Abstract
A large number of studies have shown that circular RNAs (circRNAs) are of great significance in the occurrence and development of colorectal cancer (CRC). The purpose of this study was to explore the mechanism of circ_0001535 in CRC. The expressions of circ_0001535, miR-433-3p and recombination signal-binding protein Jκ (RBPJ) mRNA and protein in CRC tissues and cells were detected by quantitative real-time polymerase chain reaction (qRT-PCR) or western blot. The effect of circ_0001535 on cell proliferation was detected using the Cell Counting Kit-8 (CCK-8) assay, colony formation assay and 5-ethynyl-2′-deoxyuridine (EdU) assay. The effects of circ_0001535 on migration, invasion, angiogenesis and apoptosis were investigated by wound healing assay, transwell assay, tube formation assay and flow cytometry, respectively. The interactions between miR-433-3p and circ_0001535 or RBPJ were studied using dual-luciferase reporter assay and RNA immunoprecipitation (RIP) assay. Xenograft tumor assay was performed to verify the role of circ_0001535 in tumor growth in vivo. The results showed that circ_0001535 and RBPJ mRNA expression levels were up-regulated and miR-433-3p was down-regulated in CRC tissues and cells. Circ_0001535 knockdown inhibited cell proliferation, migration, invasion, angiogenesis as well as promoted apoptosis in CRC cells. After analysis, it was found that circ_0001535 acted as a competing endogenous RNA (ceRNA) to inhibit miR-433-3p and then up-regulate RBPJ in CRC cells. In addition, in vivo experiment had shown that circ_0001535 knockdown inhibited tumor growth by up-regulating miR-433-3p and inhibiting RBPJ expression. The circ_0001535/miR-433-3p/ RBPJ axis plays a catalytic role in the progression of CRC, which may provide new insights into the molecular mechanism of CRC.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Colorectal cancer (CRC) is one of the malignant tumors with the highest incidence in the world, and its morbidity and mortality in China are also increasing year by year (Sung et al. 2021). It is a serious public health problem that threatens physical and mental health of people (Aran et al. 2016). The diagnosis and treatment of CRC are diversified (Binefa et al. 2014). However, due to the atypical early symptoms of CRC, most CRC patients are diagnosed in the middle and late stages, which greatly reduces the survival rate and quality of life of patients (Yang et al. 2019). Therefore, the identification of some promising biomarker molecules and their molecular mechanisms may provide additional options for the treatment of CRC.
Circular RNA (circRNA) is a type of RNA molecule with a closed circular structure, which is more stable than linear RNA and is widely distributed in various tissues (Cao et al. 2021). In recent years, studies have found that circRNA is closely related to the occurrence and development of malignant tumors, and different circRNAs show different expression specificity in tumor tissues (Zhang et al. 2018). Therefore, circRNA is expected to become novel biomarkers for early diagnosis of tumors and potential targets for drug therapy (Wang et al. 2020a). For example, circ_0004771 has been identified as a novel biomarker for the diagnosis and prognosis of CRC (Pan et al. 2019). Yang et al. found that circ_0001535 played a critical role in the growth and metastasis of CRC and might serve as a potential therapeutic target for CRC metastasis (Yang et al. 2020), whereas the function and action mechanism of the circ_0001535 in CRC are not clear, and it is worth exploring.
A large number of studies have found that circRNA can act as a miRNA sponge by targeting the binding sites of microRNA (miRNA) and thus regulating mRNA expression (Huang et al. 2020). For instance, circ_001680 promoted cell proliferation in CRC by binding miR-340 to up-regulate BMI1 (Jian et al. 2020). Circ-SMARCA5 curbed the malignant development of CRC by sponging miR-39-3p and enhancing ARID4B expression (Miao et al. 2020). This study will explore the downstream miRNA and mRNA in circ_0001535-mediated regulatory network in CRC.
In this study, we verified the role of circ_0001535 in proliferation, migration, invasion, angiogenesis and apoptosis of CRC cells. More importantly, we explored the underlying mechanisms of circ_0001535 in CRC development.
Materials and Methods
Clinical Tissue Samples
A total of 30 tumor tissues and paired adjacent tissues of patients with pathologically confirmed CRC who underwent surgical resection at Cancer Hospital of China Medical University were collected, and the tissue samples were immediately placed into -80 °C refrigerator for backup after acquisition. None of the enrolled patients received any treatment before surgery. The study has been approved by the Ethics Committee of Cancer Hospital of China Medical University, and all enrolled patients have signed informed consent.
Hematoxylin–Eosin (HE) Staining
Hematoxylin and Eosin Staining Kit (Beyotime, Shanghai, China) was used for tissue staining. The collected normal tissue and CRC tissue sections were fixed and stained using the staining solution in the kit according to the instructions. The microslides were washed in a 70% ethanol solution and then viewed and photographed under a microscope (Leica, Wetzlar, Germany).
Cell Culture and Cell Transfection
Human SW480 and HCT-116 cells and normal NCM460 cells were purchased from Cobioer (Nanjing, China). All cell lines were cultured in DMEM medium (containing 10% fetal bovine serum; GIBCO, Rockville, MD, USA) in 37 C, 5% CO2 constant temperature incubator. When the cell density reached about 80%, they were digested by trypsin for cell passage. Follow-up experiments were conducted during logarithmic growth period. According to the transfection kit instructions, Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA) was transfected with circ_0001535 small interfering RNA (siRNA) (si-circ_0001535), miR-433-3p mimic, miR-433-3p inhibitor, RBPJ overexpression plasmid (pc-RBPJ) and corresponding negative controls (si-NC, miRNA NC, inhibitor NC, pc-NC), respectively. After 48 h transfection, cells were collected for subsequent experiments.
RNA Extraction, Reverse Transcription and Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Total RNA of each group was extracted with TRIzol reagent (Sigma-Aldrich, St. Louis, MO, USA), and the concentration and purity of RNA were determined. The cDNA was synthesized with PrimeScript RT reagent Kit (TaKaRa, Dalian, China) and amplified by PCR instrument. The expression levels of circ_0001535, FAM13B, miR-433-3p and RBPJ mRNA were detected by SYBR PCR Master Mix kit (TaKaRa), with GAPDH or U6 as internal reference. All the primers used in the experiment were synthesized by TaKaRa and are listed in Table 1.
RNase R Degradation Assay
This assay was used to assess the stability of circ_0001535 and its linear isoform. A total of 2 μg isolated RNA from SW480 and HCT-116 cells was incubated with 6 U RNase R (Tiangen, Beijing, China) for 20 min. An equal RNA was then used to perform reverse transcription into cDNA, and circ_0001535 level was confirmed by qRT-PCR.
Cell Counting Kit-8 (CCK-8) Assay
The transfected cells were added to 96-well plate and cultured for 0, 24, 48 and 72 h, respectively, with 10 μL CCK-8 solution (Sigma-Aldrich) added, and cultured in an incubator with 5% CO2 at 37 °C for 2 h. Then, the absorbance value at 450 nm was measured in a microplate reader (Bio-Rad, Hercules, CA, USA).
5-Ethynyl-2'-Deoxyuridine (EdU) Assay
According to Click-iT EdU Imaging Kit (Thermo Fisher Scientific, Waltham, MA, USA) instructions, cells were incubated with EdU solution for 2 h. The cells were fixed with 4% formaldehyde (Sigma-Aldrich) for 30 min and then incubated with Triton X-100 for 10 min. The Apollo staining reaction solution and DAPI staining solution (Thermo Fisher Scientific) were added and incubated for 30 min, respectively. The experiment was observed and photographed under a fluorescence microscope (Leica).
Wound Healing Assay
The cells were added to the 12-well plate, and after transfection, the surface of the plate was scraped with the tip of a 20 μL sterile micropipette to generate wounds. The serum-free medium (GIBCO) was replaced, and the images were photographed using a microscope (× 40, Leica) at 0 h and 24 h.
Transwell Assay
The transwell chamber (BD Bioscience, Bedford, MA, USA) was first added with diluted Matrigel (BD Bioscience) before the experiment. Cells (2 × 104 cells) resuspended in serum-free medium were added into the upper chamber, and 800 μL complete culture medium was added into the lower chamber and then placed into the cell incubator for 24 h. The cells were removed and stained with crystal violet (Solarbio, Beijing, China) for 20 min at room temperature and then rinsed twice with 1 × phosphate-buffered saline (PBS; GIBCO), and the cells at the back of the cells were counted under the microscope (× 200, Leica).
Tube Formation Assay
In short, the melted Matrigel (BD Bioscience) was added to the pre-chilled 96-well plate (50 μL/well) and incubated at 37 °C for 45 min. Next, the transfected SW480 and HCT-116 cells were seeded into the well (approximately 3 × 104 cells per well) and incubated at 37 °C for 4 h. Eventually, the cells were observed and photographed under the microscope (× 200, Leica).
Flow Cytometry
Annexin V-FITC/PI double standard staining kit (BD Pharmingen, San Diego, CA, USA) was used to detect cell apoptosis. Cells were cultured in a 5% CO2 incubator at 37 °C for 48 h. Cells were collected and resuspended in 200 μL binding buffer. 10 μL Annexin V-FITC and 5 μL PI were gently mixed and reacted at room temperature for 15 min under dark conditions. Cell apoptosis was detected by flow cytometry.
Western Blot
Total proteins from tissues and cells were extracted using RIPA protein lysis solution (Beyotime). Proteins were transferred to PVDF membrane (Bio-Rad) by horizontal and vertical electrophoresis. After that, the PVDF membrane was sealed with 5% skim milk powder at room temperature for 1 h and then incubated with the primary antibodies (anti-RBPJ, 1:200 dilution, ab217683; anti-Bax, 1:1000 dilution, ab32503; anti-Bcl-2, 1:10,000 dilution, ab32124; anti-GAPDH, 1:10,000 dilution, ab181602; all from Abcam) at 4 °C overnight. Later, the second antibody (1:50,000 dilution, ab205718; Abcam) was added at room temperature for 2 h the next day. Enhanced chemiluminescent (ECL) solution (Thermo Fisher Scientific) was used to develop and observe the expression levels of target proteins. GAPDH was used as internal reference.
Dual-Luciferase Reporter Assay
The binding fragment between miR-433-3p and circ_0001535 or RBPJ 3’UTR was predicted by bioinformation prediction website, and the binding fragment was inserted into pMIR-REPORT (Thermo Fisher Scientific) to construct circ_0001535 and RBPJ 3’UTR wild plasmids, and the binding site was mutated by gene mutation technology to construct circ_0001535 and RBPJ 3’UTR mutant plasmids. Cells were transfected with WT-circ_0001535, WT-RBPJ-3’UTR, MUT-circ_0001535 or MUT-RBPJ-3’UTR and miR-433-3p mimic or miRNA NC, respectively. The dual-luciferase reporter kit (Promega, Madison, WI, USA) was used to detect the dual-luciferase activity.
RNA Immunoprecipitation (RIP) Assay
RNA immunoprecipitation assay kit (MBL, Woburn, MA, USA) was used to perform IgG RIP in SW480 and HCT-116 cells using the normal rabbit IgG antibody (anti-IgG) or perform Argonaute 2 (Ago2) RIP using anti-Ago2 (RNO03M, MBL). Cell lysis was incubated with Protein A/G Agarose beads (YEASEN, Shanghai, China) and anti-IgG or anti-Ago2 for 12 h, and co-precipitated RNAs were recovered in TRIzol (Invitrogen) for qRT-PCR.
Immunohistochemistry (IHC) Assay
The specimens were fixed by formaldehyde, embedded in paraffin and sectionalized. After dewaxing and repair, primary antibodies including anti-RBPJ (1:100 dilution), anti-Ki67 (1:100 dilution, ab92742; Abcam) or anti-MMP-9 (1:1000 dilution, ab283575; Abcam) were used overnight in a refrigerator at 4 °C. Then, the second antibody (1:10,000 dilution, ab205718; Abcam) was added and stood at room temperature for 50 min. Diaminobenzidine (DAB; Beyotime) was used for coloration, and hematoxylin (Beyotime) was used for redyeing. The field (200 × magnification) was randomly selected under the microscope (Leica) to observe and photograph.
Xenograft Models
The circ_0001535 short hairpin RNA (sh-circ_0001535) and its control (sh-NC) were designed and supplied by Sangon Biotech (Shanghai, China). The stably transfected sh-NC or sh-circ_0001535 SW480 cells were subcutaneously injected into 6 nude mice (female, 6–8 weeks old, Vital River Laboratory Animal Technology, Beijing, China) with 2 × 106 cells per mouse. The growth and state of the transplanted tumor were observed every day. The length (L) and width (W) of the tumor were measured weekly, and the volume was calculated. The calculation formula was: tumor volume = L × W2 × 0.5. After 4 weeks, the tumor was removed for further analysis. The study was approved by the Animal Research Ethics Committee of Cancer Hospital of China Medical University Hospital.
Statistical Analysis
SPSS statistical software was used to process the data. Measurement data were expressed as mean ± standard deviation, analysis of variance (ANOVA) was used for comparison among multiple groups, and Student’s t test was used for comparison between 2 groups. When P < 0.05, the difference was statistically significant, and each experiment was repeated more than 3 times.
Results
The Expression of circ_0001535 was Markedly Promoted in CRC Tissues and Cells
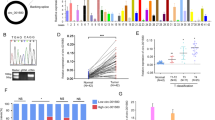
At first, HE staining of normal tissues and CRC tissues is shown in Fig. 1A. Then, we addressed whether the expression of circ_0001535 was abnormal in CRC. Compared with control tissues and cells, circ_0001535 expression was significantly elevated in 30 CRC tissues and CRC cell lines (SW480 and HCT-116) by qRT-PCR (Fig. 1B and C). Circ_0001535 was formed by back splicing of exon 4–19 (Fig. 1D). In SW480 and HCT-116 cells, the expression of FAM13B mRNA was sharp decreased and circ_0001535 level had few change after digestion by RNase R, suggesting the cyclic structure of circ_0001535 (Fig. 1E and F). Summarily, the expression of circ_0001535 was significantly increased in CRC.
The expression level of circ_0001535 in CRC tissues and cells. A HE staining of normal tissues and colorectal cancer tissues. B–C The expression level of circ_0001535 was analyzed by qRT-PCR assay in CRC tissues (n = 30) and cells (n = 3). D Circ_0001535 structure diagram was exhibited. E–F The relative levels of circ_0001535 and FAM13B were measured after treatment with RNase R by qRT-PCR (n = 3). ***P < 0.001 (Student’s t test)
The Functional Roles of circ_0001535 in Cell Proliferation, Migration, Invasion, Angiogenesis and Apoptosis of CRC Cells
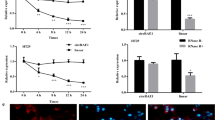
We transfected si-circ_0001535 into SW480 and HCT-116 cells. Figure 2A shows that si-circ_0001535 transfection down-regulated the expression of circ_0001535. Then, CCK-8 and EdU assays showed that circ_0001535 knockdown inhibited the viability and proliferation of SW480 and HCT-116 cells (Fig. 2B and C). It could be seen from wound healing assay and transwell assay that the migration and invasion of SW480 and HCT-116 cells were blocked due to the knockdown of circ_0001535 (Fig. 2H and I). Tube formation assay showed that circ_0001535 silencing significantly inhibited angiogenesis rate (Fig. 2F). Apoptosis of SW480 and HCT-11 cells was analyzed by flow cytometry. As shown in Fig. 2G, knockdown of circ_0001535 obviously induced the apoptosis of SW480 and HCT-11 cells. Additionally, compared with the control group, circ_0001535 knockdown decreased the expression of anti-apoptotic protein B-cell lymphoma-2 (Bcl-2) and increased the expression of pro-apoptotic protein BCL2-associated X protein (Bax) (Fig. 2H and I). In short, circ_0001535 knockdown could inhibit the development of CRC cells.
The functional roles of circ_0001535 on cell proliferation, migration, invasion, angiogenesis and apoptosis in CRC cells. SW480 and HCT-116 cells were transfected with si-circ_0001535 and si-NC. A The expression level of circ_0001535 was tested by qRT-PCR assay. B Cell viability was detected by CCK-8 assay. C Cell proliferation was measured by EdU assay. D–E Cell migration and invasion were measured by wound healing and transwell assays. F Tube formation assay was used to assess tube formation and angiogenesis rate. G Flow cytometry analysis was used to assess apoptosis of transfected SW480 and HCT-116 cells. (H-I) Western blot analysis was used to examine the expression of Bcl-2 and Bax. n = 3. ***P < 0.001 (Student’s t test)
Circ_0001535 Acted as a Sponge for miR-433-3p
In order to study the mechanism of circ_0001535 as competing endogenous RNA (ceRNA) in CRC, miRNA associated with circ_0001535 was predicted, and it was found that circ_0001535 had target sites with miR-433-3p (Fig. 3A). Firstly, SW480 and HCT-116 cells were transfected with miR-433-3p mimic, and the expression analysis showed that the level of miR-433-3p was significantly increased after transfection (Fig. 3B). We used the dual-luciferase reporter and RIP assays to observe the relationship between circ_0001535 and miR-433-3p. Luciferase report results showed that miR-433-3p significantly decreased luciferase activity in WT-circ_0001535-transfected cells, but there was no change in luciferase activity of MUT-circ_0001535-transfected cells (Fig. 3C and D). Moreover, RIP analysis showed that circ_0001535 and miR-433-3p were mainly enriched in Ago2 complexes compared with IgG complexes (Fig. 3E and F). Next, through performing qRT-PCR analysis, we found that miR-433-3p expression was reduced in CRC tumor tissues and cells (Fig. 3G and H). Furthermore, silencing of circ_0001535 significantly increased the expression of miR-433-3p (Fig. 3I). In summary, circ_0001535 acted as a sponge for miR-433-3p to inhibit the expression of miR-433-3p in CRC cells.
MiR-433-3p was a direct target of circ_0001535. A The complementary sequences between miR-433-3p and circ_0001535 were shown. B The expression level of miR-433-3p was detected by qRT-PCR after SW480 and HCT-116 cells were transfected with miR-433-3p mimic or miRNA NC (n = 3). C–F Dual-luciferase reporter and RIP assays were performed to confirm the association between miR-433-3p and circ_0001535 (n = 3). G–H The relative expression level of miR-433-3p was measured by qRT-PCR assay in CRC tissues (n = 30) and cells (n = 3). I The expression level of miR-433-3p was detected by qRT-PCR after SW480 and HCT-116 cells were transfected with si-NC or si-circ_0001535 (n = 3). ***P < 0.001 (Student’s t test)
MiR-433-3p Inhibition Recovered the Effects of circ_0001535 Knockdown on Cell Proliferation, Migration, Invasion, Angiogenesis and Apoptosis
SW480 and HCT-116 cells were transfected with miR-433-3p inhibitor, and the expression analysis showed that the level of miR-433-3p was significantly decreased after transfection with miR-433-3p inhibitor (Fig. 4A). In order to further confirm that circ_0001535 acted on the progression of CRC cells through miR-433-3p, si-circ_0001535 and miR-433-3p inhibitor were co-transfected into SW480 and HCT-116 cells. The results showed that miR-433-3p inhibitor could eliminate the inhibition in cell proliferation induced by circ_0001535 knockdown (Fig. 4B and C). Circ_0001535 knockdown significantly inhibited the invasion and migration of SW480 and HCT-116 cells, and co-transfection with miR-433-3p inhibitor partially offset the effects of circ_0001535 knockdown on the invasion and migration (Fig. 4D and E). Furthermore, tube formation assay showed that si-circ_0001535 transfection significantly reduced the angiogenesis rate in SW480 and HCT-116 cells, while miR-433-3p inhibitor transfection recovered this effect (Fig. 4F). Flow cytometry analysis indicated that knockdown of circ_0001535 significantly promoted apoptosis of SW480 and HCT-116 cells, whereas miR-433-3p inhibition restored this effect (Fig. 4G). And the effect of circ_0001535 knockdown on Bcl-2 and Bax protein expression was reversed by the co-transfection of si-circ_0001535 and miR-433-3p inhibitor (Fig. 4H and I). The overall results confirmed that miR-433-3p inhibitor restored the effects caused by circ_0001535 knockdown in CRC cells.
Circ_0001535/miR-433-3p axis regulated proliferation, migration, invasion, angiogenesis and apoptosis of CRC cells. A The expression level of miR-433-3p was detected by qRT-PCR after SW480 and HCT-116 cells were transfected with inhibitor NC or miR-433-3p inhibitor (Student’s t test). B–C CCK-8 and EdU assay was conducted to assess cell viability and proliferation level in transfected SW480 and HCT-116 cells (ANOVA). D–E The cell migration and invasion were measured by wound healing assay and transwell assays (ANOVA). F The tube formation and angiogenesis rate were detected by tube formation assay (ANOVA). G The cell apoptosis was quantified by flow cytometry assay (ANOVA). H–I Western blot analysis was performed to estimate Bcl-2 and Bax levels (ANOVA). n = 3. **P < 0.01, ***P < 0.001
RBPJ was a Target Gene of miR-433-3p
StarBase predicted that RBPJ was the target of miR-433-3p, and there was a special binding site between RBPJ 3’UTR and miR-433-3p fragment (Fig. 5A). To observe the relationship between RBPJ and miR-433-3p, we conducted dual-luciferase reporter and RIP assays. After co-transfection with miR-433-3p, the luciferase activity was decreased in the WT-RBPJ-3’UTR group, but there was no significant difference in the MUT-RBPJ-3’UTR group (Fig. 5B and C). RIP assay results displayed that circ_0001535 and miR-433-3p were significantly enriched in anti-Ago2 group (Fig. 5D and E). Meanwhile, we detected increased expression of RBPJ in CRC tumor tissues and cells (Fig. 5F–H). Moreover, transfection tests revealed that miR-433-3p could significantly decrease the expression of RBPJ protein (Fig. 5I). Hence, RBPJ was a direct target gene of miR-433-3p.
MiR-433-3p targeted RBPJ in CRC. A MiR-433-3p had the binding regions in 3’UTR of RBPJ mRNA. B–E Interaction between miR-433-3p and RBPJ was confirmed by dual-luciferase reporter gene and RIP assays (n = 3). F–H The mRNA and protein expression levels of RBPJ were determined by qRT-PCR and western blot analysis in CRC tissues (n = 30) and cells (n = 3). I qRT-PCR and western blot were used to examine RBPJ level in transfected SW480 and HCT-116 cells (n = 3). ***P < 0.001 (Student’s t test)
RBPJ Overexpression Recovered the Effects of miR-433-3p on Cell Proliferation, Migration, Invasion, Angiogenesis and Apoptosis
Firstly, we designed RBPJ overexpression plasmid to increase the expression of RBPJ protein (Fig. 6A). The cell viability and proliferation curbed by miR-433-3p were significantly recovered in miR-433-3p mimic and pc-RBPJ co-transfected cells (Fig. 6B and C). Besides, RBPJ overexpression significantly restored the cell migration and invasion inhibited by miR-433-3p (Fig. 6D and E). Increased miR-433-3p reduced the angiogenesis rate of SW480 and HCT-116 cells, but this effect was overturned after co-transfection with miR-433-3p mimic and pc-RBPJ (Fig. 6F). After transfection with pc-RBPJ, the increased apoptosis of SW480 and HCT-116 cells induced by miR-433-3p was reversed (Fig. 6G). Furthermore, the expression changes of apoptosis-related proteins in cells also confirmed the above conclusion (Fig. 6H and I). Subsequently, SW480 and HCT-116 cells were transfected with si-circ_0001535 or si-circ_0001535+miR-433-3p inhibitor, with si-NC or si-circ_0001535+inhibitor NC as the control. The results showed that transfection of si-circ_0001535+miR-433-3p inhibitor could recover the reduced expression of RBPJ caused by si-circ_0001535 introduction (Fig. 7). In conclusion, circ_0001535 played a regulatory role in CRC cells via miR-433-3p/RBPJ axis.
MiR-433-3p/RBPJ axis proliferation, migration, invasion, angiogenesis and apoptosis of CRC cells. A The expression level of RBPJ detected by qRT-PCR after SW480 and HCT-116 cells were transfected with pc-NC or pc-RBPJ (Student’s t test). B–C Viability and proliferation levels of transfected SW480 and HCT-116 cells were detected by CCK-8 and EdU assays (ANOVA). D–E Cell migration and invasion were detected by wound healing assay and transwell assay (ANOVA). F Test tube formation assay was used to detect angiogenesis rate (ANOVA). G Flow cytometry to detect apoptosis (ANOVA). H–I Western blot to detect Bcl-2 and Bax levels (ANOVA). n = 3. **P < 0.01, ***P < 0.001
Circ_0001535 sponged miR-433-3p to indirectly regulate RBPJ expression. The expression of RBPJ in SW480 and HCT-116 cells transfected with si-circ_0001535 or si-circ_0001535+miR-433-3p inhibitor was detected by western blot, and si-NC or si-circ_0001535+inhibitor NC was used as control. n = 3. **P < 0.01, ***P < 0.001 (ANOVA)
circ_0001535 Knockdown Could Inhibit the Tumor Growth In Vivo
SW480 cells transfected with sh-circ_0001535 were subcutaneously injected into nude mice to study whether circ_0001535 regulated CRC tumor growth in vivo. As shown in Fig. 8A and B, compared with the control group, the tumor volume and weight in the circ_0001535 knockdown group were observably reduced. The change of mouse body weight was showed in Supplementary file 1. Then, qRT-PCR and western blot analyses of tumor tissues showed that compared with the control group, circ_0001535 and RBPJ expression was decreased and miR-433-3p expression was increased (Fig. 8C–E). Furthermore, IHC results showed that there were fewer RBPJ, Ki67 and MMP-9 positive cells in sh-circ_0001535 group compared with the control group (Fig. 8F). In summary, circ_0001535 knockdown inhibited tumor growth in vivo.
Suppression of circ_0001535 repressed tumor growth in vivo. A After inoculation, tumor volume was recorded once a week (n = 6), and the representative images of xenografts were showed. B Tumor weight was measured after 4 weeks when the mice were killed (n = 6). (C–E) The expression of circ_0001535, miR-433-3p and RBPJ in these excised tumor tissues was measured by qRT-PCR or western blot (n = 3). (F) The expression of RBPJ, Ki67 and MMP-9 in each group of tumors was analyzed by IHC. ***P < 0.001 (Student’s t test)
Discussion
With the continuous improvement of living standards and changes in diet, the incidence and mortality of colorectal cancer (CRC) are continuously increasing (Keum and Giovannucci 2019; Patel and Ahnen 2018). Although progress has been made in the diagnosis and treatment of CRC, the overall 5-year survival rate of CRC, especially in CRC patients with metastasis and recurrence, remains low (McQuade et al. 2017). Therefore, it is very important to further explore the development, invasion and metastasis mechanism of CRC, so as to find new therapeutic targets for CRC.
CircRNA is a type of non-coding RNA, whose 5' end and 3' end are joined together to form a covalently closed circular RNA structure, which is highly conserved and tissue specific (Li et al. 2020; Kristensen et al. 2019). Numerous studies have existed reporting that circRNA can act as cancer promoters to drive cancer progression (Yu et al. 2019; Zhao et al. 2019). For example, circRNA-002178 as ceRNA could promote the development of lung adenocarcinoma (Wang et al. 2020b). Zeng et al. discovered that circHIPK3 accelerated the growth and metastasis of CRC (Zeng et al. 2018). Circ_0001535 has previously been found to be up-regulated in hepatocellular carcinoma (Zhang et al. 2020). In our research, we proved the up-regulated expression of circ_0001535 in CRC and discussed its specific effect on the progression of CRC. Furthermore, circ_0001535 knockdown significantly weakened cell proliferation, migration, invasion and angiogenesis, as well as increased apoptosis of CRC in vitro. The in vivo studies proved that silencing of circ_0001535 significantly inhibited CRC tumor growth. These findings indicated that circ_0001535 was an oncogene in CRC.
CircRNA could release the interaction between miRNA and mRNA 3’ UTR by binding miRNA to regulate the expression level of downstream target gene of miRNA (Huang et al. 2020; Panni et al. 2020). It has previously been reported that circRIMS1 acted as an oncogene for bladder cancer through miR-433-3p-mediated CCAR1 expression (Wang et al. 2020c). Li et al. confirmed that circ_0023984 knockdown inhibited the tumor occurrence of esophageal squamous cell carcinoma by targeting miR-433-3p (Li and Li 2021). Similarly, miR-433-3p was identified as a potential target for circ_0001535 in our study, and the expression of miR-433-3p was controlled by circ_0001535 in CRC cells. Additionally, miR-433-3p inhibition recovered the effects of circ_0001535 knockdown on cell proliferation, migration, invasion, angiogenesis and apoptosis. Therefore, we identified a direct targeting relationship between circ_0001535 and miR-433-3p.
It was predicted that miR-433-3p had binding sites with RBPJ. RBPJ is a crucial transcription factor in the Notch pathway (Chen et al. 2019). It has been reported that knockdown of RBPJ inhibited the growth of lung cancer (Lv et al. 2015). Xiao et al. verified that RBPJ could inhibit the migration of endometrial cancer cells through the miR-155/NF-κB/ROS pathway (Xiao et al. 2019). In this research, we discovered that the expression of RBPJ was promoted in CRC. Moreover, the expression of RBPJ in CRC cells transfected with si-circ_0001535 was decreased, while inhibition of miR-433-3p restored its expression. Furthermore, rescued experiments showed that RBPJ overexpression rescued the effects of miR-433-3p on cell proliferation, migration, invasion, angiogenesis and apoptosis. More importantly, our findings confirmed that circ_0001535 served as a miR-433-3p sponge to alleviate the inhibitory effect of miR-433-3p on RBPJ, suggesting that circ_0001535 regulated the miR-433-3p/RBPJ axis in CRC. Consequently, we suggested that circ_0001535 might be a potential therapeutic target for CRC.
Conclusion
Our results showed that circ_0001535 expression was elevated in CRC tissues. Circ_0001535 knockdown could effectively increase the expression of miR-433-3p and thus inhibit the expression of RBPJ, thereby inhibiting the development of CRC tumors. We concluded that inhibition of circ_0001535 might be a therapeutic target for CRC.
Data Availability
The analyzed data sets generated during the present study are available from the corresponding author on reasonable request.
References
Aran V, Victorino AP, Thuler LC, Ferreira CG (2016) Colorectal cancer: epidemiology, disease mechanisms and interventions to reduce onset and mortality. Clin Colorectal Cancer 15:195–203
Binefa G, Rodriguez-Moranta F, Teule A, Medina-Hayas M (2014) Colorectal cancer: from prevention to personalized medicine. World J Gastroenterol 20:6786–6808
Cao YZ, Sun JY, Chen YX, Wen CC, Wei L (2021) The roles of circRNAs in cancers: perspectives from molecular functions. Gene 767:145182
Chen ELY, Thompson PK, Zuniga-Pflucker JC (2019) RBPJ-dependent Notch signaling initiates the T cell program in a subset of thymus-seeding progenitors. Nat Immunol 20:1456–1468
Huang A, Zheng H, Wu Z, Chen M, Huang Y (2020) Circular RNA-protein interactions: functions, mechanisms, and identification. Theranostics 10:3503–3517
Jian X, He H, Zhu J, Zhang Q, Zheng Z, Liang X et al (2020) Hsa_circ_001680 affects the proliferation and migration of CRC and mediates its chemoresistance by regulating BMI1 through miR-340. Mol Cancer 19:20
Keum N, Giovannucci E (2019) Global burden of colorectal cancer: emerging trends, risk factors and prevention strategies. Nat Rev Gastroenterol Hepatol 16:713–732
Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J (2019) The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet 20:675–691
Li T, Li S (2021) Circ_0023984 facilitates esophageal squamous cell carcinoma progression by regulating miR-433–3p/REV3L Axis. Dig Dis Sci. https://doi.org/10.1007/s10620-021-06916-4
Li J, Sun D, Pu W, Wang J, Peng Y (2020) Circular RNAs in cancer: biogenesis, function, and clinical significance. Trends Cancer 6:319–336
Lv Q, Shen R, Wang J (2015) RBPJ inhibition impairs the growth of lung cancer. Tumour Biol 36:3751–3756
McQuade RM, Stojanovska V, Bornstein JC, Nurgali K (2017) Colorectal cancer chemotherapy: the evolution of treatment and new approaches. Curr Med Chem 24:1537–1557
Miao X, Xi Z, Zhang Y, Li Z, Huang L, Xin T et al (2020) Circ-SMARCA5 suppresses colorectal cancer progression via downregulating miR-39-3p and upregulating ARID4B. Dig Liver Dis 52:1494–1502
Pan B, Qin J, Liu X, He B, Wang X, Pan Y et al (2019) Identification of serum exosomal hsa-circ-0004771 as a novel diagnostic biomarker of colorectal cancer. Front Genet 10:1096
Panni S, Lovering RC, Porras P, Orchard S (2020) Non-coding RNA regulatory networks. Biochim Biophys Acta Gene Regul Mech 1863:194417
Patel SG, Ahnen DJ (2018) Colorectal cancer in the young. Curr Gastroenterol Rep 20:15
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A et al (2021) Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 71:209–249
Wang Y, Li Z, Xu S, Guo J (2020a) Novel potential tumor biomarkers: circular RNAs and exosomal circular RNAs in gastrointestinal malignancies. J Clin Lab Anal 34:e23359
Wang J, Zhao X, Wang Y, Ren F, Sun D, Yan Y et al (2020b) circRNA-002178 act as a ceRNA to promote PDL1/PD1 expression in lung adenocarcinoma. Cell Death Dis 11:32
Wang F, Fan M, Cai Y, Zhou X, Tai S, Yu Y et al (2020c) Circular RNA circRIMS1 acts as a sponge of miR-433-3p to promote bladder cancer progression by regulating CCAR1 expression. Mol Ther Nucleic Acids 22:815–831
Xiao Y, Wang X, Dong X, Zhang Y, Liu H (2019) RBPJ inhibits the movability of endometrial carcinoma cells by miR-155/NF-kappaB/ROS pathway. Onco Targets Ther 12:8075–8084
Yang W, Shi J, Zhou Y, Liu T, Zhan F, Zhang K et al (2019) Integrating proteomics and transcriptomics for the identification of potential targets in early colorectal cancer. Int J Oncol 55:439–450
Yang H, Li X, Meng Q, Sun H, Wu S, Hu W et al (2020) CircPTK2 (hsa_circ_0005273) as a novel therapeutic target for metastatic colorectal cancer. Mol Cancer 19:13
Yu T, Wang Y, Fan Y, Fang N, Wang T, Xu T et al (2019) CircRNAs in cancer metabolism: a review. J Hematol Oncol 12:90
Zeng K, Chen X, Xu M, Liu X, Hu X, Xu T et al (2018) CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7. Cell Death Dis 9:417
Zhang Z, Xie Q, He D, Ling Y, Li Y, Li J et al (2018) Circular RNA: new star, new hope in cancer. BMC Cancer 18:834
Zhang T, Jing B, Bai Y, Zhang Y, Yu H (2020) Circular RNA circTMEM45A Acts as the Sponge of MicroRNA-665 to promote hepatocellular carcinoma progression. Mol Ther Nucleic Acids 22:285–297
Zhao X, Cai Y, Xu J (2019) Circular RNAs: biogenesis, mechanism, and function in human cancers. Int J Mol Sci. https://doi.org/10.3390/ijms20163926
Acknowledgements
Not applicable.
Funding
No funding was received.
Author information
Authors and Affiliations
Contributions
DL and JX contributed to conceptualization and methodology; ZM and DL were involved in formal analysis and data curation; ZM and DL contributed to validation and investigation; ZM, DL and JX were involved in writing—original draft preparation and writing—review and editing; and all authors approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethical Approval
The present study was approved by the ethical review committee of The Third Hospital of Mianyang (Sichuan Mental Health Center). Written informed consent was obtained from all enrolled patients.
Consent for Publication
Patients agree to participate in this work
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Mao, Z., Lin, D. & Xu, J. Hsa_circ_0001535 Regulates Colorectal Cancer Progression via the miR-433-3p/RBPJ Axis. Biochem Genet 61, 861–878 (2023). https://doi.org/10.1007/s10528-022-10287-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10528-022-10287-4