Abstract
The complete mitochondrial genome (mitogenome) of Saturnia japonica (Lepidoptera: Saturniidae) was sequenced and annotated. It is a circular molecule of 15, 376 bp, composed of 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), two ribosomal RNA genes (rRNA), and an adenine (A) + thymine (T)-rich region. All protein-coding genes (PCGs) are initiated by the ATN codon except for cytochrome c oxidase subunit 1 (cox1) gene that is seemingly initiated by the CGA codon. Except for cox2 and nad4, which were terminated by incomplete stop codon T or TA, the rest were terminated by canonical stop codon TAA. The A + T-rich region is high conservative, including ‘ATAGA’ motif followed by a 19 bp poly-T stretch, a microsatellite-like element (AT)9 and also a poly-A element, with a total length of 332 bp. The Asn codon was the most frequently used codon, followed by Ile, Leu2, Lys, Met, Phe, and Tyr, while Cys was the least frequently used codon. Phylogenetic relationships analysis based on the 13 PCGs by using maximum likelihood (ML) and neighbor Joining (NJ) revealed that S. japonica belongs to the Saturniidae family. In this study, the annotation and characteristics of the mitogenome of S. japonica were resolved for the first time, which laid a foundation for species classification and the molecular evolution of Lepidoptera: Saturniidae.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Saturnia japonica (Lepidoptera) is one of the most precious wild silkworms, mainly distributed in China, Japan, and Korea, which typically consumes the leaves from walnut or chestnut (Chen et al. 2020). S. japonica can spin fluorescent and scintillating silk, which is often used as a raw material for various anti-counterfeiting signs and high-grade clothing (Li et al. 2014). In addition, S. japonica is also a kind of resource insect. Its pupa is often used as an edible insect for supplement protein.
Insect mitogenome DNA (mtDNA) is typically composed of 37 conserved genes. It is a closed-circular molecule ranging in size from 14 to 19 kilobases (kb), including the very short or non-existent interval between genes (Jiang et al. 2009). It contains 13 protein-coding genes (PCGs), 22 transfer RNAs (tRNAs), and 2 ribosomal RNAs (rrnL and rrnS) (Boore 1999; Wolstenholme 1992). In addition, there is a poorly conserved adenine (A)+thymine (T)-rich region, which contains the initiation sites of genome transcription and replication (Cameron 2014; Kim et al. 2006; Shadel and Clayton 1993; Lu et al. 2013). This control region is also considered to control transcription initiation and genome replication (Wolstenholme 1992). MtDNA is a powerful tool for species identification (Dowton et al. 2002), genomic evolution (Miya et al. 2001), phylogeography (Avise et al. 1987), and molecular evolution (Forstén 1991) based on its simple genomic organization, high rate of evolutionary, and almost clear homology (Zhang et al. 2015).
Lepidoptera is a gigantic family with more than 160000 species, which can be divided into 45–48 superfamilies (Hao et al. 2012). Saturniidae contains 800 species distributed worldwide and more than 44 species recorded in China. In addition to being an important indicator of plant pest control in agriculture and forestry, they are also valuable in the process of plant pollination (Chen et al. 2020). However, only a few species mitogenomes have been completely sequenced, which are publically available in GenBank (Table 1) (Kawaguchi and Nishida 2001, Sun et al. 2017).
It is important to understand the genetic characteristics and phylogenetic status of S. japonica for improving its comprehensive utilization. The complete mitogenome of S. japonica was sequenced, annotated, and compared with the other Lepidoptera species to elucidate molecular evolution, comparative and evolutionary genomics, phylogenetics, and population genetics (Jiang et al. 2009).
Materials and Methods
Experimental Insects and DNA Extraction
Saturnia japonica pupae were collected from the chestnut trees on the Scenic of Dabie Mountains, Anhui Province, China. Fresh specimens were preserved in 100% ethanol and stored at -80℃. The total DNA was extracted using the Genomic DNA Extraction Kit according to the manufacturer’s instructions (Sangon Biotech Co. Ltd, Shanghai, China). The extracted DNA quality was examined by 1% agarose gel electrophoresis (w/v) and then used for the PCR amplification.
Primer Design, PCR Amplification, and Sequencing
Fourteen overlapping fragments pairs of primers were designed based on the conserved nucleotide sequences of known the reported mitogenome of Lepidopteran species and synthesized by General Biosystems Co. Ltd. (Chuzhou, China). The complete list of successful primers is given in Table 2. All PCRs were performed in 50 μL reaction volume, which including 35 μL sterilized distilled water, 5 μL 10 × Taq buffer (Mg2+ plus), 4 μL dNTP (2.5 mM), 1.5 μL DNA as template, 2 μL of each primer (10 μM), and 0.5 μL (1 unit) Taq DNA polymerase (TaKaRa Biotechnology Co., Dalian, China). The PCR amplification conditions were as follows: 4 min at 94 °C, followed by 30 cycles of 30 s at 94 °C, 40 s at 46–56 °C, and 2–3 min (depending on putative length of the fragments) at 72 °C, and then a final extension step of 72 °C for 10 min. The PCR products were detected by 1% agarose gel electrophoresis (w/v) and then purified using a DNA gel extraction kit (Sangon Biotech Co. Ltd., Shanghai, China). The purified fragments were ligated into the T-vector (TaKaRa Biotechnology Co., Dalian, China) and transformed into Escherichia coli TOP10. The insert DNA-positive recombinant colonies were sequenced bidirectionally at least three times by General Biosystems Co. Ltd. (Chuzhou, China).
Sequence Assembly and Gene Annotation
Sequence annotation was performed by comparing with other Lepidoptera species sequenced previously using blast tools available from the NCBI (https://blast.ncbi.nlm.nih. gov/Blast.cgi) and SeqMan II program from the Lasergene software package (DNASTAR Inc., Madison, USA) (Guo et al. 2019). The protein-coding sequences were translated into putative proteins on the basis of the Invertebrate Mitochondrial Genetic Code. The skewness was measured by the method given by Junqueira et al and describe the base composition of the nucleotide sequence as follows: AT skew = [A–T]/[A+T], GC skew = [G–C]/[G+C] (Junqueira et al. 2004). The relative synonymous codon usage (RSCU) values were calculated by using MEGA7.0 (Tamura et al. 2011).
The tRNA genes were verified using either program tRNA scan-SE Search with the default settings (http://lowelab.ucsc.edu/tRNAscan-SE/) (Tamura et al. 2011), or by manually identifying sequences with the appropriate anticodon capable of folding into the typical cloverleaf secondary structure (Lowe and Eddy 1997). The tandem repeats in the A+T-rich region were determined by the tandem repeats finder program (http://tandem.bu.edu/trf/trf.html) (Dai et al. 2015).
Phylogenetic Analysis
To illustrate the phylogenetic relationship among Lepidoptera insects, 38 complete mitogenomes were downloaded from the GenBank DataBase (Table 1). The mitogenomes of Cucujus clavipes (GU176341.1) (Song et al. 2010) and Cucujus haematodes (KX087268.1) were downloaded as outgroups. Multiple comparison of the 13 PCGs concatenated nucleotide sequences were conducted using MEGA version 7.0. Then the 13 protein nucleotide sequences were serialized into a group for phylogenetic analysis, which were performed using Maximum Likelihood (ML) and Neighbor Joining (NJ) method based on the Kimura 2-parameter (K2P) model (Kimura et al. 1980). Evolutionary analysis was conducted in the MEGA version7.0 program (Tamura et al. 2011). The ML analysis was used to infer phylogenetic trees with 1000 bootstrap replicates, and Neighbor Joining (NJ) distance analysis was performed using PAUP4b 10 (Thompson et al. 1997). The NJ analysis was done with 1000 bootstrap replicates. The consensus tree was visualized using FigTree v1.4.0 (http://tree.bio.ed.ac.uk/Sofw-are/fgtree/).
Results and Discussions
Genome Structure, Organization and Composition
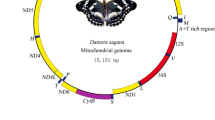
The mitogenome of S. japonica is a circular molecule of 15, 376 bp in length (Fig. 1), which is within the range observed in the entire sequence of Lepidopteran species with the size ranging from 16, 179 bp in Plutella xylostella (Plutellidae) to 15, 113 bp in Lamproptera meges (Papilionidae) (Table 1) (Swofford 2003). The sequence had been annotated and deposited into GenBank under the accession number MT614593. The mitogenome structure of S. japonica conforms to the classic 38 regions of the Lepidopteran mitogenome, including 13 protein-encoding regions, 22 tRNA-encoding regions, two rRNA-encoding regions, and a large non-coding-region with high A + T-rich composition (Table 3) (Wei et al. 2013; Rand 1993). The arrangement and orientation of genes in the mitogenome of S. japonica is trnM-trnI-trnQ, which is different from the ancestral gene order trnI-trnQ-trnM (Boore 1999). Most of the 23 genes were transcribed on the majority-coding strand (H-strand) and a few on the minority-coding strand (L-strand). The comparison of S. japonica mitogenome composition and skewness level with other sequenced Lepidoptera species is represented in Table 4. The genome composition of S. japonica is A: 39.37%, T: 41.30%, G: 7.56%, and C: 11.77%, with a total A + T content of 80.66%. This is within the scope of similar species (A + T bias of 78.26% in Tecia solanivora and 82. 56% in Leucoptera malifoliella) (Table 4) (Mcknight and Shaffer 1997; Ramirez-Rios et al. 2016). Additionally, it exhibits negative AT skewness (− 0.024) and negative GC skewness (− 0.218). The AT skewness in other Lepidopteran mitogenomes sequenced to date ranges from 0.057 (A. cinerarium) to − 0.036 (T. maculata) (Wu et al. 2012), while the GC skewness from − 0.247 (L. dispar) to − 0.177 (A. ipsilon) (Table 4) (Liu et al. 2014). Similarly, the 13 PCGs, tRNA, rRNA, and A + T-rich region of S. japonica are all within the range of the observed Lepidoptera.
Protein-Coding Genes and Codon Usage
The 13 protein-coding genes of S. japonica mitogenome are 11, 235 bp long (Appendix, Table 5) and account for 73.07% of the total nucleotides. The AT negative skewness (− 0.019) indicates the occurrence of less As than Ts. Nine of these PCGs (nad2, cox1, cox2, atp8, atp6, cox3, nad3, nad6, and cob) are coded by the H-strand, while the remaining four PCGs (nad5, nad4, nad4L, and nad1) are coded by the L-strand. In addition, twelve of these PCGs begin with ATN (four with ATA, two with ATT, four with ATG, and two with ATC) codons, while the remaining cox1 gene of S. japonica starts with CGA codon as previously documented in Leucoma salicis (Wu et al. 2015). Except that cox2 terminates with a single T and nad4 terminates with TA, the remaining 11 PCGs use typical TAA termination codon (Table 3). This phenomenon has been reported in most of the sequenced Lepidopteran mitogenomes (Sun et al. 2016). Single T-stop codon generates functional stop codons by polyadenylation of adjacent PCGs and endonuclease recognition of polycistronic pre-mRNA transcription (Liu et al. 2013).
The mitochondrial genome codon usage of ten Lepidopteran insects was analyzed and divided into eight superfamilies: three species belonging to Bombycoidea, and seven belonging to Pyraloidea, Noctuoidea, Papilionoidea, Geometroidea, Yponomeutoidea, Gelechioidea, and Hepialoidea (Fig. 2). The results revealed that Asn, Ile, Leu2, Lys, Met, Phe, and Tyr were the most frequently utilized amino acids; however, the Arg codon family was the rarest. Codon distributions of three species in Bombycoidea are in consistency, and each amino acid has equal contents in different species (Appendix, Fig. 7).
The 13 PCGs of the S. japonica mitogenome contain all codons (Appendix, Fig. 8). This is similar to A. cinerarium (Wu et al. 2012), L. dispar, T. renzhiensis (Wich et al. 1986), T. solanivora (Mcknight and Shaffer 1997), and G. timur (Cao et al. 2012), but in some Lepidopteran insects, high GC content codons are abandoned as one of the mitochondrial features (Lu et al. 2013; Chen et al.2016), such as B. mori (lack of GCG), C. suppressalis (lack of GCG & CGT), P. colligata (lack of GCG, GCG & CGT), and P. xylostella (lack of GCG) (Swofford 2003).
Ribosomal and Transfer RNA Genes
The mitogenome of S. japonica including two typical rRNA genes of Lepidoptera has been sequenced. The large ribosomal gene (rrnL) is 1407 bp in length and resided between tRNALeu (CUN) and tRNAVal, whereas the small ribosomal gene (rrnS) is only 772 bp, and located between tRNAVal and A + T-rich region (Table 3). The A + T content of the two rRNAs is 84.72%, which ranged from 83.60% (T. solanivora) to 86.09% (L. malifoliella) of Lepidopterans. In addition, AT skewness (−0.037) and GC skewness (-0.351) of the two rRNAs are negative (Appendix, Table 5), which are located within the range of reported Lepidopteran insects. This was similar to A. assama, A. ipsilon, and P. flavescens and so on.
The S. japonica mitochondrial genome harbors an entire set of 22 tRNA genes ranging from 63 bp (tRNACys) to 73 bp (tRNAAsp). 14 of the 22 tRNA genes were coded by the H-strand and remainder eight were coded by the L-strand. The A + T content of the 22 tRNA genes is 81.79%, and AT skewness (− 0.013) and GC skewness (− 0.138) of the two rRNAs are negative (Appendix, Table 5). All tRNAs exhibit typical secondary structure of clover, except for trnS1 lacking the DHU stem (Fig. 3),which is similar to several other previously sequenced Lepidopterans insects (Dai et al. 2016; Liao et al. 2010).
A total of 12 mismatched bps in the S. japonica tRNAs were identified, 7 of the 12 G-U wobble pairs scatter throughout the seven tRNAs (two in acceptor stem, four in DHU, and one in anticodon stem). A–A mismatches in the DHU of the tRNG, three U–U mismatch in acceptor stem of the trnA, trnL2, trnN, and one U–U mismatches in anticodon stem of the trnV (Fig. 3).
Overlapping and Intergenic Spacer Regions
The mitogenome of S. japonica cins 42 bp overlapping nucleotides, which are located in 12 pairs of adjacent genes with the length from 1 to 17 bp and the longest overlap (17 bp) existed between trnF and nad5. In addition, there is 194 bp intergenic nucleotides (IGN) that are distributed among 26 pairs of adjacent genes ranging from 1 to 53 bp, and the longest spacer sequence was located between trnQ and nad2, which is usually found in Lepidopteran mitogenomes (Wu et al. 2010). Surprisingly, the seven nucleotides sequence “ATGATAA” is included in the mitochondrial sequence of the ten Lepidopteran insects observed (Fig. 4), common feature across Lepidopteran mitogenomes (Zhu et al. 2013). The 22 bp spacer between trnS2 (UCN) and nad1 contains the motif ‘ATACTAA,’ which is highly conserved region and found in most insect mtDNAs, but Hepialoidea only contains the motif ‘ATACTA’ (Fig. 5A); hence, the fragment ‘ATACTA’ is highly conserved region, which may be the peptide-binding site of mitochondrial transcription termination (mtTERM protein) (Taanman 1999).
(A) Alignment of the intergenic spacer region between trnS2 (UCN) and nad1 of several Lepidopteran insects. The shaded ‘ATACTAA’ motif is conserved across the Lepidoptera order. (B) Features present in the A + T-rich region of S. japonica. The sequence is transcribed in the reverse strand. The ATATG motif is shaded. The poly-T stretch is underlined while the poly-A stretch is boldly underlined. The single-microsatellite T/A repeats sequence is indicated by dotted underlining
Phylogenetic relationships among species. Tree constructed using Maximum Likelihood (ML) and Neighbor Joining (NJ) with 1000 bootstrap replicates showed the phylogenetic relationships among 39 species, Cucujus clavipes (GU176341.1) and Cucujus haematodes (KX087268.1) were used as outgroups. This tree (ML tree), with bootstraps values from ML and NJ is at the nodes above and below, respectively
The A + T-Rich Region
The A + T-rich region of S. japonica mitogenome is located between the rrnS and trnM, with a length of 332 bp, remarkably shorter than that of T. renzhiensi (1367 bp) and longer than L. botrana (286 bp) (Appendix, Table 5). This region contains the highest A + T content (91.87%) in the mtDNA, and its AT skewness (− 0.082) and GC skewness (-0.481) are both negative (Appendix, Table 5). Several short repeating sequences scattered throughout the entire region, including the motif ‘ATAGA’ followed by a 17 bp poly-T stretch, a microsatellite-like (AT)9 element and a poly-A element upstream of trnM gene similar to other Lepidopteran mitogenomes (Fig. 5B). The length of poly-T stretch is variable (Lu et al. 2013), while ‘ATAGA’ region is highly conserved among Lepidoptera species (Cameron and Whiting 2008).
Phylogenetic Analysis
In this study, the nucleotide sequences of the 13 PCGs were concatenated and aligned to reconstruct the phylogenetic relationships among 39 Lepidoptera insects by using Maximum Likelihood (ML), Neighbor Joining (NJ) methods (Chai et al. 2012; Hassanin 2006). Species are clustered by family (Fig. 6). The phylogenetic analysis showed that the S. japonica was within the Saturniidae family (Bombycoidea), and it has a closer relationship to S. boisduvalii and S. jonasii. This is consistent with the conclusion of Kim et al. (2015) and clustered with other superfamilies, ordinal the Geometroidea, Noctuoidea, Pyraloidea, Gelechioidea, Papilionoidea, Tortricoidea, and Yponomeutoidea. The phylogenetic tree constructed by ML and NJ methods showed that Saturnia japonica, Saturnia boisduvalii, Saturnia jonasii, Eriogyna pyretorum, Antheraea assama, and Samia cynthia ricini belonged to Saturniidae, which was consistent with traditional taxonomic results. Similar phenomena also occur in other superfamilies, and the results of phylogenetic tree analysis are completely consistent with the results of traditional entomological taxonomy.
References
Avise JC, Arnold J, Ball RM, Bermingham E, Lamb T, Neigel JE, Saunders NC (1987) Intraspecific Phylogeography: The Mitochondrial DNA Bridge Between Population Genetics and Systematics. Annu Rev Ecol Syst 18(10):489–522
Boore JL (1999) Animal mitochondrial genomes. Nucleic Acids Res 27(8):1767–1780
Cameron SL (2014) Insect mitochondrial genomics: implications for evolution and phylogeny. Annu Rev Entomol 59:95–117
Cameron SL, Whiting MF (2008) The complete mitochondrial genome of the tobacco hornworm, Manduca sexta, (Insecta: Lepidoptera: Sphingidae), and an examination of mitochondrial gene variability within butterflies and moths. Gene 408(1–2):112–123
Cao YQ, Ma C, Chen JY, Yang DR (2012) The complete mitochondrial genomes of two ghost moths, Thitarodes renzhiensis and Thitarodes yunnanensis: the ancestral gene arrangement in Lepidoptera. BMC Genomics 13:276
Chai HN, Du YZ, Zhai BP (2012) Characterization of the complete mitochondrial genomes of Cnaphalocrocis medinalis and Chilo suppressalis (Lepidoptera: Pyralidae). Int J Biol Sci 8(4):561–579
Chen Y, Gan S, Shao L, Cheng C, Hao J (2016) The complete mitochondrial genome of the Pazala timur (Lepidoptera:Papilionidae: Papilioninae). Mitochondrial DNA Part A 27(1):533–534
Chen YM, Sun JW, Iqbal A, Lv R, Wang H, Zang LS (2020) An investigation of Caligula japonica (Lepidoptera: Saturniidae) egg distribution and associated parasitoids on walnut trees (Juglans regia L.) in northwestern China. Int . Pest Manage 24:1–8
Dai L, Qian C, Zhang C, Wang L, Wei G, Li J, Zhu B, Liu C (2015) Characterization of the complete mitochondrial genome of Cerura menciana and comparison with other lepidopteran insects. PLoS ONE 10(8):17
Dai LS, Zhu BJ, Qian C, Zhang CF, Li J, Wang L, Wei GQ, Liu CL (2016) The complete mitochondrial genome of the diamondback moth, Plutella xylostella (Lepidoptera: Plutellidae). Mitochondrial DNA Part A 27(2):1512–1513
Dowton M, Castro LR, Austin AD (2002) Mitochondrial gene rearrangements as phylogenetic characters in the invertebrates: the examination of genome “morphology.” Invertebr Syst 16(3):345–356
Forstén A (1991) Mitochondrial-DNA time-table and the evolution of Equus: comparison of molecular and paleontological evidence. Ann Zool Fenn 28(3/4):301–309
Guo J, Miao X, He P, Li M, Wang S, Cui J, Huang C, He L, Zhao J (2019) Babesia gibsoni endemic to Wuhan, China: mitochondrial genome sequencing, annotation, and comparison with apicomplexan parasites. Parasitol Res 118(1):235–243
Hao J, Sun Q, Zhao H, Sun X, Gai Y, Yang Q (2012) The Complete mitochondrial genome of Ctenoptilum vasava (Lepidoptera: Hesperiidae: Pyrginae) and its phylogenetic implication. Comparat Funct Genom. https://doi.org/10.1155/2012/328049
Hassanin A (2006) Phylogeny of Arthropoda inferred from mitochondrial sequences: strategies for limiting the misleading effects of multiple changes in pattern and rates of substitution. Mol Phylogenet Evol 38(1):100–116
Jiang ST, Hong GY, Yu M, Li N, Yang Y, Liu YQ, Wei ZJ (2009) Characterization of the complete mitochondrial genome of the giant silkworm moth, Eriogyna pyretorum (Lepidoptera: Saturniidae). Int J Biol Sci 5(4):351–365
Junqueira AC, Lessinger AC, Torres TT, Silva FRD, Vettore AL, Arruda P, Espin AMLA (2004) The mitochondrial genome of the blowfly Chrysomya chloropyga (Diptera: Calliphoridae). Gene 339:7–15
Kawaguchi A, Nishida M (2001) Mitogenomic exploration of higher teleostean phylogenies: a case study for moderate-scale evolutionary genomics with 38 newly determined complete mitochondrial DNA sequences. Mol Biol Evol 18(11):1993–2009
Kim I, Lee EM, Seol KY, Yun EY, Lee YB, Hwang JS, Jin BR (2006) The mitochondrial genome of the Korean hairstreak, Coreana raphaelis (Lepidoptera: Lycaenidae). Insect Mol Biol 15(2):217–225
Kim MJ, Choi SW, Kim I (2015) Saturnia jonasii Butler, 1877 on Jejudo Island, a new saturnid moth of South Korea with DNA data and morphology (Lepidoptera: Saturniidae). Zootaxa 3946(3):374–386
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16(2):111–120
Liao F, Wang L, Wu S, Li YP, Zhao L, Huang GM, Niu CJ, Liu YQ, Li MG (2010) The complete mitochondrial genome of the fall webworm, Hyphantria cunea (Lepidoptera: Arctiidae). Int J Biol Sci 6(2):172–186
Liu QN, Zhu BJ, Dai LS, Liu CL (2013) The complete mitogenome of Bombyx mori strain Dazao (Lepidoptera: Bombycidae) and comparison with other Lepidopteran insects. Genomics 101:64–73
Liu S, Xue D, Cheng R, Han H (2014) The complete mitogenome of Apocheima cinerarius (Lepidoptera: Geometridae: Ennominae) and comparison with that of other Lepidopteran insects. Gene 547(1):136–144
Li SY, Du ZJ, Qi I (2014) Wild silkworm series II-Saturnia japonica. Sci Seric 35(2):85–87
Lu HF, Su TJ, Luo AR, Zhu CD, Wu CS (2013) Characterization of the complete mitochondrion genome of Diurnal Moth Amata emma (Butler) (Lepidoptera: Erebidae) and its phylogenetic implications. PLoS ONE 8(9):48
Lowe TM, Eddy SR (1997) tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25:955–964
Mcknight ML, Shaffer HB (1997) Large, rapidly evolving intergenic spacer in the mitochondrial DNA of the salamander family Ambystomatidae (Amphibia: Caudata). Mol Biol Evol 14(11):1167–1176
Ramirez-Rios V, Franco-Sierra ND, Alvarez JC, Saldamando-Benjumea CI, Villanueva-Mejia DF (2016) Mitochondrial genome characterization of Tecia solanivora (Lepidoptera: Gelechiidae) and its phylogenetic relationship with other Lepidopteran insects. Gene 581(2):107–116
Rand DM (1993) Endotherms, ectotherms, and mitochondrial genome-size variation. J Mol Evol 37(3):281–295
Shadel GS, Clayton DA (1993) Mitochondrial transcription initiation. Variation and conservation. J Biol Chem 268(22):16083–16086
Song H, Sheffield NC, Cameron SL, Miller KB, Whiting MF (2010) When phylogenetic assumptions are violated: base compositional heterogeneity and among-site rate variation in beetle mitochondrial phylogenomics. Syst Entomol 35(3):429–448
Sun Y, Chen C, Gao J, Abbas MN, Kausar S, Qian C, Wang L, Wei G, Zhu BJ, Liu CL (2017) Comparative mitochondrial genome analysis of Daphnis nerii and other Lepidopteran insects reveals conserved mitochondrial genome organization and phylogenetic relationships. PLoS ONE 12(6):19
Sun YX, Wang L, Wei GQ, Qian C, Dai LS, Sun Y, Abbas MN, Zhu BJ, Liu CL (2016) Characterization of the complete mitochondrial genome of Leucoma salicis (Lepidoptera: Lymantriidae) and comparison with other lepidopteran insects. Sci Rep 15(6):39153
Swofford, DL (2003) PAUP*: Phylogenetic analysis using parsimony, Version 4. 0 b10. (Sinauer 2003).
Taanman JW (1999) The mitochondrial genome: structure, transcription, translation and replication. Biochim Biophys Acta 1410(2):103–123
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28(10):2731–2739
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25(24):4876–4882
Wei SJ, Shi BC, Gong YJ, Li Q, Chen XX (2013) Characterization of the mitochondrial genome of the diamondback moth Plutella xylostella (Lepidoptera: Plutellidae) and phylogenetic analysis of advanced moths and butterflies. DNA Cell Biol 32(4):173–187
Wich GN, Hummel H, Jarsch M, Bär U, Böck A (1986) Transcription signals for stable RNA genes in Methanococcus. Nucleic Acids Res 14(6):2459
Wolstenholme DR (1992) Animal mitochondrial DNA: structure and evolution. Int Rev Cytol 141(6):173
Wu LW, Lees DC, Yen SH, Lu CC, Hsu YF (2010) The complete mitochondrial genome of the near-threatened swallowtail, Agehana maraho (Lepidoptera: Papilionidae): evaluating sequence variability and suitable markers for conservation genetic studies. Entomol News 121(3):267–280
Wu QL, Cui WX, Wei SJ (2015) Characterization of the complete mitochondrial genome of the black cutworm Agrotis ipsilon (Lepidoptera: Noctuidae). Mitochondrial DNA Part A 26(1):139–140
Wu YP, Zhao JL, Su TJ, Li J, Yu F, Chesters D, Fan RJ, Chen MC, Wu CS, Zhu CD (2012) The complete mitochondrial genome of Leucoptera malifoliella Costa (Lepidoptera: Lyonetiidae). DNA Cell Biol 31(10):1508–1522
Zhang Z, Wang X, Li R, Guo R, Zhang W, Song W, Hao C, Wang H, Li M (2015) The mitochondrial genome of Dastarcus helophoroides (Coleoptera: Bothrideridae) and related phylogenetic analyses. Gene 560(1):15–24
Zhu BJ, Liu QN, Dai LS, Wang L, Sun Y, Lin KZ, Wei GQ, Liu CL (2013) Characterization of the complete mitochondrial genome of Diaphania pyloalis (Lepidoptera: Pyralididae). Gene 527(1):283–291
Acknowledgements
This work was supported by the grant from the National Natural Science Foundation of China (31472147); the fund for Anhui International Joint Research and Development Center of Sericulture Resources Utilization (2017R0101); and the foundation for Innovative Research Groups of Anhui Agricultural University (ANRC2019032).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Liu, J., Dai, J., Jia, J. et al. Characterization and Phylogenetic Analysis of the Complete Mitochondrial Genome of Saturnia japonica. Biochem Genet 60, 914–936 (2022). https://doi.org/10.1007/s10528-021-10129-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10528-021-10129-9