Abstract
A genetic component is accepted in the etiology of the glioma. Evidence from candidate genes studies and GWAS reveal that CCDC26 gene could increase the risk of glioma. We performed a systematic review and up-to-date meta-analysis to explore if polymorphisms of CCDC26 gene (rs891835, rs6470745, and rs55705857) may be a susceptibility factor in developing glioma. An online search in PubMed, Web of Science, and SCOPUS up to September 2018 was performed. The pooled odds ratios were evaluated by fixed effects model and random effects model. Analyses of the overall sample and ethnic sub-groups were performed. In all the analyses, the allelic, additive, dominant, and recessive models were used. We found an association between all polymorphisms evaluated and an increased risk for glioma in the overall population in all the models studied. In sub-group analysis, we found that rs891835 and rs6470745 increased the risk of glioma in Europeans and Caucasians. On the other hand, the rs891835 polymorphism did not reveal any statistical association in Chinese population. Taken into consideration the limitations of this study, the present findings suggest a possible participation of rs891835, rs6470745, and rs55705857 as risk factors to develop glioma. Furthermore, it is possible that the involvement of CCDC26 variants depends on ethnicity. However, we recommend to perform further studies to have conclusive outcomes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Brain tumors are relatively rare; however, they represent a serious health problem due to the very poor prognosis, high morbidity, and mortality (Adel Fahmideh et al. 2015; Cui 2015; Vaubel et al. 2017; Walsh et al. 2013a). Furthermore, the brain tumors are a leading cause of pediatric death in children, while in adults, they are among the first causes of death (Di Stefano et al. 2013; Egan et al. 2011, 2012; Walsh et al. 2013b). Gliomas are the most common type of brain tumors, they are derived from glial cells that surround and support neurons; understanding their etiology is very important to identify strategies for prevention, surveillance, and potential targets for treatments (Enciso-Mora et al. 2013; Ghasimi et al. 2016; Jenkins et al. 2011).
Consequently, the genetic predisposition of glioma risk has been widely explored. In this sense, the chromosomal region 8q24.21 has been associated with the risk of several common cancer sites (Jenkins et al. 2012; Lachance et al. 2011; Lasho et al. 2012). Likewise, these loci contain the gene that codifies the coiled-coil domain-containing protein 26 (CCDC26) in which genome wide association studies support a linkage with tumor, including low-grade glioma (Li et al. 2012, 2013; Liu et al. 2010a). Evidence suggest that CCDC26 increases apoptosis induced by death stimuli in neuroblastoma cells and in glioblastoma cells with down-regulation of telomerase activity (Liu et al. 2010b, c; Lu et al. 2015).
Due to the importance of CCDC26, the association of this gene with glioma risk has been explored (Melin 2011; Oktay et al. 2016; Rajaraman et al. 2012), providing some candidate genetic variants of CCDC26. Some of the main ones are rs891835 (ancestral allele T>allele G), rs6470745 (ancestral allele A>allele G), and rs55705857 (ancestral allele A>G allele) (Egan et al. 2011; Enciso-Mora et al. 2013; Li et al. 2012; Oktay et al. 2016). The genetic variant rs55705857 has revealed an OR of approximately 6 to IDH-mutated tumor and the histopathological subtype oligodendroglioma (Enciso-Mora et al. 2013; Oktay et al. 2016). However, the etiology of glioma has not been very well understood for many years; probably because glioma is a rare cancer and in a considerable number of the studies there a serious limitations such as small sample sizes (a very low proportion have more than 500 cases or controls) also, the heterogeneity of gliomas could cause that the studies do not have the statistical power to establish precise interactions (Schoemaker et al. 2010; Shete et al. 2009; Simon et al. 2010). Therefore, in order to address the limitations of the single association studies, we considered it necessary to perform a systematic review and up-to-date meta-analysis. Our aim is to explore the role of the genetic variants of CCDC26 (rs891835, rs6470745 and rs55705857) as a probable markers of glioma risk in the pooled combination that offers the meta-analytic technique.
Methods
Data Sources and Search
The present systematic review and meta-analysis were performed based on the reported guidelines of the Preferred Reporting Items for Systematic Review and Meta-analyses (PRISMA). Potentially related studies were searches in electronic databases of PubMed, Scopus, and Web of Sciences by two researchers (González-Castro and Tovilla-Zárate). Articles published before September 2018 were searched using the following keywords combination: “rs891835” AND “glioma” OR “glioblastoma” OR “brain tumor,” “rs6470745” AND “glioma” OR “glioblastoma” OR “brain tumor,” and “rs55705857” AND “glioma” OR “glioblastoma” OR “brain tumor.” All eligible studies were retrieved and their reference lists also were reviewed to find other relevant studies.
Inclusion/Exclusion Criteria
The selected studies had to meet the following criteria: (1) studies that address the association between rs891835, rs6470745 or rs55705857, and glioma; (2) cases: patients diagnosed with glioma, controls: cancer-free individuals (Hospital-based or Health-based); (3) reporting numbers or frequencies of alleles or genotypes in both cases and controls; (4) provide adequate data to calculate odds ratios (ORs) with 95% confidence intervals (CIs); (5) case–control design. The exclusion criteria were family-based, case reports, case series, reviews, comments, letters, and conference presentations. No restriction on ethnicity, or geographic region was imposed.
Data Extraction
The following data were collected from the included studies: (i) name of the first author, (ii) publication year, (iii) country and ethnicity of individuals studied, (iv) diagnostic, (v) number of cases and controls, (vi) allelic or genotypic distribution, (vii) ORs or p values found, and (vii) methodological data of genotyping. The same two researchers extracted the information. When necessary, the reviewers wrote to the corresponding author for extra information.
Quality Assessment
The Newcastle–Ottawa scale (NOS) was used to measure the quality of the eligible studies included (http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp). The NOS evaluated three aspects of the studies: (a) selection, (b) comparability, and (c) exposure between cases and controls. The NOS has score range of zero to nine stars, only studies with a score of 6 or more stars were included (Gonzalez-Castro et al. 2016; Hernandez-Diaz et al. 2016). Likewise the literature search and data extraction, the researchers González-Castro and Tovilla-Zarate evaluated the quality of studies independently; when there was a disagreement, it was solved in a discussion with a third reviewer (Genis-Mendoza) until consensus was reached.
Data Synthesis and Statistical Analysis
The strength of the association between rs891835, rs6470745, rs55705857, and glioma risk was estimated by ORs and 95% CIs. Moreover, the significance of the pooled ORs was determined by the Z test, a p value less than 0.05 was considered as statistically significant. Furthermore, genetic models to evaluate for the pooled OR were used: allele contrast ( m vs M), homozygote comparison (mm vs MM), heterozygote comparison (mM vs MM), dominant model (mm+mM vs MM), as well as recessive model (mm vs mM+MM). In addition, we examined the heterogeneity of the included studies assessed by χ2-based Q statistical test and I2 metric value. Heterogeneity presence based on I2 was considered as 0–25% absent, 25–50% low, 50–75% moderate, and 75–100% high; p value of Q test < 0.10 was considered significant to heterogeneity. In addition, Galbraith plot was used to visualize the impact of individual studies on the overall heterogeneity, which spotted the outlier as the possible origin of heterogeneity. Subsequently, fixed effects model was used in the absence of heterogeneity; otherwise, random effects model was selected. Likewise, the Hardy–Weinberg equilibrium (HWE) was assessed in the control group in each article included, p < 0.05 was considered as a significant disequilibrium.
On the other hand, sensitivity analysis was performed by excluding a single article each time, to detect potential influence that could have each study in the pooled ORs. Publication bias was measured by Egger linear regression test and funnel plots. All the analyses were performed using the Comprehensive Meta-analysis (CMA Version 2.0, Biostat, Inc., USA) software. Finally, sub-group analyses were conducted: overall population (rs891835, rs6470745, rs55705857), Caucasian population (rs891835, rs6470745), European population (rs891835, rs6470745), USA (rs891835), and Chinese population (rs891835).
Results
Study Characteristics
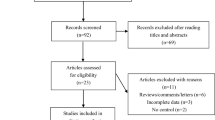
The electronic database search generated 40 records from PubMed, 37 records from Scopus, and 38 records from Web of Science. After scanning titles and abstracts and discarding duplicates, 35 studies relevant to our aim were fully scrutinized. After full text evaluation of these 35 relevant studies, we excluded abstract and comments among others (6 studies), also if they did not accomplish our particular point of interest (7 studies), we also eliminated review articles or meta-analysis (9 studies). If the records did not detail data or those for which the authors could not be contacted were also eliminated (4 studies), Fig. 1. Finally, 10 studies met the inclusion criteria for this meta-analysis. The characteristics of the included studies are presented in Table 1. These studies were performed in 9 countries (France, Germany, Sweden, UK, USA, China, Norway, Denmark, and Switzerland) published from 2009 to 2018. The number of the cases sample sizes ranged from 72 to 2564; giving a total of 11,575 cases and 17,718 controls for rs891835; 9484 cases and 15,229 controls for rs6470745; and for rs55705857 10,543 cases and 19,331 controls. Some samples were stratified depending on the population origin or place of birth. For example, the group of Shete S. 2009 performed their analysis in individuals born in France, Germany, Sweden, UK, and USA. Another example is the group of Ostrom Q.T. 2018 that performed their investigations with individuals at the Glioma International Case–Control Study (GICC), San Francisco Adult Glioma study GWAS (SFAGS-GWAS), MD Anderson Glioma GWAS (MDA-GWAS); Gliomascan: National Cancer Institue’s Gliomascan (Gliomascan). The articles by Shete S. and Ostrom Q.T. were consider for the stratified population (Ostrom et al. 2018).
Glioma Risk and CCDC26 Variants in Overall Population
Firstly, we explored the association of rs8918358, rs6470745, and rs55705857 in the overall population. Initially, the analysis of rs8918358 was statistically significant when the heterogeneity was excluded: allelic (OR 1.19, 95% CI 1.11–1.26, p value < 0.000), homozygous (OR 1.56 95% CI 1.33–1.84, p value < 0.001), heterozygous (OR 1.26, 95% CI 1.17–1.36, p value < 0.000), recessive (OR 1.46, 95% CI 1.24–1.71, p value < 0.001), and dominant (OR 1.32, 95% CI 1.23–1.42, p value < 0.001) models Fig. 2. Secondly, the analysis of rs6470745 indicated an association in all the models evaluated, even in the existence of heterogeneity between the studies (allelic: OR 1.19 95% CI 1.11–1.26, p value < 0.000; homozygous OR 1.56 95% CI 1.33–1.84, p value < 0.001; heterozygous OR 1.26 95% CI 1.17–1.36, p value < 0.001; recessive OR 1.46 95% CI 1.24–1.71, p value < 0.000; and dominant OR 1.32, 95% CI 1.23–1.42, p value < 0.000 models); Fig. 3. Thirdly, when evaluating the rs55705857, we observed an association after discarding the heterogeneity in the studies: allelic (OR 1.43, 95% CI 1.35–1.52, p value < 0.000), homozygous (OR 2.04 95% CI 1.73–2.39, p value < 0.000), heterozygous (OR 1.64, 95% CI 1.46–1.84, p value < 0.001), recessive (OR 1.80, 95% CI 1.53–2.11, p value < 0.001), and dominant (OR 1.49, 95% CI 1.40–1.60, p value < 0.001) models. No publication bias was observed, Table 2.
Meta-regression Analysis
Lastly, in order to explore the source of the heterogeneity, we performed a meta-regression based on the age of the population in the three polymorphism: first the polymorphism rs8918358 (Slope: OR 0.009, 95% CI − 0.01 to 0.02, p value 0.34; intercept -0.51), then the variant rs6470745 (Slope: OR 0.0006, 95% CI − 0.01 to 0.02, p value 0.94; intercept 0.24), and finally rs55705857 (Slope: OR -0.019, 95% CI − 0.04 to 0.008, p value 0.177; intercept 0.49); Fig. 4.
Sub-group Analysis
Glioma Risk and CCDC26 Variants in Caucasian Individuals
Due to previous results, we explored the role that rs8918358 and rs6470745 could have as markers of glioma risk in Caucasian populations. In a first step, we analyzed the participation of rs891835 by the genetic models assigned; only when studies that favored heterogeneity were excluded, we obtained statistical significance (allelic: OR 1.22 95% CI 1.14–1.32, p value < 0.001; homozygous OR 1.57 95% CI 1.32–1.86, p value < 0.001; heterozygous OR 1.28 95% CI 1.18–1.39, p value < 0.001; recessive OR 1.46 95% CI 1.24–1.73, p value < 0.001; and dominant OR 1.34, 95% CI 1.24–1.45, p value < 0.001 models). We followed the same procedure to explore the role of rs6470745 in Caucasian populations and even when there were studies that favored heterogeneity in the analysis, we observed statistical significance (allelic: OR 1.37 95% CI 1.30–1.43, p value < 0.001; homozygous OR 1.87 95% CI 1.62–2.15, p value < 0.000; heterozygous OR 1.21 95% CI 1.11–1.31, p value < 0.001; recessive OR 1.65 95% CI 1.45–1.89, p value < 0.001; and dominant OR 1.29, 95% CI 1.19–1.41, p value < 0.000 models); Fig. 3. We did not observe any publication bias, Table 2.
Glioma Risk and CCDC26 Variants in European Individuals
Due to the fact that genetic influence differs among ethnicities, we explored the participation of CCDC26 variants in European populations. Subsequently, the analysis of rs891835 polymorphism revealed a significance p value in all the models when heterogeneity was discarded: allelic (OR 1.22, 95% CI 1.11–1.34, p value < 0.000), homozygous (OR 1.53 95% CI 1.23–1.90, p value < 0.000), heterozygous (OR 1.23, 95% CI 0.11–1.38, p value < 0.000), recessive (OR 1.46, 95% CI 1.24–1.73, p value < 0.000), and dominant (OR 1.32, 95% CI 1.19–1.46, p value < 0.000) models. Likewise, the variant rs6470745 was evaluated and in the presence and absence of heterogeneity, we found an association with glioma risk (allelic: OR 1.38 95% CI 1.31–1.45, p value < 0.000; homozygous OR 2.02 95% CI 1.76–2.33, p value < 0.000; heterozygous OR 1.35 95% CI 1.26–1.45, p value < 0.000; recessive OR 1.70 95% CI 1.47–1.98, p value < 0.000; and dominant OR 1.42, 95% CI 1.33–1.51, p value < 0.000 models); Fig. 2. Any publication bias were observed in the analysis, Table 2.
Glioma Risk and CCDC26 Variants in Individuals Born in USA
In order to better understand the association of rs891835 and glioma, we performed another sub-group analysis exploring the role of this polymorphism with glioma risk in individuals born in the USA. The analysis revealed no association with glioma risk in the presence of heterogeneity (allelic: OR 1.09 95% CI 0.81–1.47, p value 0.54; homozygous OR 1.21 95% CI 0.55–2.62, p value 0.62; heterozygous OR 1.18 95% CI 0.88–1.58, p value 0.26; recessive OR 1.18 95% CI 0.66–2.09, p value 0.56; and dominant OR 1.12, 95% CI 0.76–1.66, p value 0.54 models). However, when we excluded the study that favored heterogeneity, we observed a significant association: allelic (OR 1.23, 95% CI 1.10–1.38, p value < 0.000), homozygous (OR 1.62 95% CI 1.24–2.13 p value < 0.000), heterozygous (OR 1.34, 95% CI 1.18–1.52, p value < 0.000), recessive (OR 1.46, 95% CI 1.11–1.91, p value < 0.000), and dominant (OR 1.38, 95% CI 1.22–1.56, p value < 0.000) models. Concerning publication bias, no significant p value was observed, Table 2.
Glioma Risk and CCDC26 Variants in Subjects Born in China
Similarly, we evaluated the involvement of rs891835 as a glioma risk in individuals born in China. In this case, the models that presented heterogeneity were the heterozygous (OR 1.29, 95% CI 0.91–1.84, p value 0.14) and dominant (OR 1.32, 95% CI 0.95–1.83, p value 0.09). However, even we discarded the study that favored the presence of heterogeneity (Wei et al. 2014), no association was observed: allelic (OR 1.17, 95% CI 0.99–1.38, p value 0.06), homozygous (OR 1.51 95% CI 0.86 -2.65 p value 0.14), heterozygous (OR 1.11, 95% CI 0.92–1.35, p value 0.25), recessive (OR 1.44, 95% CI 0.82–2.52, p value 0.19), and dominant (OR 1.14, 95% CI 0.95–1.37, p value 0.15) models, Fig. 1. There was no publication bias in the analyses, Table 2.
Meta-regression Based on Ages
Lastly, in order to explore the source of the heterogeneity, we performed meta-regression based on age of the populations. Firstly, the polymorphism rs8918358 (Slope: OR 0.009, 95% CI − 0.01 to 0.02, p value 0.34; intercept -0.51), then the variant rs6470745 (Slope: OR 0.0006, 95% CI − 0.01 to 0.02, p value 0.94; intercept 0.24), and finally rs55705857 (Slope: OR -0.019, 95% CI − 0.04 to 0.008, p value 0.177; intercept 0.49); Fig. 4.
Discussion
Glioma is one of the least understood and most aggressive tumors affecting humans (Wang et al. 2011, 2018). The risk factors are largely unknown but some studies have suggested a heredity factor that increases the risk of developing theses tumors (Wang et al. 2016; Wei et al. 2014). In this sense, some investigations have focused on the role of genetic polymorphisms in glioma risk; however, it remains unclear (Wibom et al. 2015; Wu et al. 2016). Therefore, our aim was to perform a systematic review and meta-analysis in order to address the possible involvement between CCDC26 variants (rs891835, rs6470745, and rs55705857) and the predisposition to glioma.
As a first step, we conducted an analysis in the overall population following the genetics models allelic, homozygous, heterozygous, recessive, and dominant. After discarding the studies that favoring the heterogeneity, we found a statistical significance in the analysis of rs891835 and rs55705857; only the rs6470745 showed a significant p value in the presence and absence of heterogeneity. Our results suggest that genetic variants of CCDC26 gene increase the risk for glioma. One explanation could be that CCDC26 has a direct or indirect participation in controlling cellular checkpoints of DNA damage recovering that may be interfering in the prompting cell cycle or maintenance of genomic stability; hence, we recommend to perform a systematic scrutiny of CCDC26 gene mutations or rearrangements in primary tumors (Liu et al. 2010b; Schoemaker et al. 2010; Simon et al. 2010; Zeng et al. 2017).
To date, GWAS research on the susceptibility of glioma has been performed (Melin et al. 2017; Richardson et al. 2017). After the identification through fine-mapping, rs55705857 represents the likely causal variant at the 8q24.21 glioma risk locus; this association was confined to IDH-mutated gliomas, in particular those with 1p/19q codeletion (Enciso-Mora et al. 2013; Jenkins et al. 2012) This positive evidence and the results of our meta-analysis suggest that CCDC26 polymorphisms may represent a predictive biomarkers for glioma. Derived from these, it could be suggested that CCDC26 variant could be a retinoic acid-dependent modulator of myeloid cell differentiation and death (Kinnersley et al. 2018). Future studies with large sample size that confirm this outcome are necessary; the detection of CCDC26 variants as biomarkers in patients with primary tumors may help to select, in future clinical trials, those patients who could benefit of target therapies, although these polymorphisms (rs891835, rs55705857, rs6470745) did not have significant eQTLs in tissues.
On the other hand, we understand that the genetic background of patients could have a considerable contribution in the genotype frequency distribution. For example, data of HapMap-CEU in a European population revealed a frequency of 0.761 for AA, 0.212 for AG, and 0.027 for GG, while data from HapMap-CHD in Asian population revealed that the frequencies go to 0.365, 0.488, and 0.147 for AA, AG, and GG, respectively; Table 3 for checking minor allele frequency. Subsequently, we consider important address the association of rs891835, rs6470745, and rs55705857 variants as glioma risk factors depend of origin of the patients. The available data allowed us to perform the following sub-group analyses: Caucasian populations, individuals born in Europe, individuals born in USA, and finally, individuals born in China. The sub-group analysis in Caucasians and Europeans revealed that rs89183 and rs64707455 showed a statistical significance for glioma risk; in the case of rs64707455, the association remained regardless of the presence of the heterogeneity. In the case of USA individuals, we only conducted an evaluation of the rs89183 variant, we excluded heterogeneity and found that the polymorphism may play a role as risk factor, but we need to take into consideration that this sub-group included a small number of studies; hence, we could not evaluate publication bias and for these reasons, we need to interpret the findings with caution.
Finally, we conducted an analysis in Chinese individuals and could not observe any relation with glioma. A possible explanation of the differences observed between Caucasians, Asians, or USA sub-groups and what we observed in Chinese people, could be the strong involvement of genetic background specific of ethnics. For example, it has been reported that incidence and survival rates of gliomas vary between different ethnicities (Barnholtz-Sloan et al. 2007; Shabihkhani et al. 2017; Wei et al. 2014). This variation has been investigated with emphasis on White and Black populations. Some studies have shown that glioma rates are highest in non-Latino Whites followed by Hispanics/Latinos and Blacks; while Asian/Pacific Islanders tend to have the lowest incidence (Barnholtz-Sloan et al. 2007; Shabihkhani et al. 2017). Derived from this, we could assume that there is a different participation of CCDC26 variants in Chinese individuals and due to their particular genetic background, the polymorphism did not have a participation in glioma development. We have to consider that may be other factors in Chinese individuals such as epigenetic regulation or cell metabolism could play irreplaceable roles in tumorigenesis (Pop et al. 2018).
Our analysis presents a statistical association of CCDC26 variants in the majority of the sub-groups; nevertheless, it essential to take into consideration some limitations. Firstly, the sample size, although the pathogenesis of glioma is poorly understood, the number of studies that have evaluated the association between glioma with CCDC26 is limited; hence, it is possible that in some sub-analysis, the power of the sample might not be enough to detect small effect of these genetic variants. Nevertheless, to our knowledge, this is the first meta-analysis that englobes three variants of CCDC26 and their role in glioma risk. Furthermore, we performed several evaluations of CCDC26 variants using several models; due to lack of data, we did not evaluate the role of these polymorphisms on different types of glioma, age, or gender among others factors that could participate in the manifestation of glioma (Simon et al. 2010; Wang et al. 2011). Therefore, we recommend that in future studies, outcomes are adjusted by the raw OR for age, gender, smoking, radiation exposure among other factors associated with tumors. There was no detailed information about gene–gene or gene–environment interactions and we were unable to perform more precise analysis. We also tried to address the problems that could cause heterogeneity between studies, but it was not completed, which may reduce the quality of the analysis.
In conclusion, based on the outcomes obtained in the present analysis, we could assume that rs891835, rs6470745, or rs55705857 variants of CCDC26 gene are possible risk biomarkers to glioma. However, we consider that rs891835 polymorphism may not participate in the pathogenesis of glioma in Chinese individuals. Nevertheless, we need to take into consideration the limitations of this study to make definite conclusions. Further studies with larger samples and different populations are necessary to fully understand the development of glioma.
References
Adel Fahmideh M et al (2015) CCDC26, CDKN2BAS, RTEL1 and TERT polymorphisms in pediatric brain tumor susceptibility. Carcinogenesis 36:876–882. https://doi.org/10.1093/carcin/bgv074
Barnholtz-Sloan JS, Maldonado JL, Williams VL, Curry WT, Rodkey EA, Barker FG 2nd, Sloan AE (2007) Racial/ethnic differences in survival among elderly patients with a primary glioblastoma. J Neurooncol 85:171–180. https://doi.org/10.1007/s11060-007-9405-4
Chen H et al (2011) Association of sequence variants on chromosomes 20, 11, and 5 (20q13.33, 11q23.3, and 5p15.33) with glioma susceptibility in a Chinese population. Am J Epidemiol 173:915–922. https://doi.org/10.1093/aje/kwq457
Cui T (2015) CCDC26 rs4295627 polymorphism and glioma risk: a meta-analysis. Int J Clin Exp Med 8:3862–3868
Di Stefano AL et al (2013) Association between glioma susceptibility loci and tumour pathology defines specific molecular etiologies. Neuro Oncol 15:542–547. https://doi.org/10.1093/neuonc/nos284
Egan KM et al (2011) Cancer susceptibility variants and the risk of adult glioma in a US case–control study. J Neurooncol 104:535–542. https://doi.org/10.1007/s11060-010-0506-0
Egan KM, Wrensch MR, Jenkins RB (2012) Rare and uncommon genetic variants may hold key to the ‘missing heritability’ in glioma CNS. Oncol 1:109–112. https://doi.org/10.2217/cns.12.19
Enciso-Mora V et al (2013) Deciphering the 8q24.21 association for glioma. Hum Mol Genet 22:2293–2302. https://doi.org/10.1093/hmg/ddt063
Ghasimi S, Wibom C, Dahlin AM, Brannstrom T, Golovleva I, Andersson U, Melin B (2016) Genetic risk variants in the CDKN2A/B, RTEL1 and EGFR genes are associated with somatic biomarkers in glioma. J Neurooncol 127:483–492. https://doi.org/10.1007/s11060-016-2066-4
Gonzalez-Castro TB, Hernandez-Diaz Y, Juarez-Rojop IE, Lopez-Narvaez ML, Tovilla-Zarate CA, Genis-Mendoza A, Alpuin-Reyes M (2016) The role of C957T, TaqI and Ser311Cys polymorphisms of the DRD2 gene in schizophrenia: systematic review and meta-analysis. Behav Brain Funct (BBF) 12:29. https://doi.org/10.1186/s12993-016-0114-z
Hernandez-Diaz Y et al (2016) Effects of paraoxonase 1 gene polymorphisms on heart diseases: systematic review and meta-analysis of 64 case–control studies. Medicine (Baltimore) 95:e5298. https://doi.org/10.1097/md.0000000000005298
Jenkins RB et al (2011) Distinct germ line polymorphisms underlie glioma morphologic heterogeneity. Cancer Genet 204:13–18. https://doi.org/10.1016/j.cancergencyto.2010.10.002
Jenkins RB et al (2012) A low-frequency variant at 8q24.21 is strongly associated with risk of oligodendroglial tumors and astrocytomas with IDH1 or IDH2 mutation. Nat Genet 44:1122–1125. https://doi.org/10.1038/ng.2388
Kinnersley B, Houlston RS, Bondy ML (2018) Genome-wide association studies in glioma. Cancer Epidemiol Biomarkers Prev 27:418–428. https://doi.org/10.1158/1055-9965.epi-17-1080
Lachance DH et al (2011) Associations of high-grade glioma with glioma risk alleles and histories of allergy and smoking. Am J Epidemiol 174:574–581. https://doi.org/10.1093/aje/kwr124
Lasho TL et al (2012) Differential distribution of CCDC26 glioma-risk alleles in myeloid malignancies with mutant IDH1 compared with their IDH2R140-mutated or IDH-unmutated counterparts. Leukemia 26:1406–1407. https://doi.org/10.1038/leu.2011.336
Li S et al (2012) Polymorphisms of TREH, IL4R and CCDC26 genes associated with risk of glioma. Cancer Epidemiol 36:283–287. https://doi.org/10.1016/j.canep.2011.12.011
Li M, Zhou Q, Tu C, Jiang Y (2013) A meta-analysis of an association between the XRCC1 polymorphisms and gliomas risk. J Neurooncol 111:221–228. https://doi.org/10.1007/s11060-012-1022-1
Liu Y et al (2010a) Polymorphisms of LIG4, BTBD2, HMGA2, and RTEL1 genes involved in the double-strand break repair pathway predict glioblastoma survival. J Clin Oncol 28:2467–2474. https://doi.org/10.1200/jco.2009.26.6213
Liu Y, Shete S, Hosking F, Robertson L, Houlston R, Bondy M (2010b) Genetic advances in glioma: susceptibility genes and networks. Curr Opin Genet Dev 20:239–244. https://doi.org/10.1016/j.gde.2010.02.001
Liu Y, Shete S, Hosking FJ, Robertson LB, Bondy ML, Houlston RS (2010c) New insights into susceptibility to glioma. Arch Neurol 67:275–278. https://doi.org/10.1001/archneurol.2010.4
Lu HW, Huang M, Wang JH, Sun XL, Ke YQ (2015) CCDC26 rs4295627 polymorphism (8q24.21) and glioma risk: a meta-analysis. Genet Mol Res 14:12074–12084. https://doi.org/10.4238/2015.october.5.20
Melin B (2011) Genetic causes of glioma: new leads in the labyrinth. Curr Opin Oncol 23:643–647. https://doi.org/10.1097/CCO.0b013e32834a6f61
Melin BS et al (2017) Genome-wide association study of glioma subtypes identifies specific differences in genetic susceptibility to glioblastoma and non-glioblastoma tumors. Nat Genet 49:789–794. https://doi.org/10.1038/ng.3823
Oktay Y et al (2016) IDH-mutant glioma specific association of rs55705857 located at 8q24.21 involves MYC deregulation. Sci Rep 6:27569. https://doi.org/10.1038/srep27569
Ostrom QT et al (2018) Sex-specific glioma genome-wide association study identifies new risk locus at 3p21.31 in females, and finds sex-differences in risk at 8q24.21. Sci Rep 8:7352. https://doi.org/10.1038/s41598-018-24580-z
Pop S, Enciu AM, Necula LG, Tanase C (2018) Long non-coding RNAs in brain tumours: focus on recent epigenetic findings in glioma. J Cell Mol Med. https://doi.org/10.1111/jcmm.13781
Rajaraman P et al (2012) Genome-wide association study of glioma and meta-analysis. Hum Genet 131:1877–1888. https://doi.org/10.1007/s00439-012-1212-0
Richardson TE et al (2017) Rapid progression to glioblastoma in a subset of IDH-mutated astrocytomas: a genome-wide analysis. J Neurooncol 133:183–192. https://doi.org/10.1007/s11060-017-2431-y
Schoemaker MJ et al (2010) Interaction between 5 genetic variants and allergy in glioma risk. Am J Epidemiol 171:1165–1173. https://doi.org/10.1093/aje/kwq075
Shabihkhani M et al (2017) Incidence, survival, pathology, and genetics of adult Latino Americans with glioblastoma. J Neurooncol 132:351–358. https://doi.org/10.1007/s11060-017-2377-0
Shete S et al (2009) Genome-wide association study identifies five susceptibility loci for glioma. Nat Genet 41:899–904. https://doi.org/10.1038/ng.407
Simon M et al (2010) Genetic risk profiles identify different molecular etiologies for glioma. Clin Cancer Res 16:5252–5259. https://doi.org/10.1158/1078-0432.ccr-10-1502
Vaubel RA et al (2017) Synchronous gemistocytic astrocytoma IDH-mutant and oligodendroglioma IDH-mutant and 1p/19q-codeleted in a patient with CCDC26 polymorphism. Acta Neuropathol 134:317–319. https://doi.org/10.1007/s00401-017-1727-5
Walsh KM et al (2013a) Analysis of 60 reported glioma risk SNPs replicates published GWAS findings but fails to replicate associations from published candidate-gene studies. Genet Epidemiol 37:222–228. https://doi.org/10.1002/gepi.21707
Walsh KM et al (2013b) Genetic variants in telomerase-related genes are associated with an older age at diagnosis in glioma patients: evidence for distinct pathways of gliomagenesis. Neuro Oncol 15:1041–1047. https://doi.org/10.1093/neuonc/not051
Wang SS et al (2011) Joint associations between genetic variants and reproductive factors in glioma risk among women. Am J Epidemiol 174:901–908. https://doi.org/10.1093/aje/kwr184
Wang X, Luo T, Ruan M, Liu P, Wang S, Zhu W (2016) Association of the CCDC26 rs4295627 polymorphism with the risk of glioma: evidence from 7,290 cases and 11,630 controls. Mol Clin Oncol 4:878–882. https://doi.org/10.3892/mco.2016.813
Wang S, Hui Y, Li X, Jia Q (2018) Silencing of lncRNA CCDC26 restrains the growth and migration of glioma cells in vitro and in vivo via targeting miR-203. Oncol Res 26:1143–1154. https://doi.org/10.3727/096504017x14965095236521
Wei XB et al (2014) CCDC26 gene polymorphism and glioblastoma risk in the Han Chinese population. Asian Pac J Cancer Prev 15:3629–3633
Wibom C et al (2015) Investigation of established genetic risk variants for glioma in prediagnostic samples from a population-based nested case–control study. Cancer Epidemiol Biomarkers Prev 24:810–816. https://doi.org/10.1158/1055-9965.epi-14-1106
Wu Q, Peng Y, Zhao X (2016) An updated and comprehensive meta-analysis of association between seven hot loci polymorphisms from eight GWAS and glioma risk. Mol Neurobiol 53:4397–4405. https://doi.org/10.1007/s12035-015-9346-4
Zeng J, Luo Y, Yu M, Li J, Liu Z (2017) CCDC26 rs4295627 polymorphisms associated with an increased risk of glioma: a meta-analysis. Adv Clin Exp Med 26:1275–1281
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Contributions
T.B.G.C., C.A.T.Z., and A.D.G.M. performed substantial contributions to conception and design; J.J.M.M., T.B.G.C., and J.M.R.P. participated in acquisition of data, or analysis and interpretation of data; I.E.J.R., M.L.L.N., and N.P.H. drafted the article or revised it critically for important intellectual content; and all the authors gave their final approval of the version to be published.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
González-Castro, T.B., Juárez-Rojop, I.E., López-Narváez, M.L. et al. Genetic Polymorphisms of CCDC26 rs891835, rs6470745, and rs55705857 in Glioma Risk: A Systematic Review and Meta-analysis. Biochem Genet 57, 583–605 (2019). https://doi.org/10.1007/s10528-019-09911-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10528-019-09911-7