Abstract
Neuronal regeneration related protein (NREP), also known as P311, has been reported to participate in multiple biological processes. The detection of tumor biomarker favored a non-invasive early entry for cancer diagnosis and disease monitoring to prevent its worsening symptoms. This study is intended to investigate the clinical roles of NREP in gastric cancer (GC) and its effect on gastric cancer cell proliferation and angiogenesis. Our results demonstrated that NREP was typically upregulated in GC tissues compared with normal control. The Kaplan–Meier analysis showed correlations between increased NREP level and poor survival, indicating the prognostic value of NREP in GC patients. The expression levels of NREP varied by races, clinical T stages, and histologic grades. NREP expression was associated with tumor-associated immune infiltration. The NREP expression was powerfully associated with clinical characteristics of GC patients, in particular, with T stage and histologic grade. Gene ontology and KEGG signaling analysis indicated that NREP-related genes were predominantly enriched in various pathways. Additionally, knockdown of NREP inhibited human gastric adenocarcinoma cell proliferation and angiogenesis. Collectively, our results suggested that NREP may be an excellent biomarker for the clinical diagnosis, prognosis, and therapy of GC.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Gastric cancer (GC), always refers to gastric adenocarcinoma, is the fourth most common type of cancer worldwide (Jemal et al. 2011), and ranks the second leading cause of cancer-related mortality causes (Espinosa-Parrilla et al. 2014; Li et al. 2018). As an aggressive form of disease in the gastric tract (Sun et al. 2010), GC could not be easily found during early stages. With poor lifestyle and eating habits becoming more common in populations, the occurrence of GC is increasing year by year and getting younger and younger. Surgical resection and chemotherapy are standard treatments for GC patients (Zhang et al. 2017; Smyth et al. 2018, Kim et al. 2019). Despite these therapeutic approaches, the mortality rate of GC remains high. Up to now, there are still limited effective treatment options to cure GC and reduce its mortality. It is known that GC patients are relatively easier cured during the early stages before the tumor become more aggressive. Hence, early detection can buy time for GC treatment and reduce mortality of GC patients.
Neuronal regeneration related protein (NREP), a highly conserved 8-kDa protein, was first discovered in developing brain and mainly existed in pathological tissues (Stradiot et al. 2018; Zhou et al. 2020a, b). NREP regulates the development of neurons and smooth muscles, and contains several PEST-like domains (Pan et al. 2002). In NREP knockout mice, the capacity for learning and memory is impaired (Sun et al. 2008; Taylor et al. 2008). Studies have shown that NREP is involved in the regulation of chemokine-induced cell migration (Mariani et al. 2001; McDonough et al. 2005; Guimaraes et al. 2015). NREP expression gradually increases in muscular tissue during embryonic pig development (Ooi et al. 2006). Overexpression of NREP in skin cells resulted in an upregulation of TGFβ1 expression and increased skin cell proliferation (Tan et al. 2010). It was also reported that NREP regulates lung redox events, cell cycle progression, and distal lung development (Liu et al. 2020). Previous studies revealed that upregulated expression of NREP was linked to cell cycle progression and development, however, the role of NREP in GC has rarely been reported. Dysregulation of gene networks has been demonstrated to be linked with proliferation, invasion, immune infiltration, and angiogenesis of GC cells (Zhou et al. 2020a, b; Du et al. 2022). In this report, we proved that NREP upregulation was responsible for cell proliferation and angiogenesis, and associated with tumor-associated immune infiltration in GC.
In this study, we systematically investigated the expression of NREP in human cancers and its diagnostic significance in GC. Besides, we further examined the associations of NREP expression with clinical variables, immune infiltration, clinical characteristics, and related pathways in GC. Above all, we comprehensively assessed the feasibility that NREP may be a potential novel marker for diagnosis, prognosis and treatment of GC.
Materials and Methods
Online Analysis
Gene expression profiling interactive analysis (GEPIA) database (http://gepia.cancer-pku.cn/index.html) was applied to analyze the gene expression of NREP in different kinds of tumor samples and paired normal samples. Besides, NREP-related genes were obtained using GEPIA database.
Patient Datasets
The Cancer Genome Atlas (TCGA) and Genotype-Tissue Expression (GTEx) databases were used to analyze the expression of NREP mRNA. The batch effects were adjusted using the "sva" R package. The number of tumor samples from TCGA database was 408. Normal tissues (n = 211) from tumor free donors (GTEx) and GC patients (TCGA) were used as the control group. The following samples were enrolled in our study: 1) the value of gene expression was not 0; and 2) has survival information. The expression of NREP in GC patients was determined using scatter plots. The cut-off value of NREP was defined as the median value of gene expression based on order statistics.
Metascape Database Analysis
Metascape database (http://metascape.org/), a gene list analysis tool, was applied to predict their functional enrichment information of NREP-related genes in GC. “Multiple Gene list” module of the Metascape tool was utilized for gene annotation, gene ontology (GO), and KEGG enrichment analyses on NREP-related genes. The threshold conditions included: Input as species and Analysis as species: homo sapiens; P-value cut-off value < 0.01; a minimum overlap of 3 and the enrichment factor > 1.5 to obtain significant statistical differences. Then, GO (Biological Processes, Cellular Components, Molecular Functions), and KEGG pathways section in our results were downloaded and visualized using GraphPad Prism 9.0 software.
Kaplan–Meier plotter Database Analysis
Kaplan–Meier plotter database (https://kmplot.com/analysis/), which designed for the prognostic value of genes in several types of human cancers, was utilized to analyze the prognostic potential of NREP in GC. All datasets used for the analysis were as follows: GSE14210, GSE15459, GSE22377, GSE 29272, GSE51105, and GSE62254. The correlation between NREP expression and overall survival (OS), post-progression survival (PPS), and first progression survival (FPS) of GC patients was studied using the Kaplan–Meier curve. The hazard ratio (HR) with 95% confidence intervals (CI) and log-rank P-value were computed, and a P-value < 0.05 was considered to be statistically significant.
TIMER Database Analysis
TIMER database (www.cistrome.shinyapps.io/timer/) is designed for systematic analysis of the clinical impact of different immune cell in certain human tumors. The data of immune infiltration in GC was downloaded from TIMER database. Then, we used TIMER database to analyze the association of NREP expression with the abundance of 24 types of immune cells, including Mast cells, Tem, natural killer (NK) cells, follicular helper T (Tfh) cells, Eosinophils, Tcm, Tgd, dendritic cell (DC), iDC, pDC, Macrophages, T cells, B cells, CD8 T cells, NK CD56bright cells, Cytotoxic cells, T helper (Th) cells, T helper 1 (Th1) cells, TReg, aDC, NK CD56dim cells, neutrophils, Th2 cells, and Th17 cells. The Boxplots, using the expression of NREP expression as the variable, were generated to determine the abundance of immune cells in GC.
Clinical Sample Collection
GC tissues and adjacent normal tissues were collected from 23 patients from The Affiliated Tumor Hospital of Zhengzhou University, Henan Cancer Hospital. These GC patients were diagnosed with GC by pathological examination. The tissues were excised and immediately stored in liquid nitrogen for reverse transcription quantitative polymerase chain reaction (RT-qPCR) assay. All the patients were signed the informed consent before surgery and our subject was approved by ethic committee of The Affiliated Tumor Hospital of Zhengzhou University, Henan Cancer Hospital.
RT-qPCR
TRIzol reagent (Life Technologies, Carlsbad, CA, USA) was applied for RNA isolation, and QIAamp DNA Mini Kit (QIAGEN, Valencia, CA, USA) was utilized for cDNA synthesis. qPCR assay was undertaken with SYBR Premix Ex TaqTM kit from Takara (Dalian, China), in the light of the directions. The relative expression of NREP was normalized to that of GAPDH using 2−△△Ct method. The sequences of the qPCR primers are as following: NREP, forward: 5′-TTG AGC GAA TGC TAC CAG AG-3′ and reverse: 5′-AGG CGA GGC TAC GGA AAG-3′; GAPDH, forward: 5′-TCA AGG CTG AGA ACG GGA AG-3′ and reverse: 5′-TCG CCC CAC TTG ATT TTG GA-3′.
Western Blotting
Total protein was extracted with a Protein Extraction Kit (Boster, Wuhan, China). Protein (30 μg) was separated on the SDS-PAGE gel and then transferred to polyvinylidene difluoride membrane (Merck Millipore, Darmstadt, Germany). After that, membranes were blocked with 5% non-fat milk for 1 h at room temperature, and then incubated with anti-NREP antibody (1: 1000; Boster) or anti-GAPDH antibody (1: 1000; Boster) overnight at 4 °C. Membranes were washed three times and then incubated with goat anti-rabbit antibody (1: 2000; Boster) for 1 h at room temperature. The protein blots were observed with an ECL-Plus reagent (Invitrogen Life Technologies, Carlsbad, CA, USA).
Cell Culture and Transfection
Two human GC cell lines (SGC-7901 and AGS) and human umbilical vein endothelial cells (HUVECs) were purchased from Procell (Wuhan, China) and cultured in RPMI-1640 or Ham’s F-12 medium (Invitrogen, Carlsbad, CA, USA) containing 10% fetal bovine serum (FBS) (Solarbio, Beijing, China) and 1% penicillin and streptomycin (Solarbio) with 5% CO2 at 37 °C. For siRNA experiments, SGC-7901 and AGS cells were seeded into 12-well plates one day in advance. When the cells reached 70 ~ 80% confluence, 25 nM of each siRNA or 800 ng vectors and 4 μL Lipofectamine™ RNA iMAX Transfection Reagent (13,778,075; Thermo Fisher) were suspended in respective 120 μL serum-free medium. After gently mixing the two components, the mixed solution was then allowed to remain still for 30 min at room temperature and subsequently added to transfected cells. Experiments were conducted depending on the experimental design. The sequence of siRNA specific for NREP (si-NREP) is as following: 5′-AGU AGU UUA UGU UCA UCA ATT-3′ (sense) and 5′-UUG AUG AAC AUA AAC UAC UTT-3′ (antisense). The sequence of negative control (si-NC) is as following: 5′-UUC UCC GAA CGU GUC ACG UTT-3′ (sense) and 5′-ACG UGA CAC GUU CGG AGA ATT-3′ (antisense). These siRNA sequences, NREP overexpression vector, and pcDNA3.1 empty vectors were obtained from GenePharma (Suzhou, China).
Cell Viability and EdU Assays
GC cell viability was tested by the CCK-8 Cell Proliferation Kit (Beyotime, Beijing) according to the manufacturer’s recommendations. After transfection, 2 × 103 cells for each well were seeded in a 96-well plate. After culture for the indicated times (24 h, 48 h, or 72 h), cells were incubated with CCK-8 solution for 4 h and absorbance was measured at 450 nm using a microplate reader (Thermo Fisher). At 48 h after transfection, EdU assays were performed by Click-iT EdU imaging Kit (Invitrogen) following the manufacturer’s instructions. SGC-7901 and AGS Cells were incubated with 10 μM EdU for 2 h before detection. Cell nuclei were stained using DAPI staining (Beyotime). EdU-positive cells were photographed and automatically quantified by ScanR image acquisition (Olympus, Tokyo, Japan).
Tube Formation Assay
After culture for 48 h, conditioned medium of transfected SGC-7901 or AGS cells was collected and stored at − 80 °C for the next experiments. The Matrigel-based tube formation assay was performed to assess the cell angiogenesis of HUVECs. HUVECs were resuspended in FBS-free medium with GC cell culture supernatant and then seeded on Matrigel-coated plates (Corning) for each 24-well. Then cells were incubated at 37 °C and 5% CO2 cell incubator for 6 h, subsequently, photographed by Olympus microscope. The number of branch points was quantified by ImageJ software.
Statistical Analysis
Our data were given as means ± standard deviation. Unpaired t test was used to investigate the difference between two groups in Fig. 1A and B, 2C, 3B–G, 6A and C, 7C and D, 8A and B The paired t test was performed in Fig. 6B. The two-way ANOVA was performed in Fig. 7A and B. The multiple comparisons in Fig. 2A and B were analyzed by one-way ANOVA followed by Tukey post hoc test with Bonferroni adjust. A received operating characteristic (ROC) curve was applied to assess the diagnostic value of NREP in GC in Fig. 1C, and the bigger the area under the ROC curve (AUC), the better the accuracy. The correlation between NREP expression and survival of GC patients was studied using the Kaplan–Meier curve and log-rank test in Fig. 1D-F. The relationship between NREP and immune infiltration in GC patients was analyzed using Pearson correlation coefficient test in Fig. 3A. In addition, multivariate Cox analysis was carried out to elucidate the impact of NREP level on overall survival rate along with other clinical variables in patients with GC in Fig. 4. Difference is considered statistically significant at the P < 0.05 level.
The diagnostic and Prognostic roles of NREP in cancer. A Analysis of mRNA expression of NREP in multiple types of human tumor samples, B The NREP expression of 408 STAD tumor tissues were compared with that of 211 control tissue samples by RT-qPCR, C The ROC curve of NREP was shown. The area under the curve was 0.723. Kaplan–Meier curves of GC patients according to expression levels of NREP in GC tissues. Survival curves: overall survival (D), First progression survival (E), and Post-progression survival (F). *P < 0.05, **P < 0.01, and ***P < 0.001
NREP expression is associated with clinical variables in patients with GC. (A) Relative NREP expression levels in different races. (B) Relative NREP expression levels in different clinical tumor T stage (T1-T4). (C) Relative NREP expression levels of GC patients in G1&G2 or G3 histological grades. *P < 0.05, **P < 0.01, and ***P < 0.001
NREP expression is correlated immune infiltration. A Different immune cells were analyzed in the NREP low-expression group and NREP high-expression group by online tools. Enrichment scores of NK cells (B), Macrophages (C), Mast cells (D), Tgd (E), NK CD56bright cells (F), and Th17 cells (G) were classified by NREP expression level. *P < 0.05 and ***P < 0.001
Results
The Diagnostic and Prognostic Roles of NREP in GC
In the present study, we compared the expression profiles of NREP in tumor tissues (n = 408; TCGA) and non-tumor tissues (n = 211; TCGA and GTEx). As shown in Fig. 1A, we found that NREP expression was upregulated in multiple types of cancer, including BRCA, CHOL, ESCA, GBM, HNSC, KIRC, KIRP, LIHC, LUAD, LUSC, and stomach adenocarcinoma (STAD) (P < 0.05). To confirm the dysregulation of NREP in GC, we further validated the expression of NREP in samples of GC and non-tumor tissues. Results showed that NREP was highly expressed in STAD (P < 0.05) (Fig. 1B). We constructed ROC curve to evaluate and compared the diagnostic capacity of NREP. The area under the curve (AUC) for NREP was 0.723 (Fig. 1C), which indicated that NREP may be used as a diagnostic indicator of GC. To gain insight into the prognostic function of NREP in GC, Kaplan–Meier (KM) analyses of overall survival (OS), first progression survival (FPS), and post-progression survival (PPS) were performed to analyze the relationship between NREP expression and survival rates of GC patients. OS, FPS, and PPS curves showed significantly worse survival in the NREP high-expression group (log-rank P < 0 0.001) (Fig. 1D–F). Above results demonstrated that the high expression of NREP portends a poor prognosis suggesting that it could be adopted as a prognostic marker in GC.
NREP Expression is Associated with Clinical Variables in Patients with GC
Further analysis within the population showed that the expression of NREP was lower in tumor tissues of the black or African America GC patients than those in the Asian and White population across different ethnicities (P < 0.05) (Fig. 2A). We further analyzed the mRNA expression levels of NREP in different clinical GC stages. Higher mRNA levels of NREP were associated with higher pathological stage (T2–T4) of GC (P < 0.01) (Fig. 2B), which reflected the strong correlation of NREP with GC progression. Besides, as shown in Fig. 2C, higher levels of NREP were observed in grade G3 GC tumors (P < 0.001), but not in grade G1 and G2 GC tumors. We concluded that the GC patients with higher expression of NREP exhibited higher aggressive pathological stage and histopathological grade.
The Correlation Between NREP Expression and Immune Infiltration
NREP expression was negatively correlated with the infiltrations of Th2, Th17, NK, and CD56 bright cells et al. but its expression was positively correlated with the infiltrations of B, T, Mast, Macrophages, NK CD56dim, aDC, Treg, and Th1 cells et al. (Fig. 3A). Our data indicated that the NREP high-expression group showed higher NK cells, Macrophages, Mast cells, and Tgd cells immune infiltration, but lower NK CD56 bright cells and Th17 cells immune infiltration compared to the NREP low-expression group (P < 0.05) (Fig. 3B–G). Therefore, we hypothesized that tumor-intrinsic activation of NREP expression was closely correlated to immune infiltration. These results indicated NREP might have important roles in immunotherapy of GC.
The Correlation Between NREP Expression and Clinical Characteristics of Patients with GC
To examine the clinical disease monitoring effect of NREP, we enrolled data from TCGA database and assessed the correlation between NREP expression and several clinical characteristics of patients with GC. Results showed that high expression of NREP was associated with race, age, T stage, tumor histological type, and histological grade (P < 0.05). There was no significant correlation between NREP expression and other clinical characteristics shown in Table 1 (All P > 0.05). The multivariable Cox regression analyses were performed to analyze the impact of NREP expression levels on overall survival rate along with other GC patients’ clinicopathological features. Results showed that high expression of NREP was an independent risk factor for poor survival in GC patients (Fig. 4).
Functional Enrichment and Analyses of NREP-Related Genes in GC
To explore whether NREP-related genes in GC were related to specific functional features, we performed gene ontology (GO) and KEGG signaling analyses. GO analysis results indicated that NREP-related genes were predominantly enriched in biological processes, molecular functions, and cellular components, and the top ten enrichment terms were shown in Fig. 5A. The most significantly enriched terms in the three categories were extracellular matrix organization, collagen trimer and collagen binding, respectively. The results of KEGG pathway enrichment analysis revealed that NREP and its related genes were primarily associated with extracellular matrix structural constituent, collagen-containing extracellular matrix, extracellular matrix, and external encapsulating structure (Fig. 5B).
NREP is Upregulated in GC Tissues
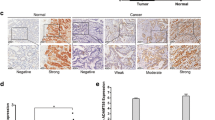
To further define the early warning role of NREP, 23 pairs of human GC tumor tissues and their corresponding adjacent non-cancerous tissues from the surgical operation were used to analyze NREP gene expression by RT-qPCR. Significant upregulation of NREP was observed in GC tissues compared to their control normal tissues (P < 0.001) (Fig. 6A and B). In comparison, tumor tissues of GC patients in T2–T4 stage showed a higher NREP expression compared to those of GC patients in T1 stage (P < 0.01) (Fig. 6C). Together, our results showed that NREP expression is significantly up-regulated in GC tissues compared with non-tumoral stomach tissues, and NREP might have important roles in GC development.
Knockdown of NREP Inhibits Human Gastric Adenocarcinoma Cell Proliferation
Based on our above results, the increased expression of NREP was associated with GC, we wondered what role NREP may play in GC. To investigate the underlying effects of NREP in GC, we decreased NREP expression in two human gastric adenocarcinoma cell lines by transfection of small interfering RNA (siRNA). Transfection of si-NREP markedly decreased NREP expression (P < 0.001) (Supplementary Fig. 1). The proliferative capacities of the SGC-7901 and AGS cells were detected by CCK-8 and EdU assays. Results of CCK-8 assay showed that knocking down NREP decreased the cell viability in SGC-7901 and AGS cells (P < 0.001) (Fig. 7A and B). Additionally, number of EdU-positive cells was significantly decreased (P < 0.001) following NREP knockdown in SGC-7901 cells (Fig. 7C), as well as in AGS cells (Fig. 7D). Moreover, NREP overexpression significantly promoted cell proliferation by increasing cell viability and number of EdU-positive cells (P < 0.05) (Supplementary Fig. 2A–D).
Knockdown of NREP inhibits human gastric adenocarcinoma cell proliferation. The proliferative capacities of the SGC-7901 and AGS cells were detected by CCK-8 and EdU assays. Cell viability was detected by CCK-8 assays after siRNAs transfection in SGC-7901 (A) and AGS (B) cells. Representative images and quantitative analysis of EdU-positive cells in SGC-7901 (C) and AGS (D) cells at 48 h post transfection were shown. ***P < 0.001
Knockdown of NREP Inhibits Tube Formation Capacity of HUVECs
Considering that enhanced tumor angiogenesis is tightly associated with tumor progression (Demircioglu et al. 2020), we further investigated the function of NREP in angiogenesis. As showcased in Fig. 8A and B, the angiogenesis of HUVECs was significantly reduced by NREP knockdown relative to the si-NC group (P < 0.01). While NREP overexpression led to increased angiogenesis of HUVECs (P < 0.05) (Supplementary Fig. 3A and B).
Discussion
Advanced GC is a malignant tumor with high mortality and poor prognosis (Wang;Jiang 2021). Early diagnosis and treatment is the prerequisite for a favorable prognosis of GC. It is important to search the efficient diagnostic and prognostic markers to improve early detection rate and prognosis of GC (Chen et al. 2021; Li et al. 2021). In recent years, although some studies have found new biomarkers for the diagnosis and prognosis of GC, such as cagA, hopQII, cytokeratin-20, and cytokeratin-7 (Guo et al. 2021; Zheng et al. 2021), more and deeper research are still urgently needed. In this study, we found a significantly higher expression of NREP in several human cancers, and NREP was expected to be a diagnostic biomarker for GC. Kaplan–Meier (KM) analyses of OS, FPS, and PPS demonstrated that the high expression of NREP portends a poor prognosis of GC patients. Further studies of relevant clinical variables suggested that NREP expression varied by race, pathological stage, and histopathologic regression grading. The high-expression level of NREP was associated with a high GC severity and disease progression stage based on T stage and histopathologic regression grading. We also showed that tumor-intrinsic activation of NREP expression was closely correlated with immune infiltration. The multivariable Cox regression analyses provided further proof of the relationships between high expression of NREP with race, age, T stage, tumor histological type, and histological grade in patients with GC.
We identified NREP as a biomarker for GC in diagnosis and prognosis. In the meantime, a deeper understanding of the pathological mechanisms of GC is necessary for the block and treatment of GC. Dysregulation of tumor-suppressor genes and oncogenes is an important cause of malignant proliferation and metastasis of GC cells (Dai et al. 2021). In this study, GO and KEGG pathway analyses indicated that NREP-related genes were predominantly enriched in biological processes, molecular functions, and cellular components, which provided further evidence for our point of view. Significant upregulation of NREP was observed in GC tissues compared to the control normal tissues, supporting that NREP could have important roles in GS progression and treatment. Our studies of potential functional effects of NREP in GC revealed that knockdown of NREP substantially suppressed GC cell proliferation and angiogenesis, reflected by impaired cell viability and decreased number of EdU-positive cells and angiogenic tubes.
It was reported that NREP regulates adipocyte development (Nunez et al. 2019) and induces the epidermal stem cells transdifferentiation (Yue et al. 2014; Li et al. 2016). In this study, for the first time, we evaluated the effect of NREP on cell viability, however, the underlying mechanisms for the roles of NREP on GC cell proliferation should be further investigated in the future. Angiogenesis plays a pivotal role in growth and metastasis of malignant tumor (Wang et al. 2021). Previous reports showed that NREP markedly promotes angiogenesis in cutaneous wound healing via the regulation of TGFβ1 and VEGF (Yao et al. 2015; Wang et al. 2017; Yang et al. 2020). Our research firstly confirmed that the NREP deficiency could result in attenuated angiogenesis in HUVECs. However, the detailed mechanisms underlying the regulation of angiogenesis remain further investigations. The carcinogenic role of NREP in GC might be related to its stimulative effect on cancer cell proliferation and angiogenesis.
Overall, our study proved that NREP might serve as a suitable diagnostic and prognostic marker for GC. Moreover, NREP might be used as a therapeutic target for regulate GC proliferation and angiogenesis in vitro, which would greatly benefit the early screening and treatment of GC. To better understanding the anti-cancer role of NREP silencing in GC, the animal experiments would be explored in future.
Data Availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Chen B, Zhang J, Ma Q (2021) The relationship between the simultaneity present of caga and hopqi genes in helicobacter pylori and the risk of gastric cancer (Noisy-le-Grand, France). Cell Mol Biol (noisy-Le-Grand) 67:121–126
Dai Y, Yang G, Yang L, Jiang L, Zheng G, Pan S, Zhu C (2021) Expression of Foxa1 gene regulates the proliferation and invasion of human gastric cancer cells. Cell Mol Biol 67:161–165
Demircioglu F, Wang J, Candido J, Costa ASH, Casado P, de Luxan Delgado B, Reynolds LE, Gomez-Escudero J, Newport E, Rajeeve V, Baker AM, Roy-Luzarraga M, Graham TA, Foster J, Wang Y, Campbell JJ, Singh R, Zhang P, Schall TJ, Balkwill FR, Sosabowski J, Cutillas PR, Frezza C, Sancho P, Hodivala-Dilke K (2020) Cancer associated fibroblast Fak regulates malignant cell metabolism. Nat Commun 11:1290
Du Y, Zhang JY, Gong LP, Feng ZY, Wang D, Pan YH, Sun LP, Wen JY, Chen GF, Liang J, Chen JN, Shao CK (2022) Hypoxia-induced Ebv-Circlmp2a promotes angiogenesis in Ebv-associated gastric carcinoma through the Khsrp/Vhl/Hif1α/Vegfa pathway. Cancer Lett 526:259–272
Espinosa-Parrilla Y, Munoz X, Bonet C, Garcia N, Vencesla A, Yiannakouris N, Naccarati A, Sieri S, Panico S, Huerta JM, Barricarte A, Menendez V, Sanchez-Cantalejo E, Dorronsoro M, Brennan P, Duarte-Salles T, B As Bueno-de-Mesquita H, Weiderpass E, Lund E, Clavel-Chapelon F, Boutron-Ruault MC, Racine A, Numans ME, Tumino R, Canzian F, Campa D, Sund M, Johansson M, Ohlsson B, Lindkvist B, Overvad K, Tjonneland A, Palli D, Travis RC, Khaw KT, Wareham N, Boeing H, Nesi G, Riboli E, Gonzalez CA, Sala N (2014) Genetic association of gastric cancer with mirna clusters including the cancer-related genes Mir29, Mir25, Mir93 and Mir106: results from the epic-eurgast study. Int J Cancer 135:2065–2076
Guimaraes EL, Stradiot L, Mannaerts I, Schroyen B, van Grunsven LA (2015) P311 modulates hepatic stellate cells migration. Liver Int 35:1253–1264
Guo X, Yin B, Wang C, Huo H, Aziziaram Z (2021) Risk assessment of gastric cancer in the presence of helicobacter pylori caga and hopqii genes. Cell Mol Biol 67:299–305
Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D (2011) Global cancer statistics. CA Cancer J Clin 61:69–90
Kim HH, Han SU, Kim MC, Kim W, Lee HJ, Ryu SW, Cho GS, Kim CY, Yang HK, Park DJ, Song KY, Lee SI, Ryu SY, Lee JH, Hyung WJ, G Korean Laparoendoscopic Gastrointestinal Surgery Study (2019) Effect of laparoscopic distal gastrectomy vs open distal gastrectomy on long-term survival among patients with stage I gastric cancer: the Klass-01 randomized clinical trial. JAMA Oncol 5:506–513
Li H, Yao Z, He W, Gao H, Bai Y, Yang S, Zhang L, Zhan R, Tan J, Zhou J, Takata M, Wu J, Luo G (2016) P311 induces the transdifferentiation of epidermal stem cells to myofibroblast-like cells by stimulating transforming growth factor beta1 expression. Stem Cell Res Ther 7:175
Li X, Pasche B, Zhang W, Chen K (2018) Association of muc16 mutation with tumor mutation load and outcomes in patients with gastric cancer. JAMA Oncol 4:1691–1698
Li M, Zheng G, Yu L, Tan LL, Li X, Li MJ, Li C, Li S, Liang J, Zhong Z, Li ZM (2021) Diagnostic value of Mri-Dwi signal intensity value combined with Serum Pgi, Pgii and Ca199 in early gastric cancer (Noisy-le-Grand, France). Cell Mol Biol (noisy-Le-Grand) 67:95–100
Liu Y, Zhou X, Hu N, Wang C, Zhao L (2020) P311 regulates distal lung development via its interaction with several binding proteins. Mech Dev 163:103633
Mariani L, McDonough WS, Hoelzinger DB, Beaudry C, Kaczmarek E, Coons SW, Giese A, Moghaddam M, Seiler RW, Berens ME (2001) Identification and validation of P311 as a glioblastoma invasion gene using laser capture microdissection. Cancer Res 61:4190–4196
McDonough WS, Tran NL, Berens ME (2005) Regulation of glioma cell migration by serine-phosphorylated P311. Neoplasia 7:862–872
Nunez S, Young C, Adebayo O, Muppuru KM, Badri KR (2019) P311, a Novel intrinsically disordered protein, regulates adipocyte development. Biochem Biophys Res Commun 515:234–240
Ooi PT, da Costa N, Edgar J, Chang KC (2006) Porcine congenital splayleg is characterised by muscle fibre atrophy associated with relative rise in Mafbx and fall in P311 expression. BMC Vet Res 2:23
Pan D, Zhe X, Jakkaraju S, Taylor GA, Schuger L (2002) P311 induces a Tgf-Beta1-independent, nonfibrogenic myofibroblast phenotype. J Clin Invest 110:1349–1358
Smyth EC, Nyamundanda G, Cunningham D, Fontana E, Ragulan C, Tan IB, Lin SJ, Wotherspoon A, Nankivell M, Fassan M, Lampis A, Hahne JC, Davies AR, Lagergren J, Gossage JA, Maisey N, Green M, Zylstra JL, Allum WH, Langley RE, Tan P, Valeri N, Sadanandam A (2018) A Seven-gene signature assay improves prognostic risk stratification of perioperative chemotherapy treated gastroesophageal cancer patients from the magic trial. Ann Oncol 29:2356–2362
Stradiot L, Mannaerts I, van Grunsven LA (2018) P311, friend, or foe of tissue fibrosis? Front Pharmacol 9:1151
Sun YG, Gao YJ, Zhao ZQ, Huang B, Yin J, Taylor GA, Chen ZF (2008) Involvement of P311 in the affective, but not in the sensory component of pain. Mol Pain 4:23
Sun W, Powell M, O’Dwyer PJ, Catalano P, Ansari RH, Benson AB 3rd (2010) Phase Ii study of sorafenib in combination with docetaxel and cisplatin in the treatment of metastatic or advanced gastric and gastroesophageal junction adenocarcinoma: ecog 5203. J Clin Oncol 28:2947–2951
Tan J, Peng X, Luo G, Ma B, Cao C, He W, Yuan S, Li S, Wilkins JA, Wu J (2010) Investigating the role of P311 in the hypertrophic scar. PLoS ONE 5:e9995
Taylor GA, Rodriguiz RM, Greene RI, Daniell X, Henry SC, Crooks KR, Kotloski R, Tessarollo L, Phillips LE, Wetsel WC (2008) Behavioral characterization of P311 knockout mice. Genes Brain Behav 7:786–795
Wang G, Jiang X (2021) Effect of precede-proceed-model-based health education on the life quality and 5-Htt gene expression of patients with gastric cancer after surgery. Cell Mol Biol 67:242–248
Wang S, Zhang X, Qian W, Zhou D, Yu X, Zhan R, Wang Y, Wu J, He W, Luo G (2017) P311 deficiency leads to attenuated angiogenesis in cutaneous wound healing. Front Physiol 8:1004
Wang C, Jiang T, Zhu Y (2021) Experimental study on anti angiogenesis of recombinant mouse angiostatin gene in mice with gallbladder carcinoma. Cell Mol Biol 67:117–124
Yang ZQ, Liu YJ, Zhou XL (2020) An integrated microarray analysis reveals significant diagnostic and prognostic biomarkers in pancreatic cancer. Med Sci Monit 26:e921769
Yao Z, Yang S, He W, Li L, Xu R, Zhang X, Li H, Zhan R, Sun W, Tan J, Zhou J, Luo G, Wu J (2015) P311 promotes renal fibrosis via Tgfbeta1/Smad Signaling. Sci Rep 5:17032
Yue MM, Lv K, Meredith SC, Martindale JL, Gorospe M, Schuger L (2014) Novel Rna-binding protein P311 binds eukaryotic translation initiation factor 3 subunit B (Eif3b) to promote translation of transforming growth factor beta1-3 (Tgf-Beta1-3). J Biol Chem 289:33971–33983
Zhang P, Zheng Z, Ling L, Yang X, Zhang N, Wang X, Hu M, Xia Y, Ma Y, Yang H, Wang Y, Liu H (2017) W09, a novel autophagy enhancer, induces autophagy-dependent cell apoptosis via activation of the Egfr-mediated Ras-Raf1-Map2k-Mapk1/3 pathway. Autophagy 13:1093–1112
Zheng X, Chen P, Liu Y, Wang B, Liu Q (2021) Determining the relationship between cytokeratin expression and prognostic factors in human gastric cancer. Cell Mol Biol 67:311–317
Zhou D, Liu T, Wang S, He W, Qian W, Luo G (2020a) Effects of Il-1beta and Tnf-alpha on the expression of P311 in vascular endothelial cells and wound healing in mice. Front Physiol 11:545008
Zhou Y, Cao G, Cai H, Huang H, Zhu X (2020b) The effect and clinical significance of Fn1 expression on biological functions of gastric cancer cells (Noisy-le-Grand, France). Cell Mol Biol 66:191–198
Acknowledgements
We would like to thank the Affiliated Cancer Hospital of Zhengzhou University and all the authors for their help.
Funding
This work was supported by the Henan Province Medical Science and Technology Tackling Program, the Foundation for 2020 Henan Province Medical Science and Technology Tackling Program (Grant No. LHGJ20200160).
Author information
Authors and Affiliations
Contributions
QL and JW: designed the study. QL and LF: performed the experiments. DW: analyzed the data. QL: The first draft of the manuscript was written. All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Ethical Approval
This study was performed in line with the principles of the Declaration of Helsinki. Approval was granted by the Ethics Committee of The Affiliated Tumor Hospital of Zhengzhou University, Henan Cancer Hospital.
Consent to participate
Informed consent was obtained from all individual participants included in the study.
Consent to Publication
The authors affirm that human research participants provided informed consent for publication of the images in Figs. 6A–C.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
10528_2022_10276_MOESM1_ESM.tif
Supplementary file1 (TIF 410 KB) Supplementary fig. 1 The results of transfection efficient in SGC-7901 and AGS cells. Western blotting was performed to validate the transfection efficient in SGC-7901 and AGS cells. NREP protein level was reduced after transfection of si-NREP. ***P <0.001.
10528_2022_10276_MOESM2_ESM.tif
Supplementary file2 (TIF 674 KB) Supplementary fig. 2 NREP overexpression promotes human gastric adenocarcinoma cell proliferation. (A and B) Cell viability was detected by CCK-8 assays after NREP overexpression vector transfection in SGC-7901 (A) and AGS (B) cells. Representative images and quantitative analysis of EdU-positive cells in SGC-7901 (C) and AGS (D) cells at 48 h post transfection were shown. *P <0.05, **P <0.01, ***P <0.001
10528_2022_10276_MOESM3_ESM.tif
Supplementary file3 (TIF 732 KB) Supplementary fig. 3 NREP overexpression promotes angiogenesis. HUVECs were treated with the culture supernatant from SGC-7901 (A) or AGS (B) cells with NREP overexpression. Representative images of angiogenesis and quantitative analysis of angiogenic tubes in HUVECs were shown. *P <0.05
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Li, Q., Fu, L., Wu, D. et al. NREP is a Diagnostic and Prognostic Biomarker, and Promotes Gastric Cancer Cell Proliferation and Angiogenesis. Biochem Genet 61, 669–686 (2023). https://doi.org/10.1007/s10528-022-10276-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10528-022-10276-7