Abstract
Plant-specific TEOSINTE-BRANCHED1/CYCLOIDEA/PROLIFERATING CELL FACTOR1 (TCP) gene family has versatile functions in diverse aspects of plants. However, less research on banana TCPs was done comprehensively. Accordingly, 48 banana TCP genes were characterized on aspects of gene structure, conserved motifs, phylogenetic relationship, and expression patterns. Members of the MaTCP gene family were unevenly distributed among 11 chromosomes and purification selection was the driving force of the MaTCP gene family. Gene duplication analysis indicated that segmental duplication is the major contributor to family expansion. Promoter analysis showed that MaTCPs might be involved in banana growth, development, and abiotic stress responses. Further, the expression of 12 MaTCPs was analyzed by real-time quantitative RT-PCR, and the protein interaction analysis showed that MaPCF10 and MaPCF13 may have an important function in banana fruit development and ripening. These results lay the foundation for further study of the functions of TCP genes in banana.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The TCP gene family, a kind of plant-specific transcription factors (TFs), has versatile functions in diverse aspects of plant growth and development. The name of the TCP gene family originated from the first four identified genes: TEOSINTE BRANCHED1 (TB1) from maize, CYCLOIDEA (CYC) from snapdragon, and PCFs (PCF1 and PCF2 proteins from rice) (Doebley et al. 1997; Cubas et al. 1999; Luo et al. 1999). Members of this family have about 60-amino-acids encoded basic Helix–Loop–Helix (bHLH) conserved regions, which performs the functions of DNA binding, protein–protein interaction, and protein nuclear localization (Cubas et al. 1999). According to the structure of the bHLH conserved region, the TCP gene family can be divided into two subfamilies, namely Class I (PCF or TCP-P) and Class II (TCP-C) (Manassero et al. 2013). Class II is further subdivided into the CINCINNATA (CIN) and CYC/TB1 subgroups. Class I TCP proteins contain only one typical TCP conserved domain and have 4 amino acid deletions, while Class II TCP members usually have unique R domains (arginine motifs with 18–20 residues) and ECE motif (a glutamate-cysteine-glutamate peptide chain), which is speculated that they may have the function of mediating protein interactions (Cubas et al. 1999; Li 2015). Moreover, the DNA binding sequence for the two classes differs slightly but partly overlaps (GGNCCCAC for class I and GTGGNCCC for class II) (Kosugi and Ohashi 2002; Masuda et al. 2008).
The TCP gene family is widely involved in the biological processes of the plant life cycle, including branching (Takeda et al. 2003; Aguilar-Martínez et al. 2007), leaf development (Kieffer et al. 2011), flower development (Kieffer et al. 2011), hormone pathways (Aguilar-Martínez et al. 2007), seed germination (Tatematsu et al. 2008), gametophyte development (Pagnussat et al. 2005), mitochondrial biogenesis (Hammani et al. 2011), regulation of the circadian clock (Giraud et al. 2010), and fruit development and ripening (Parapunova et al. 2014) in various plants. To date, genome-wide identification of the TCP proteins has been performed in a variety of plants, such as Arabidopsis (Riechmann et al. 2000), rice (Yao et al. 2007), tomato (Parapunova et al. 2014), cotton (Ma et al. 2014), apple (Xu et al. 2014), strawberry (Wei et al. 2016) and grapevine (Leng et al. 2019). However, very little is known about the TCP family in banana, which was the most consumed fruit growing around the world. Based on transcriptomic data, Song et al. (2018) initially analyzed 25 banana TCP family genes and explored the function in banana fruit ripening. However, based on RNA-seq alone, it was difficult to achieve more complete and detailed annotations of the TCP genes in banana. In 2012 and 2016, the M. acuminata doubled-haploid genotype (A genome) sequencing and annotations were completed successfully (D’Hont et al. 2012, Martin et al. 2016), which laid the foundation for the genomic-identification of the TCP gene family in banana.
Despite the possible role of TCP family genes in banana fruit ripening has been studied based on RNA-seq data, our understanding of this family still lags behind compared with other plants like Arabidopsis, rice, and maize. Due to the critical role of TCP transcription factors in the control of plant development, fruit development and ripening, and abiotic stress responses, we performed the comprehensive analysis of the MaTCP family in banana. In this study, 48 MaTCP genes were characterized in M. acuminata using the newly genome version 2 (Martin et al. 2016). Comprehensive analyses of their phylogenetic relationship, conserved motifs, gene structure, location and duplication pattern, and cis-acting elements were conducted. Then, the dynamic expression patterns of these genes were investigated in various organs, in different stages of fruit development and ripening, and response to different abiotic stresses in banana. Further, the expression of 12 MaTCP genes was analyzed by real-time quantitative RT-PCR, and the protein–protein interaction network of MaTCPs was predicted. The results provide valuable information for further functional analyses of TCP genes in M. acuminata and other Musa species.
Materials and Methods
TCP Genes Identification and Phylogenetic Analysis
Arabidopsis and Oryza sativa TCP proteins were obtained from the TAIR (http://www.arabidopsis.org) and RGAP (http://rice.plantbiology.msu.edu) databases, respectively. Banana TCP proteins were retrieved from the DH-Pahang genome database (http://banana-genome-hub.southgreen.fr/). BLAST analysis was used to identify the predicted banana TCPs with all the TCPs from A. thaliana and O. sativa queries. After removing the repeat sequences, all the candidate TCPs were confirmed to encode the conserved TCP domain with SMART (http://smart.embl-heidelberg.de/) (Schultz et al. 1998).
A neighbor-joining (NJ) phylogenetic tree was constructed for the identified M. acuminata, 24 A. thaliana and 22 O. sativa TCPs using MEGA5.0 (Tamura et al. 2011) with 1000 bootstraps. The exon/intron structures were analyzed using the online Gene Structure Display Server (GSDS) with coding sequences and genomic sequences (Hu et al. 2015a) (http://gsds.cbi.pku.edu.cn/). The MEME (http://meme-suite.org/tools/meme) and SMART were used to identify and annotate the conserved motifs of MaTCPs. To predict miR319 target sites, full-length MaTCP nucleotide sequences were analyzed using the online software psRNATarget (Dai and Zhao, 2011) (http://plantgrn.noble.org/psRNATarget).The upstream 1-kb fragments from MaTCP genes were used to predict the cis-acting elements by PlantCARE (Lescot et al. 2002).
Chromosomal Location and Gene Duplication
The chromosomal location of each TCP gene was retrieved from the DH-Pahang genome database. Paralogous TCP genes were determined by the genomic sequence identity and the following two criteria were used: > 70% alignment coverage of the longer gene and > 70% identity in the aligned region (Yang et al. 2008). In addition, to further analyze gene duplication events, the synonymous substitution rate (Ks) and nonsynonymous substitution rate (Ka) were calculated using the software DnaSp (Librado and Rozas, 2009). The divergence times of the gene duplication events were also calculated based on the formula T = Ks/2λ (λ = 4.6 × 10−9) (Lescot et al. 2008).
Plant Material and Transcriptome Analysis
BaXi Jiao (M. acuminata, Cavendish cultivar group, AAA) was used as the main plant material. Roots, leaves, flowers and fruits at 80 days after emergence from pseudostem (DAF) were sampled for the tissue-specific expression analysis. Fruits at 0, 20 and 80 DAF (0 days after harvest (DPH)), 8 DPH, and 14 DPH were collected for fruit development and ripening analysis. For studying the expression profile of the MaTCPs under abiotic stresses, the five-leaf stage banana plantlets were subjected to 200 mM mannitol or 300 mM NaCl for 7 days and 4 °C for 22 h, respectively. Transcriptome sequencing (RNA-seq) was performed as described previously (Hu et al. 2015b). Gene expression levels were calculated as FPKM. The gene expression profiles of MaTCPs were visualized by the heatmap via Multi Experiment Viewer (MeV) software (http://mev.tm4.org/).
Real-Time Quantitative RT-PCR
The qRT-PCR was performed using SYBR® Premix Ex Taq™ (TaKaRa, Shiga, Japan) on a Mx3005P™ Real-Time PCR System (Stratagene, CA, USA). The banana MaActin (EF672732) was used as a reference gene. The relative expression levels of the target genes were assessed based on the 2−ΔΔCt method (Livark and Schmittgen 2001). For each gene, three biological replicates were carried out.
Construction of the Regulatory Networks
To further explore the function of MaTCPs, the potential protein interaction and co-expression networks were identified based on the banana genome database and transcriptome analysis. The co-expression network analysis was performed as described previously (Wang et al. 2020). We extracted the co-expression network of all MaTCPs and the network connections were visualized using Cytoscape software v.3.4.0 (Shannon et al. 2003).
Results
Identification and Evolutionary Analysis of MaTCPs in Banana
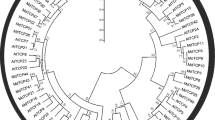
A total of 48 TCP genes were characterized by the M. acuminata genome. These TCP proteins have a peptide length ranging from 248 (MaPCF4) to 607 (MaCIN9) amino acids, with an average of 362.8 amino acids. The molecular weight (Mw) and isoelectric point (pI) varied from 25.23 (MaPCF4) to 65.18 (kDa) (MaCIN9) and from 5.25 (MaPCF7) to 10.12 (MaTB2), respectively. The other detailed information was shown in Supplementary Table S1. The alignment analysis showed that MaTCPs shared high homology in the TCP domain (Fig. 1A). Banana TCP proteins could be divided into two subfamilies. There was a four amino acid deletion in the basic helix–loop–helix-type motif of Class I compared with that of class II proteins. The two subfamilies, class I and class II, contained 27 and 21 genes, respectively. Moreover, the class II subfamily is differentiated into two subclades, CYC/TB1 and CIN (Figs. 1A and 2). The CYC/TB1 subclade contained three banana genes, MaTB1, MaTB2, and MaTB3 and CIN included 18 members, MaCIN1–MaCIN18 (Figs. 1A and 2). In M. acuminata, all three genes, MaTB1, MaTB2, and MaTB3, in CYC/TB1 and 5 genes, MaCIN1, MaCIN5, MaCIN6, MaCIN8, and MaCIN13, in the CIN subclade encoded proteins with the R-domain (Fig. 1B). This result was consistent with the previous research in Arabidopsis (Yao et al. 2007). Meanwhile, 5 genes in the CIN subclade (MaCIN1, MaCIN5, MaCIN6, MaCIN8 and MaCIN13) harbored putative target site of miR319 (Fig. 1C). Similarly, there were five CIN family members (AtTCP2, AtTCP3, AtTCP4, AtTCP10 and AtTCP24) contained miR319 binding sites in Arabidopsis (Palatnik et al. 2003). Phylogenetic tree showed that MaCIN1, MaCIN5, MaCIN6, MaCIN8 and MaCIN13 are grouped with AtTCP2 and AtTCP24 of A. thaliana (Fig. 2). This result was also found in cassava (Lei et al. 2017), P. mume (Zhou et al. 2016) and cotton (Ma et al. 2014). It is revealed that the miR319 target sites were retained during the evolution and diversification of plants.
Phylogenetic relationships of TCP transcription factors from banana, Arabidopsis and rice. The unrooted phylogenetic tree was constructed using MEGA5.0 by Neighbor-Joining method and the bootstrap test was performed with 1000 iterations. Red, blue, and green lines indicate the PCF, CIN and CYC/TB1 clades, respectively (Color figure online)
To study the evolutionary relationships of TCP family proteins, a phylogenetic tree was constructed by aligning 48, 24 and 22 TCP proteins from banana, Arabidopsis and rice. Two individual subfamilies (Classes I and II) were generated in Fig. 2. Proteins in Class II were further divided into 2 subclades: CYC/TB1 and CIN. All of the Arabidopsis and rice TCPs fell in the same Class or clade as previously reported (Manassero et al. 2013), confirming the reliability of our phylogenetic tree. Comparing the family gene number in banana, Arabidopsis and rice, the gene number in banana was the 2.0-fold size of that in Arabidopsis and rice. In addition, the biased expansion in M. acuminata was found. It occurred mainly in Class I and in CIN subclade, and the CYC/TB1 clade remained the same size as in Arabidopsis and rice (Table 1). Moreover, in comparison with other polyploidy crops, the gene member of MaTCPs was much higher than that of potato (23) and shortly higher than that of Chinese cabbage (39) and turnip (39), but it was significantly lower than that of wheat (66) (Table 1). It showed that species that experienced whole genome duplication or polyploidization seemed to have more duplication genes than species that did not experience such events (Liu et al. 2019).
Gene Structure and Conserved Motif Analysis of MaTCP Genes
A new phylogenetic tree of banana TCP genes was constructed, together with their exon/intron structure and conserved motif analysis (Fig. 3). Two genes (MaPCF14 and MaCIN9) had 1–2 introns, while the others had no introns (Fig. 3B). A total of 20 conserved motifs were identified in banana TCP proteins (Fig. 3C and Supplementary Table S2). In short, their distribution was found to be almost consistent with the phylogenetic tree. Motif 1, which was identified as the TCP domain, was found in all the banana TCP proteins. Motif 2 was only found in all class I (PCF class). Motif 11 was only found in all class II. Motif 4 was only found in all CIN subclade. Eight proteins from class II (3 genes in CYC/TB1 and 5 genes in CIN subclade) were found to have the R domain (motif 16). Moreover, some proteins from the two classes had several special motifs, respectively. For example, 5 genes in CIN subclade (MaCIN1, MaCIN5, MaCIN6, MaCIN8 and MaCIN13) harbored motifs 9, 10 and 13, while 8 genes in PCF class (MaPCF1, MaPCF5, MaPCF7, MaPCF8, MaPCF25, and MaPCF27) had motifs 7 and 15 (Fig. 3C).
Genomic structure and motif composition of banana TCPs. a Phylogenetic tree of M. acuminata TCP proteins. b Genomic structure of banana TCP genes. Exons, introns, and UTRs are indicated with yellow boxes, black lines and blue boxes, respectively. c Motif composition of banana TCP proteins. Conserved motifs in the banana TCP proteins are indicated by colored boxes (Color figure online)
Chromosomal Location and Gene Duplication Analysis
Based on the genomic data, 48 TCP genes were unevenly distributed on 10 of 11 banana chromosomes. The number of genes on each chromosome ranged from 0 (Chr 9) to 8 (Chr 7). The distribution of MaTCP genes on the 10 banana chromosomes was shown in Fig. 4 and the exact position of each gene on the banana chromosome was given in Supplementary Table S1. Thirty-four duplication events, including 20 gene pairs from PCF class and 14 pairs from Class II, were identified, accounting for 70.83% of the total TCP genes (Fig. 4 and Supplementary Table S3). In addition, 22 genes involved in two or more segmental duplication events (MaPCF2/MaPCF15/MaPCF21, MaPCF3/MaPCF9/MaPCF26, MaPCF6/MaPCF16/MaPCF17, MaPCF7/MaPCF8/MaPCF19/MaPCF20/MaPCF25/MaPCF27 and MaCIN2/MaCIN10/MaCIN14/ MaCIN9/MaCIN16/ MaCIN17/MaCIN18).
Two or more adjacent homologous genes within 200 kb on a single chromosome were defined as tandem duplications (Guo et al. 2013). According to the definition, no tandem duplication event was identified. Thirty-four pairs of TCP genes associated with segmental duplication, indicating that segmental duplications had a vital role in MaTCPs expansion in the banana genome. Furthermore, the Ka/Ks values of paralogous genes were calculated. Interestingly, three pairs (MaPCF1/MaPCF5, MaPCF13/MaPCF22 and MaTB2/MaTB3) exhibited Ka/Ks > 1.00 values, signifying positive selection, whereas the remaining underwent purifying selection with values less than 1.00. Finally, we also calculated the divergence time for these duplicated genes. The results showed that these gene pairs were estimated to diverge at approximately 5.80 ~ 100.58 million year ago (MYA) (Supplementary Table S3).
Expression Profiles of MaTCPs in Different Organs
The relative transcript abundance of MaTCPs in roots, leaves, flowers and fruits is shown in Fig. 5A and Supplementary Table S4. Generally, 39 out of 48 MaTCPs were detected in at least one tissue. A total of 10, 10, 11 and 8 genes had high expression (value > 10) in roots, leaves, flowers, and fruits, respectively. Notably, three MaTCPs (MaPCF2, MaPCF4 and MaPCF24) exhibited high transcript levels (value > 10) in four organs. However, the representatives of class II TCPs were expressed mostly in leaves. Most of them were not expressed in roots and fruits. MaCIN2 and MaCIN11 expressed highly (value > 10) in leaves. MaCIN1 and MaTB1 expressed highly (value > 10) in flowers and fruits, respectively.
Expression patterns of MaTCPs: a in the roots, leaves, flowers and fruits, b in different stages of fruit development and ripening, and c in response to cold, salt, and osmotic stresses in banana. The heat map a and b were created based on the FPKM value of MaTCPs. The heat map c was created based on log2-based values of MaTCPs. Differences in gene expression are shown in color as the scale
Expression Profiles of MaTCPs in the Fruit Development and Ripening
The expression profiles of MaTCPs in fruit development and ripening were also studied. Generally, 38 out of the 48 genes showed expression in different stages of fruit development and ripening (Fig. 5B, Supplementary Table S5). Five genes (MaPCF2, MaPCF4, MaPCF11, MaPCF19 and MaPCF24) expressed highly (values > 10) at 0, 20 and 80 DAF. Notably, MaPCF2 and MaPCF10 expressed highly (value > 10) at 8 and 14 DPH of the fruits, suggesting that they might have an extensive and vital role in the fruit development and ripening.
Moreover, banana TCP genes with close phylogenetic relationships showed both similar and divergent expression patterns. For example, MaPCF13 and MaPCF22 were both expressed at a high level at 0 and 20 DAF and at a low level at 80 DAF, 8 and 14 DPH and the expression level of MaPCF13 was higher than that of MaPCF22 in each tissue we tested. With respect to MaPCF7 and MaPCF8, however, divergent expression patterns were found at 0, 20, 80 DAF and 8 DPH, respectively (Supplementary Table S5).
Expression Profiles of MaTCPs Under Cold, Salt and Osmotic Stresses
Under the three stress conditions, the expression levels of most genes (34 out of the 48 MaTCPs) were modulated (Fig. 5C, Supplementary Table S6). Among them, 16, 12 and 14 genes showed increased expression while 18, 22 and 20 genes were downregulated by cold, salt and osmotic stress, respectively. Intriguingly, there were 6 (MaCIN15, MaPCF1, MaPCF5, MaPCF8, MaPCF21 and MaPCF22), 5 (MaCIN15, MaPCF1, MaPCF14, MaPCF21 and MaPCF23) and 5 (MaPCF1, MaPCF5, MaPCF9, MaPCF14, and MaPCF21) MaTCP genes that were notably induced (value > 1) in response to cold, salt or osmotic stress, respectively. In addition, 3 MaTCP genes (MaPCF1, MaPCF5 and MaPCF21) were upregulated in all of the stress conditions, suggesting they have major functions in response to various stress conditions.
Validation of the MaTCP Genes by qRT-PCR
Based on transcriptome sequencing data analysis, 12 differentially expressed MaTCPs were selected for qRT-PCR analysis. Primers for the quantification assay were shown in Supplementary Table S7. Four genes (MaPCF4, MaPCF12, MaPCF13 and MaPCF19) were tested in various organs and 4 genes (MaPCF2, MaPCF8, MaPCF10 and MaPCF26) were tested at different stages of fruit development and ripening. Four MaTCP genes (MaPCF1, MaPCF5, MaPCF21 and MaCIN15) that were upregulated in response to each of the stress treatments were tested. The results showed that the expression patterns of the selected MaTCPs had the same trend and consistent results between RNA-seq data and qRT-PCR data (Fig. 6).
Relative expression of MaTCPs in BX by qRT-PCR. a–d expression patterns of MaPCF4, MaPCF12, MaPCF13 and MPCF19 in different organs. The mRNA fold difference was relative to that of BX root samples used as calibrator. e–h expression patterns of MaPCF2, MaPCF8, MaPCF10 and MaPCF26 in different stages of fruit development and ripening. The mRNA fold difference was relative to that of BX 0DAF samples used as calibrator. i–l expression patterns of MaPCF1, MaPCF5, MaPCF21 and MaCIN15 in response to cold, salt and osmotic stresses. The mRNA fold difference was relative to that of untreated samples used as calibrator. Data are means ± SD of n = 3 biological replicates
Putative Cis-Elements Analysis in the MaTCP Promoters
From promoter analysis, three groups of cis-elements, including plant growth and development, phytohormone responses and abiotic and biotic stress responses, were detected (Fig. 7 and Supplementary Table S8). For plant growth and development, three types of cis-elements (Skn-1_motif and GCN4_motif, CCGTCC box and O2-site) are involved in endosperm expression, meristem specific activation and zein metabolism regulation respectively were found in the most MaTCP promoters. The as-2-box and CAT-box, involved in light-responsive and meristem expression respectively, were detected in the promoter of six diverse MaTCPs. In addition, the HD-Zip1 and HD-Zip2 motifs, involved in leaf development were only presented in the MaPCF1 promoter. In hormone responses, 2 MeJA-responsive elements (CGTCA-motif and TGACG-motif), salicylic acid-responsive element (TCA-element) and 2 ABA-responsive elements (ABRE, CE3) were found in 27, 25 and 19 MaTCPs respectively. Moreover, ethylene-responsive element (ERE), 2 auxin-responsive elements (AuxRR-core and TGA-element) and 3 gibberellin-responsive elements (TATC-box, P-box, and GRAE-motif) were also detected in 3, 7 and 9 MaTCPs, respectively. In stress responses, 10 stress-relative elements were detected. In details, 50.0% (24/48) and 52.1% (25/48) MaTCP promoter regions had drought-inducible element (MBS) and anaerobic induction element (ARE), respectively. Additionally, 29.2% (14/48), 25% (12/48) and 20.8% (10/48) MaTCP promoter regions had TC-rich repeats involved in defense and stress responses, fungal elicitor responsive element (Box-W1) and heat stress-responsive element (HSE), respectively. Low temperature-responsive element (LTR) and GC-motif involved in anoxic specific inducibility were only found in 7 and 6 MaTCPs, respectively.
Kinds and numbers of the cis-acting element analysis found in the promoter regions of banana TCP genes. a A number of each cis-acting element in the promoter region of MaTCPs. b Statistics for the total number of MaTCPs, including the corresponding cis-acting elements (red dot) and the total number of cis-acting elements in MaTCPs (black box) (Color figure online)
The Interaction Network of MaTCP Genes
To further explore the function of MaTCPs, the potential protein interaction and co-expression networks were identified using Cytoscape. Two MaTCP-mediated networks were constructed, and 23 and 47 interactive proteins for MaPCF10 and 13, respectively, were detected (Fig. 8, Supplementary Table S9 and S10).
The 47 key proteins that interacted with MaPCF13 were divided into 46 classes including the most abundant TF family bHLH (20), followed by the MYB (17), HB (12), C2H2 (11), SBP (8) and MIKC (8) with single-member for NAC, C3H, ARF, C2C2.CO.like, GNAT, etc. The 23 key proteins that interacted with MaPCF10 were divided into 21 classes, including bHLH (3), HB (3) and C2H2 (3), followed by NAC (2), ERF (2), PHD (2), MYB (2), GRAS (2) and bZIP (2), with single members for HSF, G2-like, NF-YC, OFP, WRKY, C2C2-GATA, etc. Upon comparison of the proteins interacting with MaPCF10 and 13, 16 kinds of proteins (bHLHs, bZIP, MADS-box, MYB, HB, C2H2, OFP, ERF, WRKY, PHD, C2C2-GATA, G2-like, NAC, LOB, HSF and Orphans) were found to be very same, showing that these proteins may be fundamental for banana fruit development and ripening. Another 6 kinds of proteins (NF.YC, LIM, GRAS, Jumonji, NF.X1 and Trihelix) were found to specifically interact with MaPCF10, suggesting that these proteins may interact with MaPCF10 to participate in banana fruit ripening. In addition, 30 kinds of proteins (SET, PLATZ, AUX.IAA, C3H, AP2, C2C2-Dof, VOZ, SWI.SNF.BAF60b, HMG, ARF, IWS1, SBP, zf.HD, C2C2.CO.like, GRF, BBR.BPC, GANT, BES1, C2C2.YABBY, E2F.DP, SNF2, CPP, NF.YB, ARID, TRAF, ARR.B, GRAS, B3, TUB and TCP) were found to specifically interact with MaPCF13, indicating that these proteins together with MaPCF13 may play a role in the banana fruit development.
Discussion
The TCP proteins are a kind of plant-specific TFs that are involved in regulating plant biological processes such as branch, leaf and flower development, hormone signaling and stress response. It is widely distributed in plant and the memmber of this gene family varies greatly among various species. To date, genome-wide identification of the TCP proteins has been performed in a variety of plants. Song et al. (2018) initially identified 25 banana TCP sequences based on banana transcriptome data related to fruit ripening. In our study, a total of 48 TCP genes were detected in M. acuminata genome based on the latest released banana genome database version 2 (Martin et al. 2016). 48 TCPs identified in our study included the 25 TCPs reported by Song et al. (2018) and the corresponding information was listed in Supplementary Table S1. It is significantly higher than that reported by Song et al. (2018). It is more comprehensive and accurate for gene family identification using the whole-genome information. Compared to other plants, the number of TCP genes in M. acuminata in our study is similar to that in Malus domestica (52) (Xu et al. 2014) and much higher than that in grape (18) (Leng et al. 2019), strawberry (19) (Wei et al. 2016), Arabidopsis (24) and rice (22) (Martín-Trillo and Cubas 2010). The number of TCP genes in M. acuminata is 2.0 times that in Arabidopsis, which is consistent with the fact that the protein-coding genes in the M. acuminata genome (36,542 genes) is about 1.4 times that in Arabidopsis (25,498 genes) (Arabidopsis Genome Initiative 2000; D’Hont et al. 2012).
Banana TCP gene encodes a protein sequence length ranging from 248 to 607 amino acids. Phylogenetic analysis shows that the family can be divided into two major classes, Class I and Class II, namely PCFs and CYC/TB1, which are consistent with the results of Arabidopsis and rice TCP proteins (Fig. 2). These TCPs in the same subclass had similar gene structures and protein motifs (Fig. 3), which implied that they may play similar molecular roles. Here, we found that most of the genes (95.83%) had no intron. It is in concert with the research that plants tend to retain the genes with no intron or a short intron (Mattick and Gagen 2001). Furthermore, all the proteins carried a highly conserved TCP domain (motif 1). Proteins in Class I and Class II also have different specific motifs respectively. The R domain (motif 16), with the function to mediate protein interactions, was also found in 8 proteins from class II (3 genes in CYC/TB1 and 5 genes in CIN subclade). It is speculated that these members may play a key role in protein interaction.
Gene duplication events are of great significance in gene family expansion and evolution. Segmental, tandem, and whole-genome duplication (WGD) are the main modes for the gene family expansion (Leister 2004). In this study, we found that most of the banana TCP genes were involved in segmental duplication in the banana genome, indicating that segmental duplication may be the significant driving force for the banana TCP gene family expansion. It is similar to the results of Arabidopsis, rice and maize (Yao et al. 2007; Chai et al. 2017). Based on synonymous substitution rate (Ks) analysis, the banana genome has undergone three WGDs during evolution, namely the α+ß WGD events (Ks≈0 ~ 0.45) and the γ WGD event (Ks≈0.45 ~ 0.85) that occurred 64.8 million years ago (Mya) and 96Mya respectively (D’Hont et al. 2012). The Ks value of 22 paralogous gene pairs was ranged from 0.0534 to 0.8180, showing the divergence time for these duplicated genes was ranged from 5.80 to 88.91 MYA. It suggested that WGD is also responsible for the expansion of the MaTCP gene family. This result was similar to MaERF and MaWRKY gene family (Lakhwani et al. 2016; Goel et al. 2016). In addition, we identified 2 TCP gene paralogs (MaPCF7/MaPCF19 and MaCIN4/MaCIN15 with Ks > 0.85,) attributing probably to an older duplication event. The Ka/Ks value of most of the paralogous gene pairs ranged from 0.15 to 0.67, showing the purifying selection of these genes (Supplementary Table S3). In summary, the banana TCP gene family may originate from more ancient duplication events and has undergone multiple WGDs. The combined effects of segmental duplication and WGDs may be the direct reason for the expansion of the TCP gene family in M. acuminata.
The expression pattern of TCP TFs in different tissues has been reported in plants, such as Arabidopsis (Martin-Trillo and Cubas, 2010), maize (Chai et al. 2017), apple (Xu et al. 2014) and strawberry (Wei et al. 2016). There are few studies on the dynamic and spatially expression profile of MaTCP genes in banana. Based on RNA-seq data, the tissue-specific expression of MaTCP was analyzed (Fig. 5A and Supplementary Table S4). It was found that the genes from two subclasses had a divergence tissue-specific expression. A previous study had reported that 8 CIN-type genes (AtTCP2, AtTCP3, AtTCP4, AtTCP5, AtTCP10, AtTCP13, AtTCP17 and AtTCP24) may regulate leaf growth in Arabidopsis (Palatnik et al. 2003; Ori et al. 2007). In this study, 55% of the CIN-type (Class II) were highly expressed in the leaf. However, about 60% of the PCF genes (Class I) were detected in all of the organs and were predominantly expressed in the root, leaf and flower. We speculate that Class I MaTCP genes might be involved in housekeeping functions during banana growth and development.
Previous research showed that Class I TCP proteins were involved in fruit development and ripening (Parapunova et al. 2014; Pillet et al. 2015; Wei et al. 2016). For example, FaTCP11 and FvTCP9 were found to play a role in fruit development respectively in strawberry (Pillet et al. 2015; Wei et al. 2016). Three tomato TCPs (SlTCP12, 15 and 18) were reported to be preferentially expressed in tomato fruit and their promoter elements could be bound by the ripening–related proteins such as RIN, CNR and SlAP2a, respectively (Parapunova et al. 2014). Our results also showed that MaTCPs are likely to play a role in fruit development and ripening. Five genes (MaPCF2, MaPCF4, MaPCF11, MaPCF19 and MaPCF24) showed highly expression during the stage of fruit development (0, 20 and 80 DAF). Eight genes (MaPCF1, MaPCF7, MaPCF12, MaPCF13, MaPCF20, MaPCF22, MaPCF25 and MaPCF27) high expressed during the early stage of fruit development (0 and 20 DAF) (Fig. 5B and Supplementary Table S4). It is indicated that these genes may have a function in banana fruit development. Particularly, MaPCF2 and MaPCF10 were significantly expressed at 8 and 14 DPH, which was verified by qRT-PCR (Fig. 5). As for CIN-type, only MaCIN1 showed high expression in banana fruit development and ripening. Song et al. (2018) reported that MaTCP5 and MaTCP20 promoted the transcription of MaXTH10/11 that may play a role in fruit softening during banana ripening, whereas MaTCP19 repressed their transcription, by directly binding to their promoters. Combined with the research, MaTCP5, MaTCP19 and MaTCP20 are equivalent to MaCIN1, MaPCF10 and MaPCF18 respectively in our study. These results indicated that MaPCF10 might have a vital role in banana fruit ripening.
Moreover, most MaTCP genes showed apparent responses to different kinds of abiotic stresses (Fig. 5C). Numerous cis-acting elements involved in plant growth, development, hormone responses as well as stress responses were also found in the promoter region of the TCP genes (Fig. 6), implying that MaTCPs are also involved in abiotic stress signaling or tolerance, which is worthy of further study.
References
Aguilar-Martínez JA, Poza-Carrión C, Cubas P (2007) Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. Plant Cell 19(2):458–472. https://doi.org/10.1105/tpc.106.048934
Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408(6814):796–815. https://doi.org/10.1038/35048692
Bao S, Zhang Z, Lian Q, Sun Q, Zhang R (2019) Evolution and expression of genes encoding TCP transcription factors in Solanum tuberosum reveal the involvement of StTCP23 in plant defence. BMC Genet 20:91. https://doi.org/10.1186/s12863-019-0793-1
Chai W, Jiang P, Huang G, Jiang H, Li X (2017) Identification and expression profiling analysis of TCP family genes involved in growth and development in maize. Physiol Mol Biol Plants 23(4):779–791. https://doi.org/10.1007/s12298-017-0476-1
Cubas P, Lauter N, Doebley J, Coen E (1999) The TCP domain: a motif found in proteins regulating plant growth and development. Plant J 18(2):215–222. https://doi.org/10.1046/j.1365-313x.1999.00444.x
Dai X, Zhao P (2011) psRNATarget: A plant small RNA target analysis server. Nucleic Acids Res. 39:W155–W159. https://doi.org/10.1093/nar/gkr319.P
D’Hont A, Denoeud F, Aury JM, Baurens FC, Carreel F, Garsmeur O et al (2012) The banana (Musa acuminata) genome and the evolution of monocotyledonous plants. Nature 488(7410):213–217. https://doi.org/10.1038/nature11241
Doebley J, Stec A, Hubbard L (1997) The evolution of apical dominance in maize. Nature 386(6624):485–488. https://doi.org/10.1038/386485a0
Giraud E, Ng S, Carrie C, Duncan O, Low J, Lee CP, Van Aken O, Millar AH, Murcha M, Whelan J (2010) TCP transcription factors link the regulation of genes encoding mitochondrial proteins with the circadian clock in Arabidopsis thaliana. Plant Cell 22(12):3921–3934. https://doi.org/10.1105/tpc.110.074518
Goel R, Pandey A, Trivedi PK, Asif MH (2016) Genome-wide analysis of the Musa WRKY gene family: evolution and differential expression during development and stress. Front Plant Sci 7:299. https://doi.org/10.3389/fpls.2016.00299
Guo R, Xu X, Carole B, Li X, Gao M, Zheng Y, Wang X (2013) Genome-wide identification, evolutionary and expression analysis of the aspartic protease gene superfamily in grape. BMC Genom 14:554. https://doi.org/10.1186/1471-2164-14-554
Hammani K, Gobert A, Hleibieh K, Choulier L, Small I, Giegé P (2011) An Arabidopsis dual-localized pentatricopeptide repeat protein interacts with nuclear proteins involved in gene expression regulation. Plant Cell 23(2):730–740. https://doi.org/10.1105/tpc.110.081638
Hu B, Jin J, Guo AY, Zhang H, Luo J, Gao G (2015a) GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31(8):1296–1297. https://doi.org/10.1093/bioinformatics/btu817
Hu W, Zuo J, Hou X, Yan Y, Wei Y, Liu J, Li M, Xu B, Jin Z (2015b) The auxin response factor gene family in banana: genome-wide identification and expression analyses during development, ripening, and abiotic stress. Front Plant Sci 6:742. https://doi.org/10.3389/fpls.2015.00742
Kieffer M, Master V, Waites R, Davies B (2011) TCP14 and TCP15 affect internode length and leaf shape in Arabidopsis. Plant J 68(1):147–158. https://doi.org/10.1111/j.1365-313X.2011.04674.x
Kosugi S, Ohashi Y (2002) DNA binding and dimerization specificity and potential targets for the TCP protein family. Plant J 30(3):337–348. https://doi.org/10.1046/j.1365-313x.2002.01294.x
Lakhwani D, Pandey A, Dhar YV, Bag SK, Trivedi PK, Asif MH (2016) Genome-wide analysis of the AP2/ERF family in Musa species reveals divergence and neofunctionalisation during evolution. Sci Rep 6:18878. https://doi.org/10.1038/srep18878
Leister D (2004) Tandem and segmental gene duplication and recombination in the evolution of plant disease resistance genes. Trends Genet 20(3):116–122. https://doi.org/10.1016/j.tig.2004.01.007
Lei N, Yu X, Li S, Zeng C, Zou L, Liao W, Peng M (2017) Phylogeny and expression pattern analysis of TCP transcription factors in cassava seedlings exposed to cold and/or drought stress. Sci Rep 7:10016. https://doi.org/10.1038/s41598-017-09398-5
Leng X, Wei H, Xu X, Ghuge SA, Jia D, Liu G, Wang Y, Yuan Y (2019) Genome-wide identification and transcript analysis of TCP transcription factors in grapevine. BMC Genom 20(1):786. https://doi.org/10.1186/s12864-019-6159-2
Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouzé P, Rombauts S (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30(1):325–327. https://doi.org/10.1093/nar/30.1.325
Lescot M, Piffanelli P, Ciampi AY, Ruiz M, Blanc G, Leebens-Mack J, da Silva FR, Santos CM, D’Hont A, Garsmeur O, Vilarinhos AD, Kanamori H, Matsumoto T, Ronning CM, Cheung F, Haas BJ, Althoff R, Arbogast T, Hine E, Pappas GJ Jr, Sasaki T, Souza MT Jr, Miller RN, Glaszmann JC, Town CD (2008) Insights into the Musa genome: syntenic relationships to rice and between Musa species. BMC Genomics 9:58. https://doi.org/10.1186/1471-2164-9-58
Li S (2015) The Arabidopsis thaliana TCP transcription factors: a broadening horizon beyond development. Plant Signal Behav 10(7):e1044192. https://doi.org/10.1080/15592324.2015.1044192
Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25(11):1451–1452. https://doi.org/10.1093/bioinformatics/btp187
Liu Y, Guan X, Liu S, Yang M, Ren J, Guo M, Huang Z, Zhang Y (2018) Genome-wide identification and analysis of TCP transcription factors involved in the formation of leafy head in Chinese cabbage. Int J Mol Sci 19(3):847. https://doi.org/10.3390/ijms19030847
Liu MM, Wang MM, Yang J, Wen J, Guo PC, Wu YW, Ke YZ, Li PF, Li JN, Du H (2019) Evolutionary and comparative expression analyses of TCP transcription factor gene family in land plants. Int J Mol Sci 20(14):3591. https://doi.org/10.3390/ijms20143591
Livark KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−△△CT method. Methods 25(4):402–408. https://doi.org/10.1006/meth.2001.1262
Luo D, Carpenter R, Copsey L, Vincent C, Clark J, Coen E (1999) Control of organ asymmetry in flowers of Antirrhinum. Cell 99(4):367–376. https://doi.org/10.1016/s0092-8674(00)81523-8
Ma J, Wang Q, Sun R, Xie F, Jones DC, Zhang B (2014) Genome-wide identification and expression analysis of TCP transcription factors in Gossypium raimondii. Sci Rep 4:6645. https://doi.org/10.1038/srep06645
Manassero NG, Viola IL, Welchen E, Gonzalez DH (2013) TCP transcription factors: architectures of plant form. Biomol Concepts 4(2):111–127. https://doi.org/10.1515/bmc-2012-0051
Martín-Trillo M, Cubas P (2010) TCP genes: a family snapshot ten years later. Trends Plant Sci 15(1):31–39. https://doi.org/10.1016/j.tplants.2009.11.003
Martin G, Baurens FC, Droc G, Rouard M, Cenci A, Kilian A, Hastie A, Dolezel J, Aury JM, Alberti A, Carreel F, D’Hont A (2016) Improvement of the banana “Musa acuminata” reference sequence using NGS data and semi-automated bioinformatic methods. BMC Genom 17:243. https://doi.org/10.1186/s12864-016-2579-4
Masuda HP, Cabral LM, De Veylder L, Tanurdzic M, de Almeida EJ, Geelen D, Inzé D, Martienssen RA, Ferreira PC, Hemerly AS (2008) ABAP1 is a novel plant Armadillo BTB protein involved in DNA replication and transcription. EMBO J 27(20):2746–2756. https://doi.org/10.1038/emboj.2008.191
Mattick JS, Gagen MJ (2001) The evolution of controlled multitasked gene networks: the introns and other noncoding RNAs in the development of complex organisms. Mol Biol Evol 18(9):1611–1630. https://doi.org/10.1093/oxfordjournals.molbev.a003951
Ori N, Cohen AR, Etzioni A, Brand A, Yanai O, Shleizer S, Menda N, Amsellem Z, Efroni I, Pekker I, Alvarez JP, Blum E, Zamir D, Eshed Y (2007) Regulation of LANCEOLATE by miR319 is required for compound- leaf development in tomato. Nat Genet 39(6):787–791. https://doi.org/10.1038/ng2036
Pagnussat GC, Yu HJ, Ngo QA, Rajani S, Mayalagu S, Johnson CS, Capron A, Xie LF, Ye D, Sundaresan V (2005) Genetic and molecular identification of genes required for female gametophyte development and function in Arabidopsis. Development 132(3):603–614. https://doi.org/10.1242/dev.01595
Palatnik JF, Allen E, Wu X, Schommer C, Schwab R, Carrington JC, Weigel D (2003) Control of leaf morphogenesis by microRNAs. Nature 425(6955):257–263. https://doi.org/10.1038/nature01958
Parapunova V, Busscher M, Busscher-Lange J, Lammers M, Karlova R, Bovy AG, Angenent GC, de Maagd RA (2014) Identification, cloning and characterization of the tomato TCP transcription factor family. BMC Plant Biol 14:157. https://doi.org/10.1186/1471-2229-14-157
Pillet J, Yu HW, Chambers AH, Whitaker VM, Folta KM (2015) Identification of candidate flavonoid pathway genes using transcriptome correlation network analysis in ripe strawberry (Fragaria × ananassa) fruits. J Exp Bot 66(15):4455–4467. https://doi.org/10.1093/jxb/erv205
Riechmann JL, Heard J, Martin G, Reuber L, Jiang C, Keddie J, Adam L, Pineda O, Ratcliffe OJ, Samaha RR, Creelman R, Pilgrim M, Broun P, Zhang JZ, Ghandehari D, Sherman BK, Yu G (2000) Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290(5499):2105–2110. https://doi.org/10.1126/science.290.5499.2105
Schultz J, Milpetz F, Bork P, Ponting CP (1998) SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci 95(11):5857–5864. https://doi.org/10.1073/pnas.95.11.5857
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13(11):2498–2504. https://doi.org/10.1101/gr.1239303
Song CB, Shan W, Yang YY, Tan XL, Fan ZQ, Chen JY, Lu WJ, Kuang JF (2018) Heterodimerization of MaTCP proteins modulates the transcription of MaXTH10/11 genes during banana fruit ripening. Biochim Biophys Acta Gene Regul Mech 1861(7):613–622. https://doi.org/10.1016/j.bbagrm.2018.06.005
Takeda T, Suwa Y, Suzuki M, Kitano H, Ueguchi-Tanaka M, Ashikari M, Matsuoka M, Ueguchi C (2003) The OsTB1 gene negatively regulates lateral branching in rice. Plant J 33(3):513–520. https://doi.org/10.1046/j.1365-313x.2003.01648.x
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28(10):2731–2739. https://doi.org/10.1093/molbev/msr121
Tatematsu K, Nakabayashi K, Kamiya Y, Nambara E (2008) Transcription factor AtTCP14 regulates embryonic growth potential during seed germination in Arabidopsis thaliana. Plant J 53(1):42–52. https://doi.org/10.1111/j.1365-313X.2007.03308.x
Wang Z, Jia C, Wang JY, Miao HX, Liu JH, Chen C, Yang HX, Xu B, Jin Z (2020) Genome-Wide Analysis of Basic Helix-Loop-Helix Transcription Factors to Elucidate Candidate Genes Related to Fruit Ripening and Stress in Banana (Musa acuminata L. AAA Group, cv. Cavendish). Front Plant Sci 11:650. https://doi.org/10.3389/fpls.2020.00650
Wei W, Hu Y, Cui MY, Han YT, Gao K, Feng JY (2016) Identification and transcript analysis of the TCP transcription factors in the diploid woodland strawberry Fragaria vesca. Front Plant Sci 7:1937. https://doi.org/10.3389/fpls.2016.01937
Xu R, Sun P, Jia F, Lu L, Li Y, Zhang S, Huang J (2014) Genome wide analysis of TCP transcription factor gene family in Malus domestica. J Genet 93(3):733–746. https://doi.org/10.1007/s12041-014-0446-0
Yang S, Zhang X, Yue JX, Tian D, Chen JQ (2008) Recent duplications dominate NBS-encoding gene expansion in two woody species. Mol Genet Genom 280(3):187–198. https://doi.org/10.1007/s00438-008-0355-0
Yao X, Ma H, Wang J, Zhang D (2007) Genome-wide comparative analysis and expression pattern of TCP gene families in Arabidopsis thaliana and Oryza sativa. J Integr Plant Biol 49(6):885–897. https://doi.org/10.1111/j.1744-7909.2007.00509.x
Zhao J, Zhai Z, Li Y, Geng S, Song G, Guan J, Jia M, Wang F, Sun G, Feng N, Kong X, Chen L, Mao L, Li A (2018) Genome-wide identification and expression profiling of the TCP family genes in spike and grain development of wheat (Triticum aestivum L.). Front in Plant Sci 9:1282. https://doi.org/10.3389/fpls.2018.01282
Zhou Y, Xu Z, Zhao K, Yang W, Cheng T, Wang J, Zhang Q (2016) Genome-wide identification, characterization and expression analysis of the TCP gene family in Prunus mume. Front Plant Sci 7:1301. https://doi.org/10.3389/fpls.2016.01301
Acknowledgements
This research was supported by the Natural Science Foundation of Hainan Province (318MS090), the National Natural Science Foundation of China (31501043, 31872161), the National Natural Science Foundation of Hainan Province (2019CXTD412) and the Earmarked Fund for Modern Agro-industry Technology Research System (CARS-31).
Author information
Authors and Affiliations
Contributions
JHL, ZQJ and BYX conceived the study. JYW, ZW and CHJ performed the experiments and carried out the analysis. XHM and JBZ carried out the analysis. JYW designed the experiments and wrote the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors claim no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, J., Wang, Z., Jia, C. et al. Genome-Wide Identification and Transcript Analysis of TCP Gene Family in Banana (Musa acuminata L.). Biochem Genet 60, 204–222 (2022). https://doi.org/10.1007/s10528-021-10100-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10528-021-10100-8