Abstract
Endothelial Nitric Oxide Synthase (eNOS) is an indispensable regulator of blood pressure through producing Nitric Oxide (NO). There is some evidence to suggest that eNOS gene polymorphisms are associated with Essential Hypertension (EHT). In this study, the potential association between eNOS 4a/4b, A922G, G894T, T786C gene polymorphisms and EHT as individual risk factors and as haplotypes are examined in the southern population of Iran (Bandar-Abbas). In this study, 200 EHT patients and 200 normotensive subjects which were matched for age and sex were included. Genotyping was performed by either utilizing Polymerase Chain Reaction (PCR) or PCR followed by Restriction Fragment length Polymorphism (RFLP) method. Our results demonstrated statistically significant associations between T786C, G894T, and 4a/4a and EHT (p < 0.05); however, A922G had no significant association with EHT (p > 0.05). Haplotype analysis also suggested that − 786C/− 922A/4a, − 786C/− 922A/4b and − 786C/− 922G/4a haplotypes were more frequent in EHT group than control group, hypothesizing a positive association with EHT. The present study has identified that the eNOS genetic variations are associated with EHT in southern population of Iran (Bandar-Abbas). These findings also suggested that a number of haplotypes of eNOS gene may be a driving factor for EHT susceptibility in respected population.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Essential hypertension (EHT) is a multifactorial disorder in which interaction of not only genetic but also epigenetics as well as environmental and life style factors could affect its onset and development (Salvi et al. 2012). This complex trait is one of the major risk factors for cardiovascular diseases (CVD). It has been estimated that by the year 2025, approximately 29% of the adult population (1.56 billion people) will suffer from hypertension worldwide (Rodrigo et al. 2013; Chandra et al. 2014). Both human and animal studies have shown onset of hypertension as a result of loss of Nitric Oxide (NO) (Kingah et al. 2010). NO produced from l-Arginine through the function of endothelial nitric oxide synthase (eNOS) plays a vital role in the preservation of endothelium homeostasis and proliferation of Vascular Smooth Muscle Cells (VSMCs), regulation of vasomotor tone, and eventually controlling blood pressure (Colomba et al. 2008). eNOS gene encoding the eNOS protein introduce itself as a potential candidate for investigating susceptibility to hypertension (Shankarishan et al. 2014). The human eNOS gene located on 7q35-36 consists of 26 exons spanning a genome region of 21 Kb (Li et al. 2007). Several polymorphisms within eNOS gene have been identified among which intron 4a/4b Variable Number Tandem Repeat (VNTR), T786C (rs2070744), G894T (rs1799983), A922G (rs1800779) have been found to increase EHT prevalence (Nejatizadeh et al. 2008). eNOS polymorphisms have variable prevalence among various ethnic groups, leading to uncertainty regarding this gene as a prerequisite for developing hypertension (Rahimi et al. 2013; Li et al. 2011).
T786C (rs2070744) polymorphism located in the 5′ flanking region of the eNOS gene, results in the substitution of Thymidine by Cytosine at nucleotide 786 (Hyndman et al. 2002). T786C has been reported to be associated with an increased risk of EHT in the Tunisian population (Jemaa et al. 2011). G894T (rs1799983) is a prevalent variant in exon 7 of eNOS gene that corresponds to Glutamate–Aspartate substitution at codon 298 and contributes to some CVDs such as Coronary Artery Disease (CAD), Myocardial Infarction (MI) as well as EHT (Jíra et al. 2011). Several studies have reported the prevalent of this polymorphism with occurrence of hypertension (Niu and Qi 2011; Men et al. 2011). Moreover, it has been indicated that variations in intron 4 of the eNOS gene including the 4a/4b (VNTR) polymorphism have a significant effect on the EHT incidence (Deng et al. 2007). This polymorphism encompasses two alleles: 4b allele has five tandem repeats of 27 nucleotide fragment and the 4a allele has four tandem repeats of the same fragment (Shah et al. 2013). Previous studies have failed to reach a consensus conclusion regarding these polymorphism and hypertension incidence (Zarmakhi and Hashemi 2012; Jemaa et al. 2009; Moe et al. 2006; Benjafield and Morris 2000). Abundant evidence has suggested that eNOS polymorphisms can have crucial effect of NO formation. Hence haplotype analysis can be a better approach to elucidate the role of these polymorphisms in occurrence of hypertension (Kitsios and Zintzaras 2010). To the best of our knowledge, the association of eNOS − 922A > G, − 786 T > C, intron 4 b/a VNTR and 894 G > T polymorphisms with EHT have not been investigated in southern population of Iran where the occurrences of EHT is more frequent. Therefore, the aim of this study is to evaluate the correlation between the A922G, G894T, T786C, and 4a/4b polymorphisms individually and as haplotypes with EHT in the mentioned population.
Materials & Methods
Study Population
This case–control study comprises 200 healthy subjects and 200 individuals who were suffering from EHT. Control specimens were obtained from Bandar-Abbas blood transfusion organization and general public and the patient group was obtained from Shahid Mohammadi hospital. All participants were chosen from southern Iran population. This study is approved by the Hormozgan University of Medical Sciences Ethics Committee. EHT was confirmed by clinicopathological findings in all the members of the patients group. All participants were handed an informed consent and were properly informed of the process. The participants were matched for age and sex. The following criteria must have been met before enrolling an individual in our control group: each individual must have been aged between 30 and 60 years, their blood pressure must have been lower than 140/90 mmHg, they should not have taken antihypertensive medication, and finally they must have not had a history of diabetes and general illness. Prior inclusion of anyone in patient group, subjects must have met the following criteria: all participants must have been aged between 30 and 60 years and must have had a confirmed blood pressure of higher or equal of 140/90 mmHg (based on JNC VIII). All subjects who had history of taking antihypertensive, coronary artery disease, vascular disease, stroke, secondary hypertension, diabetes mellitus, renal diseases and women who were pregnant or were taking oral contraceptive were excluded from this study. All participants were interviewed and demographic data, life style, family and occupational history were collected using a standard questionnaire.
Sample Collection and Genotyping
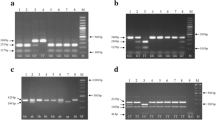
Peripheral blood samples were collected in standard EDTA tube and were stored at − 80 °C until further analysis. Genomic DNA was extracted from 5 ml blood using salting-out procedure as previously was described by Miller et al. (1988). The T786C, G894T, and A922G polymorphisms were genotyped using RFLP method and the 4a/4b polymorphism was genotyped by utilizing a conventional PCR procedure. To genotype the intended polymorphisms, target regions were amplified using specific primers sets. The 4a/4b polymorphism was simply genotyped by performing a simple PCR; since the 4a variant contained 4 copy and the 4b contained 5 copy, it was relatively simple to identify this variation by only a PCR reaction. As for T786C, G894T, and A922G SNPs, a further treatment with endonuclease was performed. Finally, all specimens were electrophoresed on agarose gel. All PCR reactions were containing 100 ng of genomic DNA, 12.5 μl of 2X PCR Master Mix (Amplicon, Denmark), 10 pmol of each primers, and double distilled water. Final volume of each reaction was 25 μL. Table 1 shows the list of primers and required restriction enzymes.
Statistical and Haplotype Analysis
All statistical analysis were performed using SPSS software version 16.0 (SPSS Inc., Chicago, IL, USA) and all continuous variables were analyzed as mean ± SD. Student’s t test and Chi-square test were used to compare quantitative data. Deviations of genotype frequencies from Hardy–Weinberg Equilibrium (HWE), differences in genotype distributions, and allele frequencies were analyzed among cases and controls using Chi-square test; odds ratios with 95% confidence intervals (CI) were calculated. P values less than 0.05 were considered statistically significant. Haplotype analysis and linkage disequilibrium between various genotypes of studied polymorphisms were analyzed using the SHEsis online software available on (https://analysis.bio-x.cn/myAnalysis.php). Linkage disequilibrium (LD) structure was measured by D′ and r2.
Results
Demographic Data and Clinical Characteristics
The demographic and clinical data of all subjects are listed in Table 2. Our results showed that BMI variable had no significant correlation with the disease (p > 0.05) (Table 2). Likewise, we found that the mean of Systolic Blood Pressure (SBP) and Diastolic Blood Pressure (DBP) in cases were significantly higher compared with controls (Table 2). Moreover, regarding smoking history, we observed a significant difference between two studied groups (p < 0.05) (Table 2). Eventually, our results revealed that the rate of physical activity in case subjects was significantly lower than that in healthy controls (p < 0.05) (Table 2).
Genotypic and Allelic Frequencies Analysis
Genotypic distribution of eNOS − 922A > G, − 786 T > C, 894 G > T and intron 4 b/a VNTR polymorphisms in the studied population are shown in Table 3. Our results revealed that eNOS − 786 T > C, 894 G > T and intron 4 b/a VNTR polymorphisms were significantly higher in EHT patients compared with healthy controls (p < 0.05) (Table 2). However, the rates of genotype frequencies for eNOS − 922A > G polymorphism was not statistically different between two groups (Table 3). The heterozygote, homozygote, and mutant allele frequencies for all studied eNOS gene polymorphisms are displayed in Table 3. Among the cases, 20 individuals featured the eNOS 4a4a genotype, whereas 5 individuals among the controls had such genotype. This difference is statistically significant (OR 4.3, 95%CI 1.5–11.8, P = 0.003) (Table 3). We did not observe a significant difference between homozygote genotypes of eNOS − 922A > G, − 786 T > C, 894 G > T polymorphisms and EHT occurrence (Table 3). Nevertheless, in regards to analysis of heterozygote genotypes frequencies of the all studied eNOS gene polymorphisms between two groups, we demonstrated that the frequency of heterozygote genotypes for eNOS − 922A > G and intron 4 b/a VNTR polymorphisms were higher in cases compared with controls although the difference was not significant (Table 3). Moreover, the frequency of heterozygote genotypes for eNOS − 786 T > C and 894 G > T polymorphisms were statistically higher in EHT patients compared with controls (Table 3). We also revealed that the frequencies of eNOS − 786 C, intron 4 a VNTR, and 894 T mutant alleles in cases were statistically higher than that in controls (Table 3). Our results showed that the eNOS − 922 G allelic frequency in patients with EHT was not significantly higher than that in healthy controls( OR 1.22, 95%CI 0.83–1.62, P = 0.3 (Table 3).
Linkage Disequilibrium and Haplotype Analysis
To evaluate the combined effect of eNOS gene polymorphisms and related haplotypes with EHT susceptibility, pairwise, ternary and foursome haplotypes of all investigated polymorphisms were analyzed. Our results revealed that the combined genotypes − 786C/− 922A, − 786C/− 922G, − 786C/4a, − 786C/894 T, − 922A/4a, and − 922G/4a were significantly higher in cases compared with controls (Table 4). However, the combined genotypes − 786 T/− 922A, − 786 T/− 922G, − 786 T/894 T and − 922A/4b were significantly higher in controls indicating their negative association with EHT (Table 4). Among twenty ternary haplotypes, five haplotypes showed a significant difference between case and control groups (p < 0.05) (Table 4). The − 786C/− 922A/4a, − 786C/− 922A/4b and − 786C/− 922G/4a haplotypes were more frequent in cases suggesting a positive association with EHT, while − 786 T/− 922G/4a and − 786 T/− 922G/894 T haplotypes had negative association with the disease since they were more frequent in controls (Table 4). Regarding foursome haplotypes, we observed a statistically significant difference between, − 786 T/− 922G/894 T/4b haplotype between studied populations (OR 0.2, 95%CI 0.02–1.3, P < 0.05) (Table 4). This haplotype was more frequent among controls suggesting its protective role in EHT development. Finally, we calculated pairwise LD estimates for eNOS − 922A > G, − 786 T > C, 894 G > T and intron 4 b/a VNTR loci; the D′ was used to measure LD between regions and alleles (Table 5). In this respect, we observed strong LD between loci − 786 T > C/− 922 A > G, − 786 T > C/4 b/a and − 922 A > G/4 b/a in patients with EHT (Table 5).
Discussion
In spite of some discrepancies upon the influence of eNOS polymorphisms on EHT, a relevant role of NO metabolism alteration in the pathogenesis of hypertension has been indicated by several studies (Zintzaras et al. 2006). Being one of the most important metabolic determinants in humans, eNOS mediates restraining blood pressure by approximately 300 mmHg through affecting soluble guanylate cyclase (GS) signaling cascade, leukocyte adhesion prevention, and NO generation (Gamboa et al. 2007). Through affecting these mechanisms, eNOS can successfully maintain basal vascular tone. Studies indicated that reduction of basal NO levels can in turn contribute to hypertension predisposition. The present study investigated eNOS polymorphisms and hypertension predisposition. Therefore, the present study aimed at investigating the association between some selected eNOS polymorphisms which are believed to reduce NO levels and hypertension predisposition in a specific population of Iran, for the first time.
It has been reported that T786C polymorphism (T > C substitution) in the promoter region can mediate significant reduction of eNOS transcription rate even up to 50% which eventually result in nitrite/nitrate serum level reduction both in hypoxia and normal conditions (Miyamoto et al. 2000; Nakayama et al. 1999). We found significantly different allele distribution between case and control groups where C allele was more frequent among patients supporting its association with the disease. The carrier status for this allele is highly associated with the disease. However, some previous studies failed to reveal such associations. Two independent studies conducted by Kajiyama et al. and Tsujita et al. investigated T786C SNP in Japanese population, where no association could be found between this SNP and EHT. Moreover, R. Li et al. and J. Li et al. studied T786C in African Americans and Han population of China, respectively, also detected no significant association between T786C SNP and hypertension (Li et al. 2011,2004; Kajiyama et al. 2000; Tsujita et al. 2001). There exist a hypothesis proposing that low expression of eNOS might seriously influence the metabolism contributing to severe health problems beyond blood pressure. This can explain the rarity of CC genotype in the studied population by (Li et al. 2011).
Regarding the investigation of G894T polymorphism, we revealed its significant association with the disease. T allele was significantly more frequent among patients suggesting its role in hypertension predisposition and also the carriers of this allele were associated with EHT. In parallel to our study, Shankarishan et al. reported a significant association between TT genotype and the risk of hypertension in Indian population (Shankarishan et al. 2014). The G > T substitution is positioned within a loop with no apparent interference with enzyme dimerization or active sites suggesting to impose no influence on eNOS catalysis (Hingorani 2001); however, this polymorphism on exon 7 causes Glu298 to change to 298Asp which alters the enzyme structure and affects its activity and ultimately leads to decreased production of NO and increased blood pressure (Yoshimura et al. 1998). Other studies also revealed significantly higher odds for T allele to be associated with hypertension and high levels of blood pressure (Cooke et al. 1997; Shoji et al. 2000; Miyamoto et al. 1998). However, Lacolley et al. in 1998 has reported the association of 894G allele with an elevated risk of hypertension in Caucasians (Lacolley et al. 1998). In some populations, it has also been reported that this polymorphism has no association with hypertension (Zintzaras et al. 2006; Kato et al. 1999; Kishimoto et al. 2004). In contrast, Li et al. found the association of this polymorphism with hypertension in female Hans of southwestern China (Li et al. 2011). Despite controversial reports, our results, in agreement with mentioned studies suggesting a significant association of 894 T allele with hypertension, indicate the predisposing influence of this SNP. Furthermore, to the best of our knowledge, there are few studies investigating the association of A922G with hypertension, and similar to them, according to our statistical analysis, we established no significant association between this polymorphism and hypertension. Although 4a/4b polymorphism is an intronic one, it has been reported that intron 4ab polymorphism can modulate eNOS mRNA transcription, translation efficiency, stability, and ultimately enzyme levels (Tsukada et al. 1998). In the present study, a statistically significant association between 4a/4b VNTR and hypertension has been demonstrated. 4a/4a genotype is highly associated with the disease and expectedly 4a allele is significantly frequent among patients. A meta-analysis conducted by Zintzaras et al. in 2006 including 35 genetic studies has also demonstrated a significant association between this polymorphism and hypertension (Zintzaras et al. 2006). Shankarishan et al. suggested that a genotype is associated with increased risk of hypertension comparing to ab and bb genotypes (Shankarishan et al. 2014). Colomba et al. studied eNOS polymorphisms and cardiovascular damage in hypertensive Italian subjects, and reported 4aa genotype to be present in 2.5% of hypertensive subjects while being completely absent in normotensives (Colomba et al. 2008). Detrimental effects of an allele have been indicated in a meta-analysis where 22% of the increased risk for hypertension was demonstrated in the presence of an allele (Zintzaras et al. 2006). Although interesting results were obtained by Kitsios and Zintzaras indicating a reverse association for this allele, taken into account the influences of potential confounders with a multivariate logistic regression model, the association was not significant. They also demonstrated no significant genotype-smoking interplays (Kitsios and Zintzaras 2010) considering previous studies suggesting smoking-dependent effects of 4a/b polymorphism (Wang et al. 1996; Rios et al. 2007).
Considering aforementioned points, significant associations between studied polymorphisms of eNOS gene and EHT are illuminated; however, relying only on individual polymorphisms may not be highly informative and the results are not usually consistent in multifactorial diseases such as hypertension. Instead, haplotypes can bring about more powerful and accurate tool in the analysis of genetic association studies. The key determinant of disease susceptibility can be the interaction between multiple genetic markers in combination with each other as haplotypes rather than individual polymorphisms (Kitsios and Zintzaras 2010; Cardon and Abecasis 2003). This approach is believed to be more powerful than single-marker analysis especially when the markers are in functional regions of DNA and can represent biological roles (Zintzaras and Kitsios 2006; Zintzaras and Lau 2008). Thus, in the present study, we provided pairwise, ternary, and foursome haplotypes to assess their associations with EHT and compare their power as risk assessing markers. Among studied pairwise haplotypes, six haplotypes were significantly associated with EHT, whereas four haplotypes were in reverse association with EHT indicating their protective effects. According to our statistical analysis, 786C/922A, 786C/922G, 786C/4a, 786C/894 T, 922A/4a, and 922G/4a were associated with high risk of EHT development, while 786 T/922A, 786 T/922G, 786 T/894 T, and 922A/4b were reversely associated. As one may consider, although haplotype analysis indicate that 786C/922A, 786C/922G, 922A/4a, and 922G/4a haplotypes are positively associated with EHT, it seems that major informative polymorphisms are 786C and 4a in these haplotypes since irrespective of both forms of A922G SNP; these haplotypes demonstrate positive associations, and hence, 786C/922A, 786C/922G, 922A/4a, and 922G/4a haplotypes cannot be considered powerful markers. Similarly, taking into account the reversely associated haplotypes, 786 T/922A and 786 T/922G cannot be considered useful haplotypes in risk assessment based on the previous description. Marked interethnic variations in polymorphisms and haplotypes of eNOS gene and also their associations with susceptibility to cardiovascular diseases have been described by Tanus Santus et al. in a study in Caucasians and African Americans as well as white and black Brazilians (Tanus-Santos et al. 2001; Marroni et al. 2005). Shankarishan et al. investigated eNOS gene polymorphisms and risk of hypertension in an Indian population considered a pairwise analysis of three eNOS gene polymorphisms and reported significant linkage disequilibrium between intron 4 and G894T polymorphisms. 894 T/4a, 894G/4a, 894 T/4b, 786 T/4a, 894 T/786 T, and 894 T/786C were reported haplotypes to be significantly associated with the risk of hypertension (Shankarishan et al. 2014).
Ternary haplotype analysis revealed that 786C/922A/4a, 786C/922A/4b, and 786C/922G/4b haplotypes are significantly associated with hypertension, while 786 T/922G/4a and 786 T/922G/894 T haplotypes demonstrate protective effects against hypertension. It has been reported by Sandrim et al. in that 786C/894G/4b haplotype indicates protective effect, while 786C/894 T/4b represents a positive association with susceptibility to hypertension (Sandrim et al. 2006a). The same group also reported a protective role for 786C/894G/4b and positive association for 786C/894 T/4b haplotypes when investigating the development of hypertension in patients with or without type 3 diabetes mellitus (Sandrim et al. 2006b). These studies are not in favor of the results of our study. Previous studies have reported different results regarding haplotypes association with hypertension. 786 T/894G/4a has been reported to have protective effects against hypertension (Kitsios and Zintzaras 2010), while others have reported their significant association with hypertension (Kumar et al. 2009). In the present study, we found neither protective role nor pathogenic role for this haplotype and EHT.
In regards with foursome haplotypes of investigated polymorphisms, in this study, only 786 T/922G/894 T/4b haplotype was statistically significant which conferred protective role against EHT.
Conclusion
In conclusion, we mentioned certain individual polymorphisms of eNOS gene to be associated with EHT where investigating them as haplotypes suggested them as more powerful markers. Although the current study does not include functional studies of investigated polymorphisms, the results for potential association of them with hypertension are supported based on with previous studies. Hypertension as a multifactorial disorder is under the influence of complex interplays between genetic predispositions and environmental factors; however, the exact mechanisms are not yet fully understood. The coexistence of genetic susceptibility and adverse environmental factors contribute to the initiation and development of hypertension. Therefore, future studies should embrace investigation of environmental factors in the background of genetic variations in larger populations to reveal risk factors more thoroughly.
References
Benjafield AV, Morris BJ (2000) Association analyses of endothelial nitric oxide synthase gene polymorphisms in essential hypertension. Am J Hypertens 13(9):994–998
Cardon LR, Abecasis GR (2003) Using haplotype blocks to map human complex trait loci. Trends Genet 19(3):135–140
Chandra S, Narang R, Sreenivas V, Bhatia J, Saluja D, Srivastava K (2014) Association of angiotensin II type 1 receptor (A1166C) gene polymorphism and its increased expression in essential hypertension: a case–control study. PLoS ONE ONE 9(7):e101502
Colomba D, Duro G, Corrao S, Argano C, Di Chiara T, Nuzzo D et al (2008) Endothelial nitric oxide synthase gene polymorphisms and cardiovascular damage in hypertensive subjects: an Italian case–control study. Immun Ageing 5(1):4
Cooke M, John P, Dzau M, Victor J (1997) Nitric oxide synthase: role in the genesis of vascular disease. Annu Rev Med 48(1):489–509
Deng F, Hu Q, Tang B, He F, Guo S, Chen J et al (2007) Endothelial nitric oxide synthase gene intron 4, 27 bp repeat polymorphism and essential hypertension in the Kazakh Chinese population. Acta Biochim Biophys Sin 39(5):311–316
Gamboa A, Shibao C, Diedrich A, Choi L, Pohar B, Jordan J et al (2007) Contribution of endothelial nitric oxide to blood pressure in humans. Hypertension 49(1):170–177
Hingorani AD (2001) Polymorphisms in endothelial nitric oxide synthase and atherogenesis. Atherosclerosis 154(3):521–527
Hyndman ME, Parsons HG, Verma S, Bridge PJ, Edworthy S, Jones C et al (2002) The T-786→C mutation in endothelial nitric oxide synthase is associated with hypertension. Hypertension 39(4):919–922
Jemaa R, Ali SB, Kallel A, Feki M, Elasmi M, Taieb SH et al (2009) Association of a 27-bp repeat polymorphism in intron 4 of endothelial constitutive nitric oxide synthase gene with hypertension in a Tunisian population. Clin Biochem 42(9):852–856
Jemaa R, Kallel A, Sediri Y, Omar S, Feki M, Elasmi M et al (2011) Association between-786TC polymorphism in the endothelial nitric oxide synthase gene and hypertension in the Tunisian population. Exp Mol Pathol 90(2):210–214
Jíra M, Závodná E, Honzíková N, Nováková Z, Vasku A, Hollá LI et al (2011) Association of eNOS gene polymorphisms T-786C and G894T with blood pressure variability in man. Physiol Res 60(1):193
Kajiyama N, Saito Y, Miyamoto Y, Yoshimura M, Nakayama M, Harada M et al (2000) Lack of association between T-786→ C mutation in the 5′-flanking region of the endothelial nitric oxide synthase gene and essential hypertension. Hypertens Res 23(6):561–565
Kato N, Sugiyama T, Morita H, Nabika T, Kurihara H, Yamori Y et al (1999) Lack of evidence for association between the endothelial nitric oxide synthase gene and hypertension. Hypertension 33(4):933–936
Kingah PL, Luu HN, Volcik KA, Morrison AC, Nettleton JA, Boerwinkle E (2010) Association of NOS3 Glu298Asp SNP with hypertension and possible effect modification of dietary fat intake in the ARIC study. Hypertens Res 33(2):165–169
Kishimoto T, Misawa Y, Kaetu A, Nagai M, Osaki Y, Okamoto M et al (2004) eNOS Glu298Asp polymorphism and hypertension in a cohort study in Japanese. Prev Med 39(5):927–931
Kitsios GD, Zintzaras E (2010) An NOS3 haplotype is protective against hypertension in a Caucasian population. Int J Hypertens 2010:865031
Kumar R, Nejatizadeh A, Arif E, Akhtar S, Gupta M, Tyagi S et al (2009) Multi-locus interactions of vascular homeostasis genes in essential hypertension: a gender-based study. Clin Chim Acta 405(1):87–93
Lacolley P, Gautier S, Poirier O, Pannier B, Cambien F, Benetos A (1998) Nitric oxide synthase gene polymorphisms, blood pressure and aortic stiffness in normotensive and hypertensive subjects. J Hypertens 16(1):31–35
Li R, Lyn D, Lapu-Bula R, Oduwole A, Igho-Pemu P, Lankford B et al (2004) Relation of endothelial nitric oxide synthase gene to plasma nitric oxide level, endothelial function, and blood pressure in African Americans. Am J Hypertens 17(7):560–567
Li A, Song B, Zheng H, Zhang X, He Y, Xu Y (2007) Association between the variable number of tandem repeat polymorphisms of endothelial nitric oxide synthase and ischemic cerebrovascular diseases in Henan Han ethnicity. Life Sci J 4:26–29
Li J, Cun Y, Tang W, Wang Y, Li S, Ouyang H et al (2011) Association of eNOS gene polymorphisms with essential hypertension in the Han population in southwestern China. Genet Mol Res 10(3):2202–2212
Marroni AS, Metzger IF, Souza-Costa DC, Nagassaki S, Sandrim VC, Correa RX et al (2005) Consistent interethnic differences in the distribution of clinically relevant endothelial nitric oxide synthase genetic polymorphisms. Nitric Oxide 12(3):177–182
Men C, Tang K, Lin G, Li J, Zhan Y (2011) ENOS-G894T polymorphism is a risk factor for essential hypertension in China. Indian J Biochem Biophys 48(3):154
Miller S, Dykes D, Polesky H (1988) A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 16(3):1215
Miyamoto Y, Saito Y, Kajiyama N, Yoshimura M, Shimasaki Y, Nakayama M et al (1998) Endothelial nitric oxide synthase gene is positively associated with essential hypertension. Hypertension 32(1):3–8
Miyamoto Y, Saito Y, Nakayama M, Shimasaki Y, Yoshimura T, Yoshimura M et al (2000) Replication protein A1 reduces transcription of the endothelial nitric oxide synthase gene containing a–786T→ C mutation associated with coronary spastic angina. Hum Mol Genet 9(18):2629–2637
Moe K, Lim S, Wong P, Chua T, DeSilva D, Koh T et al (2006) Association analysis of endothelial nitric oxide synthase gene polymorphism with primary hypertension in a Singapore population. J Hum Hypertens 20(12):956
Nakayama M, Yasue H, Yoshimura M, Shimasaki Y, Kugiyama K, Ogawa H et al (1999) T− 786→ C mutation in the 5′-flanking region of the endothelial nitric oxide synthase gene is associated with coronary spasm. Circulation 99(22):2864–2870
Nejatizadeh A, Kumar R, Stobdan T, Goyal A, Sikdar S, Gupta M et al (2008) Endothelial nitric oxide synthase gene haplotypes and circulating nitric oxide levels significantly associate with risk of essential hypertension. Free Radical Biol Med 44(11):1912–1918
Niu W, Qi Y (2011) An updated meta-analysis of endothelial nitric oxide synthase gene: three well-characterized polymorphisms with hypertension. PLoS ONE ONE 6(9):e24266
Rahimi Z, Aghaei A, Rahimi Z, Vaisi-Raygani A (2013) Endothelial nitric oxide synthase (eNOS) 4a/b and G894T polymorphisms and susceptibility to preeclampsia. J Reprod Infertil 14(4):184
Rios DL, D’Onofrio LO, Souza JK, Queiroz AM, Raduy-Maron L, Silva-Neto N et al (2007) Smoking-dependent and haplotype-specific effects of endothelial nitric oxide synthase gene polymorphisms on angiographically assessed coronary artery disease in Caucasian-and African–Brazilians. Atherosclerosis 193(1):135–141
Rodrigo R, Libuy M, Feliú F, Hasson D (2013) Oxidative stress-related biomarkers in essential hypertension and ischemia-reperfusion myocardial damage. Dis Markers 35(6):773–790
Salvi E, Kutalik Z, Glorioso N, Benaglio P, Frau F, Kuznetsova T et al (2012) Genome-wide association study using a high-density SNP-array and case-control design identifies a novel essential hypertension susceptibility locus in the promoter region of eNOS. Hypertension 59(2):248
Sandrim VC, Coelho EB, Nobre F, Arado GM, Lanchote VL, Tanus-Santos JE (2006a) Susceptible and protective eNOS haplotypes in hypertensive black and white subjects. Atherosclerosis 186(2):428–432
Sandrim VC, Yugar-Toledo JC, Desta Z, Flockhart DA, Moreno H Jr, Tanos-Santos JE (2006b) Endothelial nitric oxide synthase haplotypes are related to blood pressure elevation, but not to resistance to antihypertensive drug therapy. J Hypertens 24(12):2393–2397
Shah V, Cheema B, Kohli H, Sharma R, Khullar M, Bhansali A (2013) Endothelial nitric oxide synthase gene polymorphism and the risk of diabetic neuropathy in asian indian patients with type 2 diabetes. J Diabetes Metab 4(2):243
Shankarishan P, Borah PK, Ahmed G, Mahanta J (2014) Endothelial nitric oxide synthase gene polymorphisms and the risk of hypertension in an Indian population. BioMed Res Int 2014:793040
Shoji M, Tsutaya S, Saito R, Takamatu H, Yasujima M (2000) Positive association of endothelial nitric oxide synthase gene polymorphism with hypertension in northern Japan. Life Sci 66(26):2557–2562
Tanus-Santos JE, Desai M, Flockhart DA (2001) Effects of ethnicity on the distribution of clinically relevant endothelial nitric oxide variants. Pharmacogenet Genomics 11(8):719–725
Tsujita Y, Baba S, Yamauchi R, Mannami T, Kinoshita M, Yamamoto R et al (2001) Association analyses between genetic polymorphisms of endothelial nitric oxide synthase gene and hypertension in Japanese: the Suita Study. J Hypertens 19(11):1941–1948
Tsukada T, Yokoyama K, Arai T, Takemoto F, Hara S, Yamada A et al (1998) Evidence of association of the ecNOS gene polymorphism with plasma NO metabolite levels in humans. Biochem Biophys Res Commun 245(1):190–193
Wang XL, Sim AS, Badenhop RF, Mccredie RM, Wilcken DE (1996) A smoking–dependent risk of coronary artery disease associated with a polymorphism of the endothelial nitric oxide synthase gene. Nat Med 2(1):41–45
Yoshimura M, Yasue H, Nakayama M, Shimasaki Y, Sumida H, Sugiyama S et al (1998) A missense Glu298Asp variant in the endothelial nitric oxide synthase gene is associated with coronary spasm in the Japanese. Hum Genet 103(1):65–69
Zarmakhi L, Hashemi M (2012) The analysis of endothelial nitric oxide synthase gene polymorphism in intron 4 with hypertension disease. J Paramed Sci 3(1):2–6
Zintzaras E, Kitsios G, Stefanidis I (2007) Response to endothelial nitric oxide synthase polymorphisms and susceptibility to hypertension: genotype versus haplotype analysis. Hypertension 49(1):E2
Zintzaras E, Lau J (2008) Synthesis of genetic association studies for pertinent gene–disease associations requires appropriate methodological and statistical approaches. J Clin Epidemiol 61(7):634–645
Zintzaras E, Kitsios G, Stefanidis I (2006) Endothelial NO synthase gene polymorphisms and hypertension. Hypertension 48(4):700–710
Acknowledgments
Authors would like to express their sincerest appreciation to all subjects for participating in this study.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Farbood, Z., Sabeti Aghabozorgi, A., Nejatizadeh, A. et al. Endothelial Nitric Oxide Synthase Gene Polymorphisms (− 922A > G, − 786 T > C, Intron 4 b/a VNTR and 894 G > T) and Essential Hypertension: An Association Study with Haplotypes Analysis. Biochem Genet 58, 518–532 (2020). https://doi.org/10.1007/s10528-020-09953-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10528-020-09953-2