011, Abcam), and calnexin (1:20,000, ab92573, Abcam) for one night at 4 °C and secondary antibody HRP-conjugated goat anti-rabbit IgG (H + L) (1:3000, GB23303, Servicebio) at 25 °C for 2 h. Protein results were observed with Ultra sensitive ECL Chemiluminescence Kit (Servicebio).
Transwell Assay
Cell migration was assessed by transwell chambers and the invasion was measured by transwell chambers pre-coated with 8 µM Matrigel (Corning, Bedford, MA, USA). 100 µl 5 × 105 cell/ml was seeded in the upper compartment without serum, and the nether chamber was filled with 600 µl EMEM containing 10% FBS. The chamber was cleaned with PBS (Merck) after 24 h and the moving cells were immobilized with methanol (Merck) for 30 min and dyed with 0.1% crystal violet (Merck) for 20 min. The unmoved cells were wiped with cotton swabs, and the moving cells were observed and counted in 5 fields randomly under a microscope of 400 magnification.
Dual-Luciferase Reporter Assay
Circ_0006646 and RRM2 3′UTR wild-type (WT) fragments containing miR-758-3p seed sequences and their site-directed mutant (MUT) sequences were cloned into pmirGLO vectors (Promega, Madison, WI, USA) to build luciferase reporter vectors named circ_0006646-WT, RRM2 3′UTR-WT, circ_0006646-MUT, and RRM2 3′UTR-MUT. After transfection, Dual-Lumi™ Luciferase Assay Kit (Beyotime) was engaged for measuring the luciferase activity.
RIP Assay
The EZMagna RIP kit (Merck) was for RIP assay. In brief, 1 × 107 SiHa and C33A cells were fostered with the kit-provided RIP lysis buffer and hatched with rat anti-human argonaute 2 antibody (Anti-Ago2; Merck) or the negative control goat anti-human IgG antibody (Anti-IgG; Merck) at 4 °C for 3 h, then magnetic beads were mixed with cell lysate for another 3 h. Then the beads were collected and hatched with proteinase K (20 mg/ml) (Beyotime). Then RNA was separated and the levels of circ_0006646, miR-758-3p, and RRM2 was quantified by qRT-PCR.
RNA Pull Down Assay
Then biotin-labeled miR-758-3p probes (Bio-miR-758-3p) or control probes (Bio-NC) constructed from Sangon (Shanghai, China) were transfected into 2 × 105 SiHa and C33A cells. After 24 h of culture, cell lysis buffer and magnetic beads were added into the cells and cultivated at 4 °C for 3 h. The reaction tube was placed on the magnetic bead rack, and the beads were collected and fostered with proteinase K (Beyotime) to remove the protein. The RNA was isolated, and the level of circ_0006646 was estimated through qRT-PCR.
In Vivo Experiment in Mice
Twelve female 5-week-old nude mice from Vital River (Beijing, China) were discretionarily divided into two groups. 1.5 × 105 SiHa cells transfected with sh-NC or sh-circ_0006646 were subcutaneously injected into nude mice. The tumors volume was measured once a week for 5 weeks, and was calculated according to the formula: volume (mm3) = width2 × length/2. At the fifth week, the mice were performed euthanasia operation, the tumor weight was detected, and the tumor morphology was recorded. Tissues were used for isolating RNA and protein to detect circ_0006646, miR-758-3p, and RRM2 expression. Paraffin sections of tumor tissue were prepared for immunohistochemical (IHC) staining to detect the number of RRM2 positive cells. The Animal Research Ethics Committee of Puren Hospital Affiliated to Wuhan University of Science and Technology gave a license to the animal experiments.
Samples Collection
Fifty-five healthy controls and CC patients were recruited from Puren Hospital Affiliated to Wuhan University of Science and Technology. The plasma of each subject was collected and stored at −80 °C. The research was approbated by the Ethics Committee of Puren Hospital Affiliated to Wuhan University of Science and Technology.
Exosome Isolation and Treatment
Exosome Isolation Kit (Umibio, Shanghai, China) was used to isolate exosome from the plasma of healthy controls (n = 55) and CC patients (n = 55). Exosome morphology was observed by transmission electron microscope (TEM, StarJoy, Japan), and nanoparticle tracking analysis (NTA) instrument (ZetaView, Particle Metrix, Meerbusch, Germany) was used to detect the size and concentration of exosome.
Statistical Analysis
Results were analyzed with GraphPad Prism 8.0 software (GraphPad Inc., LaJolla, California, USA) and exhibited as mean ± standard deviation. The differences between two groups or multiple sets of data were evaluated by Student’s t-test or ANOVA. P < 0.05 indicated significant differences in data.
Results
Circ_0006646 Expression Was Enhanced in CC Cells
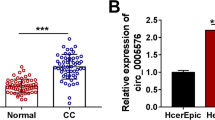
Circ_0006646 is located at chr8 and is derived from the back-splicing of exons 1–2 of PTK2 gene (Fig. 1A). The content of circ_0006646 was obviously increased in CC cell lines (SiHa, CaSki, C33A, and HeLa) (Fig. 1B). Subsequently, subcellular localization experiments were designed to determine the location of circ_0006646, and the results showed that circ_0006646 was mainly existed in cytoplasm (Fig. 1C, D). Together, the data suggested that circ_0006646 was highly expressed in CC.
Knockdown of Circ_0006646 Inhibited the Growth and Metastasis of CC Cells
We performed qRT-PCR assay to detect the inhibitory efficiencies of si-circ_0006646#1 and si-circ_0006646#2 on circ_0006646 expression. As shown in Fig. 2A, both si-circ_0006646#1 and si-circ_0006646#2 significantly downregulated circ_0006646 expression in SiHa and C33A cells. CCK8 assay results showed that the viability was greatly decreased in SiHa and C33A cells after si-circ_0006646#1 or si-circ_0006646#2 transfection (Fig. 2B, C). And EdU assay disclosed that both si-circ_0006646#1 and si-circ_0006646#2 evidently inhibited the proliferation of SiHa and C33A cells (Fig. 2D). Besides, both si-circ_0006646#1 and si-circ_0006646#2 markedly enhanced the apoptosis of SiHa and C33A cells (Fig. 2E). Furthermore, the migration and invasion abilities of SiHa and C33A cells were prominently restrained by si-circ_0006646#1 or si-circ_0006646#2 addition (Fig. 3A, B). And N-cadherin and Vimentin protein expression were suppressed by si-circ_0006646#1 or si-circ_0006646#2, while the protein expression of E-cadherin was elevated, indicating that EMT suppression occurred in SiHa and C33A cells after circ_0006646 downregulation (Fig. 3C). In a word, circ_0006646 knockdown blocked CC cell growth and metastasis.
Circ_0006646 silencing confined CC cell proliferation and apoptosis. A The transfection efficiencies of si-circ_0006646#1 and si-circ_0006646#2 were measured by qRT-PCR. B and C Cell viability was measured by CCK8 assay. D EdU assay was carried for cell proliferation. E Flow cytometry was used for cell apoptosis. *P < 0.05
Circ_0006646 silencing confined CC cell metastasis. A and B Cell migration and invasion of SiHa and C33A cells were measured by transwell assay after si-circ_0006646#1 or si-circ_0006646#2 introduction. C N-cadherin, Vimentin, and E-cadherin expression in SiHa and C33A cells transfected with si-circ_0006646#1 or si-circ_0006646#2 were detected by western blot assay. *P < 0.05
MiR-758-3p Was Directly Regulated by Circ_0006646
Circinteractome (https://circinteractome.nia.nih.gov/), StarBase 3.0 (https://starbase.sysu.edu.cn/) and circBank (http://www.circbank.cn/) were used to screen the targets of circ_0006646 (Fig. 4A), and the results showed that miR-758-3p was the target of circ_0006646, and their binding sites were shown in Fig. 4B. QRT-PCR results proved that miR-758-3p overexpression notably augmented the expression of miR-758-3p in SiHa and C33A cells (Fig. 4C). The results of dual-luciferase reporter assay disclosed that compared with the miR-NC group, miR-758-3p overexpression could dramatically reduce the luciferase activity of circ_0006646-WT reporter vector in SiHa and C33A cells, while was not able to decrease that of circ_0006646-MUT vector (Fig. 4D, E). The data of RIP assay revealed that both circ_0006646 and miR-758-3p were enriched in Anti-Ago2 complexes with respect to Anti-IgG complexes (Fig. 4F, G). It was also discovered that circ_0006646 was remarkably enriched in Bio-miR-758-3p group compared to Bio-NC group (Fig. 4H). What’s more, we found that miR-758-3p expression was drastically restrained in CC cell lines (Fig. 4I). These results proved that miR-758-3p was the target of circ_0006646.
MiR-758-3p was negatively regulated by circ_0006646. A The targets of circ_0006646 were screened by circinteractome, StarBase 3.0, and circBank. B The binding sequence between circ_0006646 and miR-758-3p was predicted by StarBase 3.0. C The overexpression efficiency of miR-758-3p was tested by qRT-PCR. D and E Dual-luciferase reporter assay was used to detect the luciferase activity of circ_0006646-WT and -MUT reporter vectors in SiHa and C33A cells with miR-758-3p transfection. F and G The enrichment of miR-758-3p and circ_0006646 by Ago2 in SiHa and C33A cells was tested by RIP assay with Anti-Ago2. IgG RIP was performed as the negative control using Anti-IgG. H RNA pull down assay was used to detect circ_0006646 enrichment by Bio-miR-758-3p or negative control Bio-NC in SiHa and C33A cells. I MiR-758-3p expression in CC cells was tested by qRT-PCR. *P < 0.05
The Effects of Circ_0006646 Knockdown on CC Cell Progression Were Overturned by miR-758-3p Downregulation
The transfection efficiency of anti-miR-758-3p was tested, and the data proved that miR-758-3p expression was conspicuously retarded by anti-miR-758-3p introduction (Fig. 5A). As presented in Fig. 5B–D, the suppressive effects of circ_0006646 knockdown on SiHa and C33A cell proliferation were partly reversed by miR-758-3p inhibitor. In addition, the deletion of miR-758-3p partially restored the promoting effects of circ_0006646 silencing on apoptosis in SiHa and C33A cells (Fig. 5E). Besides, the repressive effects of circ_0006646 depletion on migration and invasion of SiHa and C33A cells were regained by miR-758-3p insufficiency (Fig. 5F, G). And the effects of circ_0006646 deficiency on N-cadherin, Vimentin, and E-cadherin expression were weakened after anti-miR-758-3p transfection (Fig. 5H). In brief, downregulation of miR-758-3p counteracted the effects of circ_0006646 knockdown on CC cell progression.
Downregulation of miR-758-3p abolished the effects of circ_0006646 silencing on CC cells growth. A QRT-PCR was used to detect the transfection efficiency of anti-miR-758-3p. B–H The si-NC, si-circ_0006646#2, si-circ_0006646#2 + anti-miR-NC or si-circ_0006646#2 + anti-miR-758-3p was transfected into SiHa and C33A cells. B and C The cell viability was detected by CCK8 assay. D Cell proliferation was tested by EdU assay. E Cell apoptosis was measured by flow cytometry. F and G Cell migration and invasion were determined by transwell assay. H Western blot assay was used to analyze N-cadherin, Vimentin, and E-cadherin expression. *P < 0.05
RRM2 Was Negatively Regulated by miR-758-3p
StarBase 3.0 software predicted that RRM2 was the target of miR-758-3p, and the binding sequences between miR-758-3p and RRM2 were exhibited in Fig. 6A. The luciferase activity of RRM2 3′UTR-WT vectors was substantially impeded by miR-758-3p mimic rather than miR-NC, but the luciferase activity of RRM2 3′UTR-MUT vectors was hardly changed (Fig. 6B, C). Besides, RIP assay suggested that enrichments of RRM2 and miR-758-3p were apparently elevated in Anti-Ago2 group in contrast to Anti-IgG group (Fig. 6D, E). Then we found that RRM2 mRNA and protein expression in SiHa and C33A cells was confined by miR-758-3p overexpression and expedited by miR-758-3p downregulation (Fig. 6F, G). In addition, RRM2 was confirmed to be highly expressed in CC cells (Fig. 6H–I). Collectively, miR-758-3p regulated the expression of RRM2 in CC.
RRM2 was the downstream gene of miR-758-3p. A The target sites between RRM2 and miR-758-3p were predicted by StarBase 3.0. B and C Luciferase activity of RRM2 3′UTR-WT and -MUT reporter vectors were tested by dual-luciferase reporter assay in SiHa and C33A cells with miR-758-3p transfection. D and E RIP assay was used to evaluate the expression of miR-758-3p and RRM2 in Anti-Ago2 or Anti-IgG complexes. F and G The content of RRM2 was estimated by qRT-PCR and western blot after miR-758-3p was overexpressed or downregulated. H–I The level of RRM2 in CC cells was evaluated by qRT-PCR and western blot. *P < 0.05
RRM2 Overexpression Attenuated the Effects of miR-758-3p Overexpression on CC Cell Progression
We first determined the overexpression efficiency of pcDNA-RRM2 transfection, and the data uncovered that RRM2 expression in SiHa and C33A cells was abnormally reinforced by pcDNA-RRM2 addition (Fig. 7A, B). The results of CCK8 assay demonstrated that miR-758-3p overexpression blocked the proliferation of SiHa and C33A cells, and the effect was counteracted by pcDNA-RRM2 (Fig. 7C–E). Besides, the positive effect of miR-758-3p on the apoptosis of SiHa and C33A cells was abolished by RRM2 overexpression (Fig. 7F). Moreover, overexpression of miR-758-3p confined the migration and invasion of SiHa and C33A cells, and the impacts were abrogated by pcDNA-RRM2 (Fig. 7G, H). And upregulated miR-758-3p curbed N-cadherin and Vimentin expression, and accelerated E-cadherin expression, while the increased of RRM2 ameliorated these impacts (Fig. 7I). In a word, the effects of miR-758-3p on CC cell process were counteracted by RRM2 overexpression.
RRM2 increase overturned the impacts of miR-758-3p upregulation on CC cells growth. A and B The transfection efficiency of pcDNA-RRM2 was analyzed by qRT-PCR and western blot. C–I SiHa and C33A cells were transfected with miR-NC, miR-758-3p, miR-758-3p + pcDNA or miR-758-3p + pcDNA-RRM2. C and D CCK8 assay was used to measure cell viability. E and F EdU and flow cytometry were performed to detect cell proliferation and cell apoptosis, respectively. G and H Transwell assay was carried for testing cell migration and invasion. I The levels of N-cadherin, Vimentin, and E-cadherin were estimated by western blot assay. *P < 0.05
Circ_0006646 Regulated RRM2 Expression Through miR-758-3p
As displayed in Fig. 8A, B, silence of circ_0006646 hampered RRM2 expression, while this prohibitive effect was neutralized by miR-758-3p depletion. The data depicted that circ_0006646 promoted the expression of RRM2 by sponging miR-758-3p in CC cells.
Knockdown of Circ_0006646 Blocked the Growth of Tumors In Vivo
We verified the effect of circ_0006646 silencing on tumor growth in mice by in vivo experiments, the data speculated that downregulation of circ_0006646 repressed the volume and weight of tumors in mice (Fig. 9A–C). Besides, the content of circ_0006646 in sh-circ_0006646 group was decreased in comparison to sh-NC group (Fig. 9D), and the mRNA and protein expression of RRM2 in sh-circ_0006646 group was aberrantly restrained (Fig. 9E, F). Also, IHC staining results confirmed that RRM2 positive cells were reduced in the tumor tissues of the sh-circ_0006646 group (Fig. 9G). The data above all hinted that silence of circ_0006646 confined CC tumors growth in vivo.
Circ_0006646 downregulation repressed tumors growth in vivo. Xenograft tumor experiment was launched using transfected SiHa cells in nude mice (n = 6/group) and was divided into sh-NC and sh-circ_0006646 groups. A Tumors volume was tested every week. B Tumor size was shown. C Tumors weight was detected on the fifth week. D–E QRT-PCR was used for examining circ_0006646 and RRM2 expression in tumors. F Western blot was used to analyze the expression of RRM2. G IHC was used to assess RRM2 positive cells. *P < 0.05
Circ_0006646 Was Upregulated in the Exosomes of CC Patients
Exosome, small vesicles containing RNA and proteins, was secreted by all cultured cells (Schageman et al. 2013). We extracted exosomes from the CC patients and healthy controls, and the morphology and concentration of exosome were identified by TEM and NTA, respectively (Fig. 10A, B). And the results of western blot displayed that CD83 and TSG101 in exosome derived from CC patients and healthy controls were significantly fortified, indicating that exosome was successfully isolated (Fig. 10C). Through qRT-PCR, circ_0006646 was confirmed to be highly expressed in the exosomes of CC patients (Fig. 10D). According to relative expression of circ_0006646 in exosomes, the AUC value was 0.8691, suggesting that the population of CC patients and healthy control could be distinguished according to exosomal circ_0006646 expression (Fig. 10E). In addition, we found that the content of exosomal circ_0006646 did not change significantly after being placed at different room temperatures or treated with NaOH and HCl, which confirmed the stability of exosomal circ_0006646 (Fig. 10F, G). Therefore, these results demonstrated that exosomal circ_0006646 might be used as a diagnostic indicator of CC.
Circ_0006646 was increased in exosomes of CC patients. A The morphology of exosome was observed by TEM. B The concentration and size of exosome were observed by NTA. C The protein expression of exosome surface markers was determined by western blot assay. D QRT-PCR was used to measure circ_0006646 expression in CC patients and healthy controls. E AUC curve was used to analyze the used to evaluate the value of exosomal circ_0006646 as a diagnostic indicator of CC. F–G Detection of the stability of exosomal circ_0006646. *P < 0.05
Discussion
CC was the most pervasive gynecological malignant tumor of female reproductive tract (Beharee et al. 2019). In recent years, the screening, prevention, and treatment of CC have made good progress, resulting in a decrease in the prevalence of CC (Shangguan et al. 2020). Targeted therapy has become a research hotspot in recent years, providing a new idea for the treatment of CC (Crafton and Salani 2016).
The abnormal expression of circRNAs is very important in the development of CC (Tornesello et al. 2020). In our present study, we determined the role and mechanism of circ_0006646 in CC. Consistent with previous study (Li et al. 2020), we confirmed that circ_0006646 had elevated expression in CC cells. Functional results showed that knockdown of circ_0006646 drastically restrained CC cell growth and metastasis. Moreover, circ_0006646 depletion hindered CC tumor growth in vivo. All the results disclosed that circ_0006646 facilitated CC progression. Importantly, we successfully isolated the exosomes from CC patients, and found that circ_0006646 expression was increased in CC patients. These results provided evidence that exosomal circ_0006646 might be a diagnostic target for CC patients.
Circinteractome, StarBase 3.0, and circBank were important tools for screening circRNAs targets, and we found the direct targeting between circ_0006646 and miR-758-3p by using these tools. Similar to the results of previous study (Ding et al. 2021; Xie et al. 2021), we confirmed that miR-758-3p was downregulated in CC cells. Studies had suggested that miR-758-3p restrained the malignant of CC by retarding the CC cell proliferation and metastasis (Xie et al. 2021; Ding et al. 2021). In our study, we illustrated that miR-758-3p overexpression could block CC cell growth and metastasis. Furthermore, the effects of circ_0006646 deficiency on CC cell development were rescued by miR-758-3p decrease, confirming that circ_0006646 promoted CC malignant progression via targeting miR-758-3p.
StarBase 3.0 predicted the target of miR-758-3p to improve the mechanism of circ_0006646 in regulating CC process. And RRM2 was passively regulated by miR-758-3p in CC. RRM2 was identified as a biomarker for CC and it was connected with the inferior overall survival of CC patients (Yi et al. 2019). Downregulated RRM2 had been shown to limit the growth of CC in vivo and in vitro (Zhao et al. 2019). In this, we expounded that RRM2 was abnormally aggrandized in CC cells, and overexpression of RRM2 partly ameliorated the impacts of miR-758-3p on CC cell growth and metastasis, confirming that miR-758-3p suppressed CC cell progression by targeting RRM2. In addition, we found that circ_0006646 upregulated RRM2 expression by binding miR-758-3p. In brief, circ_0006646 induced the growth of CC by regulating miR-758-3p/RRM2 axis.
Conclusions
To sum up, this study was the first to study the function and mechanism of circ_0006646 in CC. Our research showed that circ_0006646 promoted CC malignant progression depending on the regulation of miR-758-3p/RRM2 axis. In addition, exosomal circ_0006646 might be a potential biomarker for the diagnosis of CC. Our study provides a potential target for the treatment of CC and has important clinical significance.
Data Availability
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
References
Barbato S, Solaini G, Fabbri M (2017) MicroRNAs in oncogenesis and tumor suppression. Int Rev Cell Mol Biol 333:229–268
Beharee N, Shi Z, Wu D, Wang J (2019) Diagnosis and treatment of cervical cancer in pregnant women. Cancer Med 8(12):5425–5430
Biewenga P, van der Velden J, Mol BW, Stalpers LJ, Schilthuis MS, van der Steeg JW et al (2011) Prognostic model for survival in patients with early stage cervical cancer. Cancer 117(4):768–776
Chen D, Ma W, Ke Z, Xie F (2018) CircRNA hsa_circ_100395 regulates miR-1228/TCF21 pathway to inhibit lung cancer progression. Cell Cycle 17(16):2080–2090
Chen J, Xu Z, Yu C, Wu Z, Yin Z, Fang F et al (2019) MiR-758-3p regulates papillary thyroid cancer cell proliferation and migration by targeting TAB1. Pharmazie 74(4):235–238
Chen L, Zhang X, Wang S, Lin X, Xu L (2020) Circ_0084927 facilitates cervical cancer development via sponging miR-142-3p and upregulating ARL2. Cancer Manag Res 12:9271–9283
Crafton SM, Salani R (2016) Beyond chemotherapy: an overview and review of targeted therapy in cervical cancer. Clin Ther 38(3):449–458
Ding Y, Yuan X, Gu W (2021) Circular RNA RBM33 contributes to cervical cancer progression via modulation of the miR-758-3p/PUM2 axis. J Mol Histol 52(2):173–185
Jin CY, Du L, Nuerlan AH, Wang XL, Yang YW, Guo R (2020) High expression of RRM2 as an independent predictive factor of poor prognosis in patients with lung adenocarcinoma. Aging (albany NY) 13(3):3518–3535
Juan C, Hua Q, Ruping Z, Tingting W (2018) miRNA-489 as a biomarker in diagnosis and treatment of cervical cancer. Bratisl Lek Listy 119(5):278–283
Li X, Ma N, Zhang Y, Wei H, Zhang H, Pang X et al (2020) Circular RNA circNRIP1 promotes migration and invasion in cervical cancer by sponging miR-629-3p and regulating the PTP4A1/ERK1/2 pathway. Cell Death Dis 11(5):399
Liu S, Li B, Li Y, Song H (2021) Circular RNA circ_0000228 promotes the malignancy of cervical cancer via microRNA-195-5p/ lysyl oxidase-like protein 2 axis. Bioengineered 12(1):4397–4406
Ma C, Luo H, Cao J, Gao C, Fa X, Wang G (2020) Independent prognostic implications of RRM2 in lung adenocarcinoma. J Cancer 11(23):7009–7022
Nordlund P, Reichard P (2006) Ribonucleotide reductases. Annu Rev Biochem 75:681–706
Pardini B, De Maria D, Francavilla A, Di Gaetano C, Ronco G, Naccarati A (2018) MicroRNAs as markers of progression in cervical cancer: a systematic review. BMC Cancer 18(1):696
Pérez-Herrero E, Fernández-Medarde A (2015) Advanced targeted therapies in cancer: drug nanocarriers, the future of chemotherapy. Eur J Pharm Biopharm 93:52–79
Revathidevi S, Murugan AK, Nakaoka H, Inoue I, Munirajan AK (2021) APOBEC: a molecular driver in cervical cancer pathogenesis. Cancer Lett 496:104–116
Schageman J, Zeringer E, Li M, Barta T, Lea K, Gu J et al (2013) The complete exosome workflow solution: from isolation to characterization of RNA cargo. Biomed Res Int 2013:253957
Shangguan H, Feng H, Lv D, Wang J, Tian T, Wang X (2020) Circular RNA circSLC25A16 contributes to the glycolysis of non-small-cell lung cancer through epigenetic modification. Cell Death Dis 11(6):437
Shen S, Zhang S, Liu P, Wang J, Du H (2020) Potential role of microRNAs in the treatment and diagnosis of cervical cancer. Cancer Genet 248–249:25–30
Sun H, Yang B, Zhang H, Song J, Zhang Y, Xing J et al (2019) RRM2 is a potential prognostic biomarker with functional significance in glioma. Int J Biol Sci 15(3):533–543
Sun Q, Qi X, Zhang W (2021) Knockdown of circRNA_0007534 suppresses the tumorigenesis of cervical cancer via miR-206/GREM1 axis. Cancer Cell Int 21(1):54
Tang Q, Chen Z, Zhao L, Xu H (2019) Circular RNA hsa_circ_0000515 acts as a miR-326 sponge to promote cervical cancer progression through up-regulation of ELK1. Aging (albany NY) 11(22):9982–9999
Tang X, Ren H, Guo M, Qian J, Yang Y, Gu C (2021) Review on circular RNAs and new insights into their roles in cancer. Comput Struct Biotechnol J 19:910–928
Tornesello ML, Faraonio R, Buonaguro L, Annunziata C, Starita N, Cerasuolo A et al (2020) The role of microRNAs, long non-coding RNAs, and circular RNAs in cervical cancer. Front Oncol 10:150
Wang Q, Zhuo Z (2021) Circ_0019435 exerts its functions in the cellular process of cervical cancer via epigenetically silencing DKK1 and PTEN. Reprod Sci 28(10):2989–2999
Wang J, Yi Y, Chen Y, Xiong Y, Zhang W (2020) Potential mechanism of RRM2 for promoting cervical cancer based on weighted gene co-expression network analysis. Int J Med Sci 17(15):2362–2372
Xie H, Wang J, Wang B (2021) Circular RNA circ_0003221 promotes cervical cancer progression by regulating miR-758-3p/CPEB4 axis. Cancer Manag Res 13:5337–5350
Yang Y, Lin J, Guo S, Xue X, Wang Y, Qiu S et al (2020) RRM2 protects against ferroptosis and is a tumor biomarker for liver cancer. Cancer Cell Int 20(1):587
Yi Y, Liu Y, Wu W, Wu K, Zhang W (2019) Reconstruction and analysis of circRNA-miRNA-mRNA network in the pathology of cervical cancer. Oncol Rep 41(4):2209–2225
Zhao H, Zheng GH, Li GC, Xin L, Wang YS, Chen Y et al (2019) Long noncoding RNA LINC00958 regulates cell sensitivity to radiotherapy through RRM2 by binding to microRNA-5095 in cervical cancer. J Cell Physiol 234(12):23349–23359
Acknowledgements
None
Funding
None.
Author information
Authors and Affiliations
Author notes
Fen Yu and Fang Luo are Co-first authors.
Contributions
QH designed and supervised the study. FY conducted the experiments and drafted the manuscript. FL collected and analyzed the data. XZ contributed the methodology and edited the manuscript. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Ethical Approval
The research was approbated by the Ethics Committee of Puren Hospital Affiliated to Wuhan University of Science and Technology.
Consent to Participate
The Animal Research Ethics Committee of Puren Hospital Affiliated to Wuhan University of Science and Technology gave a license to the animal experiments.
Consent for Publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yu, F., Luo, F., Zhang, X. et al. Circ_0006646 Accelerates the Growth and Metastasis of Cervical Cancer by Elevating RRM2 Through miR-758-3p. Biochem Genet 61, 1300–1318 (2023). https://doi.org/10.1007/s10528-022-10320-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10528-022-10320-6