Abstract
Cytochrome P450 1A1 (CYP1A1) and Glutathione S-transferase P1 (GSTP1) genes are involved in the metabolism of many carcinogens. Polymorphisms in these genes with altered enzyme activity have been reported. The present study evaluated the synergistic effect between CYP1A1 and GSTP1 gene polymorphisms and smoking on development of HCV-related liver disease and hepatocellular carcinoma (HCC). The patients group comprised 40 patients with HCC and 40 patients with liver cirrhosis. The control group comprised 40 healthy subjects having no history of malignancy. The genetic polymorphisms were studied using polymerase chain reaction restriction fragment length polymorphism (PCR RFLP) technique on blood samples. The number of current or former smoker among HCC and cirrhotic patients as well as the median Pack/year of cigarette smoked were significantly higher in HCC and liver cirrhotic patients than in control group. Subjects with CYP1A1 gene variants (m1 and m3) had no significant risk to develop cirrhosis or HCC compared to control group. Individuals carrying the Ile/Val genotype of GSTP1 had a significant increased risk of HCC (OR of 2.2, 95 % CI 1.143–4.261) and had larger tumor size. No significant risk was observed on combining both genes variants or on combining smoking with variants of both genes. In conclusion, the GSTP1 Ile/Val genotype and Val allele are associated with an increased risk of HCC. CYP1A1 and GSTP1 genes variants interaction did not increase the risk of HCC.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Hepatocellular carcinoma (HCC) is the sixth most prevalent malignant tumor worldwide and ranks the third as a cancer killer, causing about half million deaths annually (Jemal et al. 2011). In Egypt, an overall increase in the relative frequency of all liver-related cancers (>95 % as HCC), from approximately 4 % in 1993 to 7.3 % in 2003 has been reported (El-Zayadi et al. 2005).
Human hepatocarcinogenesis is a multistage process with multiple risk factors. Chronic hepatitis B and C play a major role in HCC etiology. Other risk factors including alcohol consumption, male gender, obesity, aflatoxin, or other chemicals may be involved (El-Serag and Rudolph 2007; Gross-Steinmeyer and Eaton 2012).
Tobacco smoke contains many carcinogens. Most of which require metabolic activation mainly through aryl hydrocarbon hydroxylase activity encoded by phase I drug-metabolizing enzymes, such as cytochrome P450s (CYPs). Cytochrome P-450 1A1 (CYP1A1) (cytochrome P450, family1, subfamily A, polypeptide1) is the most active among the CYPs enzyme in the metabolism of pro-carcinogenic tobacco components (Nerurkar et al. 2000) into hydrophilic, reactive, electrophilic intermediate forms capable of DNA binding that may initiate the carcinogenic process (National Research Council 1983).
Four genotypes of CYP1A1, resulting from various point mutations in the CYP1A1 gene, have been characterized and designated m1, m2, m3, and m4. Both m1 (rs4646903) variant, a T → C substitution at nucleotide 3801 and m3 (rs4986883) variant, nucleotide 3205 T → C, create an MspI restriction site in the 3′-non-coding region (Cascorbi et al. 1996).
Tobacco metabolic intermediates are further detoxified by phase II enzymes such as glutathione S-transferase (GST) (Smith et al. 1995) that protect normal cells against damage induced by hepatitis virus or aflatoxin. The family is composed of eight isoforms (Katoh et al. 2008). GSTP1 plays a central role in the inactivation of toxins and carcinogens (Hengstler et al. 1998). A single nucleotide polymorphism in the GSTP1 gene causes substitution of isoleucine to valine at amino acid codon 105 (Ile105Val) is reported (Allan et al. 2001).
Polymorphisms in genes encoding CYP1A1 or GSTP1 enzymes, possibly by altering their expression and function, may increase or decrease carcinogen activation or detoxification and modulate DNA repair, and are implicated in various cancers (Yu et al. 1999; Kweekel et al. 2008), However, results from other studies have revealed inconsistent findings and remain conflicting rather than conclusive (Silvestri et al. 2003; Li et al. 2009; Huang et al. 2013).
These inconsistent findings justify the need for additional studies to further evaluate the role of CYP1A1 and GSTP1 polymorphism—and their possible interaction as risk factors for HCC development among Egyptians and its relation to smoking. Hence, this case–control study was planned.
Materials and Methods
Subjects and Specimen Collection
This study was conducted on 40 patients with HCC and 40 patients with liver cirrhosis. Patients were recruited from the Department of Tropical medicine, Mansoura University Hospital between March 2013 and August 2014. The diagnosis of HCC was confirmed by α-fetoprotein elevation (>400 ng/ml) combined with one positive imaging study [Magnetic resonance imaging (MRI) or computerized tomography (CT)] or two positive imaging studies with normal α-fetoprotein level or a pathological examination when needed (Ryder 2003).
Diagnosis of chronic hepatitis and cirrhosis was based on biochemical markers, liver serology, imaging studies, and histopathological scoring assessment. Forty control subjects were included in the study. The selection criteria for control subjects included the absence of current or previous history of any kind of malignancy and age matching to HCC cases. Patients and control subjects gave their written informed consent to participate in the study. The Ethical Research Committee of Mansoura University approved the study protocol.
For all cases and controls, full clinical history and thorough clinical examination were carried out. Clinical history included history of smoking, alcohol consumption, and liver disease symptoms.
Subjects who smoked more than one cigarette per day for more than 1 year were classified as smokers. Others were defined as non-smokers. The average number of cigarettes smoked per day and the total number of years of smoking were used to calculate cumulative smoking dose as ‘pack-years’ (PY) [(cigarettes per day/20) × years smoked]. Subjects were also categorized as light (<30 PY) or heavy (≥30 PY) smokers, based on the mean tobacco consumption in the control group.
Venous blood samples were collected on EDTA tube (4 ml) and plain tube (4 ml) from all participants. Sera were separated from the plain tubes and distributed into aliquots that were used for the detection of serum HBV markers, antibodies to hepatitis C virus [DiaSorin ELISA (Italy) commercial kits], liver biochemical profile, and alpha-fetoprotein. Serum albumin, bilirubin, ALT, and AST were tested using automated chemistry analyzer Cobas C111 (Roche-Germany). Alpha-fetoprotein estimation was done by electrochemiluminescence technique by automated analyzer Cobas e (Roche, Germany). EDTA blood samples were used for complete blood count and immediately stored at −80 °C for the extraction of genomic DNA and detection of genetic polymorphism in the CYP1A1 and GSTP1 genes using PCR RFLP. For each patient in liver cirrhosis or HCC group, calculation of Child-Pugh score was carried out to assess the prognosis of chronic liver disease. The score employs five clinical measures (serum total bilirubin, serum albumin, prothrombin time, the presence of ascites and hepatic encephalopathy). Each measure is scored 1–3, with 3 indicating most severe derangement (Cholongitas et al. 2005). For all HCC patients, BCLC score was assessed (Forner et al. 2014).
Genomic DNA Extraction
Genomic DNA was extracted from whole blood using Gene JET whole-blood genomic DNA purification Mini Kit (#k0781). The kit utilizes silica-based membrane technology in the form of a convenient spin column.
Polymerase Chain Reaction–Restriction Fragment Length Polymorphism (PCR–RFLP)
The first mutation analyzed in the CYP1A1 gene was 6235 T → C transition 1194 bp downstream of axon 7 (m1 mutation). The other mutation analyzed was 5639 T → C transition downstream of axon 7 (m3 mutation). For GSTP1 gene, the mutation analyzed was 313A → G transition (rs1695, GSTP1 Ile I05 Val).
Of CYP1A1 and GSTP1 genes, an 899-bp and 440-bp DNA fragment was separately amplified using 1 unit Taq polymerase, 10 μmol/l of primers M3F 5′-GGCTGAG CAATCTGACCCTA, and P80, 5′-TAGGAGTCTTGTCTCATGCCT, for m1 and m3 CYP1A1 or of primers 5′-ACG CAC ATC CTC TTC CCC TC 3′ and 5′-TCA TTC TTG GCT GGT TGA TGT CC 3′ for GSTP1, 0.2 mmol/l deoxynucleotide triphosphate, and 2.4 mmol/l MgCl2 in a total volume of 30 μl. All primers were obtained from Thermo Fisher Scientific Inc. PCR reactions were carried out with a Perkin-Elmer Applied Biosystems 9600 thermal cycler. PCR conditions for CYP1A1 were initial denaturation for 3 min at 95 °C followed by 35 cycles of 0.5 min at 94 °C, 1 min at 63 °C, and 1 min at 72 °C. Final extension was at 72 °C for 10 min, while that for GSTP1 were initial denaturation for 3 min at 95 °C followed by 30 cycles of 30 s at 95 °C, 30 s at 57 °C, and 30 s at 72 °C. Final extension was at 72 °C for 7 min.
The PCR product was digested with 50 units MspI or BsmA1 (Thermo Scientific Fast digest restriction enzyme), for CYP1A1 and GSTP1 genes, respectively, generating smaller fragments in case of the mutation. Thermo Scientific Msp1 enzyme restriction site is
while Thermo Scientific BsmA1 enzyme restriction site is
The following reaction components were combined at room temperature: nuclease-free water (17 μl), 10× fast digest green buffer (2 μl), amplified PCR product (10 μl), and fast digest enzyme (1 μl) in a total volume of 30 μl. The components were mixed and incubated at 37 °C in a heat block for 15 min. The PCR product and the fragments of restriction enzyme digestion were evaluated on an ethidium bromide-stained 2 % agarose gel.
The sizes of PCR products were estimated according to the migration pattern of a 100 and 50 bp DNA ladder for CYP1A1 and GSTP1 genes, respectively (Thermo Scientific Gene ruler plus DNA ladder).
Genotypes for CYP1A1 gene were determined (Fig. 1) as homozygous for the wild-type allele (w/w; 899 bp), heterozygous (w/m1; 899, 693, 206 bp or w/m3; 802, 97, 899), or homozygous for mutant allele (m1/m1 693, 206 bp or m3/m3; 802, 97).
Agarose gel electrophoresis stained with ethidium bromide. Showing bands of CYP1A1 gene digested PCR product with Msp1 restriction enzyme using 100 bp ladder marker, showing wild type with fragment length 899 bp; ml heterozygous (wild/m1 mutant) with fragments 899, 693, and 206 bp; m3 heterozygous (wild/m3 mutant) with fragments 802, 97, 899; and m1 homozygous (m1/m1) with fragments 693, 206 bp
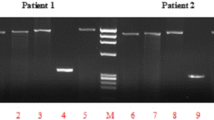
The A to G polymorphism of GSTP1 introduces a restriction site recognized by the BsmA1 restriction enzyme results either in retention of the 440 bp product or complete digestion to 212 and 228 bp fragments corresponding to individuals homozygous for the Ile or Val alleles, respectively. The presence of all three fragments corresponded to individuals heterozygous (Ile/Val), at codon 105 (Fig. 2).
Agarose gel electrophoresis stained with ethidium bromide. Showing bands of GSTP1 gene digested PCR product with BsmA1 restriction enzyme using 50 bp ladder marker (Lane 12), showing wild type (Ile/Ile) with fragment length 440 bp (Lanes 1, 3 and 5); heterozygous (Ile/Val) with fragments 440, 212, and 228 bp (Lanes 2, 6, 7, 8, 9, and 11); and homozygous (Val/Val) with fragments 212 and 228 bp (Lanes 4 and 10). NB: Fragments 212 and 228 are overlapped in this figure
Statistical Analysis
The statistical analysis of data was done using statistical package for social science program (SPSS, Inc, Chicago, IL) version 20. The normality of data distribution was tested by Kolmogorov–Smirnov test. Qualitative data were presented as frequency and percentage. Chi-square and Fisher exact tests were used to compare groups. Quantitative data were presented as minimum, maximum, median or mean and standard deviation. For comparison between groups, student t test and Mann–Whitney and Kruskal–Wallis tests were used as appropriate. Deviations from Hardy–Weinberg equilibrium expectations (HWE) were determined using logistic regression analysis. Bonferroni’s correction was used for multiple corrections. Odds ratio and 95 % confidence interval were calculated. Gene–gene interaction analysis was conducted, using as a reference group the homozygous wild-type individuals; while for gene–environment interaction analyses, the reference group was wild-type homozygous non-smoker. Multifactorial dimensional reduction method by MDR software3.0.2 was used to construct gene–gene interaction or gene–environment interaction. Kaplan–Meier test was used for survival analysis and the statistical significance of differences among curves was determined by Log-Rank test. P is significant if ≤0.05 at confidence interval 95 %.
Results
Characteristics of the Study Population
The demographic, laboratory, clinical, and radiologic characteristics of patients and control groups are shown in Table 1. Except for AFP level, there was a non-significant difference in distribution of smoking index and also in the median TLC, hemoglobin, platelets, albumin, bilirubin, ALT, AST, and INR between the HCC and liver cirrhosis patients. On the other hand, these parameters except for TLC, INR, and ALT were significantly different between each of HCC and liver cirrhosis patients as compared to control group.
Among HCC patients, 27 cases (67.5 %) had tumor size ≥3 cm and 15 (37.5 %) had multiple tumor masses. The number and percentages of HCC patients with BCLC stages A, B, C, and D were 18(45 %), 10(25 %), 6(15 %), and 6(15 %), respectively. The median follow-up time was 12 months (ranging from 2 to 18 months). By the last follow-up, 88.6 % of patients were dead, Cumulative Overall Survival (proportion surviving at 12 months) was 11.4 %, and the mean survival time for the entire cohort of patients was 6.664 (5.198–8.131) months (data not shown in the table).
Table 2 reports the distribution of the polymorphisms in CYP1A1 and GSTP1 among HCC, cirrhotic patients, and controls. This sample of individuals was selected randomly, but they were unrelated. Applying HWE revealed that GSTP1 and CYP1A1 genotypes in LC, HCC cases and control subjects were independent (i.e., they are in HWE). There is no evidence to reject the assumption of HWE in the sample (P > 0.05 for each). The frequencies of Ile/Val genotype and Val allele are more common among HCC (37.5 and 23.8 %) than cirrhotic patients (15 and 10 %) corresponding to an OR of 2.2 and 2.8 (95 % CI 1.14–4.26 and 1.15–6.85) after correction for age and gender and to an OR of 2.18 and 1.65 (95 % CI 1.12–4.24 and 1.05–2.61) after bonferroni correction. So, Ile/Val genotype was risky for development of HCC within LC patients even after adjustment for age and sex.
On the other hand, the frequency of the variant genotypes m1 and m3 of CYP1A1 was not significantly different between studied groups. The selected polymorphisms of CYP1A1 cannot be considered as a risk for HCC development. Since the allelic frequencies for m1 and m3 were relatively of low numbers, we combined m1 and m3 together. The percentage of patients carrying CYP1A1 wild-type gene among HCC and cirrhotic patients were 77.5 % (31/40) and 87.5 % (35/40), respectively. All control subjects and liver cirrhotic patients carry the wild CYP1A1 genotype apart from 5 (12.5 %) of cirrhotic patients who carried m3 heterozygote’s (Table 2). m1 heterozygous and m3 heterozygous genotypes were detected among 2(5 %) and 6(15 %) of patients with HCC. Only one patient (2.5 %) with m1/m1 variant (m1 homozygote) was detected among HCC patients (data not shown in the table).
The baseline demographic, laboratory, clinical, and radiologic data versus CYP1A1 and GSTP1 genotypes were studied in HCC patients. There were no differences between patients carrying wild and variant genotypes of CYP1A1 and GSTP1 except for age, as HCC patients with variant CYP1A1 were significantly older than those with wild type (P, 0.026) with mean ages of 60.7 and 54.1, respectively, also Ile/Val and Val/Val genotypes were significantly associated with larger tumor size (P, 0.046) as 100 and 86.7 % of HCC patients with Ile/Val and Val/Val genotypes had tumor size larger than 3 cm. On the other hand, gender, Child-Pugh, BCLC scores, and overall survival were insignificantly different between variants and wild genotypes carrying HCC patients (data not shown in the tables).
Gene–smoking interaction analysis was conducted, using the wild-type homozygous, non-smokers as a reference group; while for gene–gene interaction analysis, the homozygous wild-type individuals for both genes were the reference group. Statistical analysis combining different genetic distributions for estimating the interaction effect between the GSTP1 and CYP1A1 gene polymorphisms on the susceptibility to HCC (Table 4), and also with smoking (Tables 3 and 5) was performed. MDR analyses were also performed and shown in Fig. 3 in the form of bars representing hypothetical distributions of multifactor combination of CYP1A1, GSTP1, and smoking in control subjects, LC and HCC patients in comparison with each other. High-risk genotype, smoking interaction with ratio greater than one was observed when CYP1A1 mutants genotypes combined with smoking were compared between cirrhotic and control subjects or between cirrhotic and HCC subjects. For GSTP1 and smoking combination, high risk was reported when Val/Val genotype with smoking was compared between cirrhotic and control subjects or between cirrhotic and HCC subjects, also the ratio is greater than one when Il/Val genotype with smoking was compared between cirrhotic and HCC patients. No cases were observed to carry the combined CYP1A1 and GSTP1 mutation among control or cirrhotic subjects, while the combination between Il/Val or Val/Val genotypes with CYP1A1 mutants was risky as shown by the increase in the ratio between HCC and cirrhotic subjects. The only available data to test CYP1A1, GSTP1 smoking interaction was between CYP1A1 mutants, Il/Val genotype and smoking which are risky between HCC and cirrhotic patients. In spite of this increase in patients to control ratio, there is no statistical significant increase in risk on combining both variants (Table 4) and also on combining both variants with smoking (Table 5). Smoking in the presence of Ile/Ile genotype was considered a risk factor for LC development (P, 0.004) with OR of 1.37 and 95 % CI of 1.05–12.09. The same findings are shown in the presence of smoking with wild CYP1A1 (P, 0.022) with OR of 3.56 and 95 % CI of 1.34–9.46 which is attributed to smoking (Table 3).
MDR analysis for gene–gene and gene–environmental interaction. Bars represent hypothetical distributions of (1) control (left) and LC (right); (2) LC (left) and HCC (right) with each multifactor combination. Dark shaded cells represent genotype combinations with increased odds ratio, while light shaded cells represent genotype combinations with decreased odds ratio. No shading or white cells represent genotype combinations for which no data were observed. Smoking is divided into 0 (non-smoker) and 1 (smoker); CYP1A1 is divided into 1 (wild) and 2 (variant); GSTP1 is divided into 1 (Il/Il), 2 (Il/Val) and 3 (Val/Val)
Discussion
Most environmental carcinogens require prior metabolic activation in order to elicit their effects. Many of the enzymes involved in carcinogen metabolism exhibit genetic polymorphisms resulting in variability in enzyme activity (Salaspuro and Salaspuro 2004).
This study analyzed the influence of smoking as well as CYP1A1, GSTP1 polymorphic variants, and their interaction on the development of HCV-related HCC. The number of current or former smokers among HCC and cirrhotic patients as well as the median Pack/year of cigarette smoked was significantly higher in HCC and liver cirrhotic patients than in control group (Table 1). The most important finding was that individuals carrying the Ile/Val genotype had a significant increased risk of HCC (Table 2). This finding is consistent with previous reports demonstrating GSTP1 polymorphisms as a contributing factor to the increased risk and poor outcome of many cancers, including esophageal cancer (Li et al. 2010), breast cancer (Yang et al. 2005), and HCC in Taiwanese patients younger than 57 years (Chen et al. 2010). It was also observed that altered expression of GSTP1 existed in liver cancer cell lines (Ding et al. 2004), and in more than 77.8 % of HBV-associated HCC tissues (Zhong et al. 2002). Li et al. (2012) indicated carriage of GSTP1 Val/Val genotypes to have roles in susceptibility to HCC with OR (95 % CI) of 1.93 (1.32–2.95). Moreover, Li et al. (2013) found that GSTP1 protein and mRNA expression in HCC patients were significantly decreased when compared to chronic hepatitis B (CHB) patients. GST family acts as a part of the protection system against a wide range of potentially harmful cytotoxic compounds. Therefore, our results may support the idea that GSTP1 variants are associated with decreased GSTP1 activity that takes part in the development of HCC following HCV infection which is accompanied by massive ROS production.
The present study reported no increased risk of development of cirrhosis (P = 0.110) with CYP1A1 gene variants (m1 and m3) and did not show significant association (P = 0.478) with risk of HCC development among cirrhotic patients (Table 2).
In accordance with this, Boccia et al. (2015) reported that CYP1A1 MspI polymorphism did not significantly influence susceptibility to HCC. However, Silvestri et al. (2003) reported that the frequencies of the CYP1A1 highly inducible alleles, MspI m2 and Val, were increased in HCV-related liver disease compared with healthy subjects, in particular with asymptomatic carriers. No association with HCC was found, and no relationship with tobacco smoke was seen. Similar observation was obtained in previous studies on hepatitis B carriers (Yu et al. 1999, 2015. Several studies have suggested the CYP1A1 polymorphisms to be associated with elevated risks of prostate cancer (Ding et al. 2013), esophageal cancer (Gong et al. 2014), and head and neck cancer (Liu et al. 2013). However, no significant associations between the CYP1A1 polymorphisms and risks for gastric cancer (And and Guo 2012) and colorectal cancer were found (Zheng et al. 2012).
The contradictory findings among different studies may be explained by the difference in ethnicity or by the possibility that polymorphisms of CYP1A1 might exert different effects in different types of tissues, an idea that needs to be confirmed. Our results revealed that the mean age of patients with CYP1A1 variants (60.7 years) was higher than those with wild CYP1A1 type (54.1 years) and so these variants may evoke their effect among elderly individuals.
In the current study, we estimated the effect of these CYP1A1 and GSTP1 gene polymorphisms on the clinical status, such as clinical stage, tumor size, Child-Pugh grade, overall survival (OS), and the serum levels of liver-related biochemical markers, such as alpha-fetoprotein, AST, and ALT in HCC patients. Except for tumor size with GSTP1, a lack of association between the gene polymorphisms and those estimated factors was found. This study shows that HCC patients with GSTP1 variants had larger tumor size than patients with wild type (P = 0.046). Similar results were reported previously (Chen et al. 2010).
Analyses of gene–gene interactions in relation to susceptibility to tobacco carcinogens were carried out. In this study, there is an increased ratio of number of smokers with CYP1A1 mutation or Val/Val genotype of GSTP1 in cirrhotic than control subjects and in HCC than cirrhotic patients. The ratio of smokers with Ile/Val genotype of GSTP1 was also higher in HCC patients than cirrhotic patients. Moreover, the combination between Il/Val or Val/Val genotypes of GSTP1 with CYP1A1 mutants or between CYP1A1 mutants, Il/Val genotype and smoking was risky as shown by the increased ratio between cirrhotic and HCC subjects (Fig. 3). In spite of this increase in patients to control ratio, there are no statistically significant synergistic gene–gene, gene smoking or gene–gene–smoking interactions in association to HCC development (Tables 4 and 5). Similar results were obtained with lung cancer by previous authors (López-Cima et al. 2012). However, Boccia et al. (2015) observed a borderline increased risk for HCC among carriers of combined CYP1A1 and SULT1A1 variant alleles as compared to the double wild-type homozygous. To the best of our knowledge, this is the first study to analyze the role of interaction of CYP1A1 and GSTP1 gene polymorphism as a risk for HCC among Egyptian patients chronically infected with HCV.
The cumulative overall survival (OS) in HCC patients included in this study was analyzed with the possible role of polymorphic CYP1A1 and GSTP1 in determining survival outcome (Fig. 4). No significant differences were found in OS between HCC patients carrying wild and variant CYP1A1 as well as GSTP1 genotypes (P = 0.32 and 0.38, respectively). Contrary to this finding, Li et al. (2012) found that GSTP1 IIe/Val or GSTP1 Val/Val genotypes had better survival outcomes. Also Qu et al. (2015) found that patients with GSTP1, WV+VV genotypes of SNP rs4147581 had a longer median survival time as compared to those with WW genotype. It is to be noted that the age, sex, smoking behavior, liver cirrhosis, Child-Pugh score, AFP, and tumor size differ among these studies.
Conclusion
This study shows that the risk of developing HCC in Egyptian population can be partly explained by genetic polymorphisms in GSTP1 gene. On the other hand, no significant increase in risk of HCC development was noted with CYP1A1 m1 and m3 gene variants. Both genes variants have no association with survival. No statistically significant gene–gene or gene–smoking interaction was reported in association to HCC risk with only increase in ratio of combined CYP1A1 gene mutants, GSTP1 Il/Val and smoking which need extension of the study on large number of patients.
References
Allan JM, Wild CP, Rollinson S, Willett EV, Moorman AV, Dovey GJ, Roddam PL, Roman E, Cartwright RA, Morgan GJ (2001) Polymorphism in Glutathione S-transferase P1 is associated with susceptibility to chemotherapy—induced leukemia. Proc Natl Acad Sci USA 98:11592–11597
And GR, Guo X (2012) Quantitative assessment of the associations between CYP1A1 polymorphisms and gastric cancer risk. Tumor Biol 33:1125–1132
Boccia S, Miele L, Panic N, Turati F, Arzani D, Cefalo C, Amore R, Bulajic M, Pompili M, Rapaccini G, Gasbarrini A, La Vecchia C, Grieco A (2015) The effect of CYP, GST, and SULT polymorphisms and their interaction with smoking on the risk of hepatocellular carcinoma. Hindawi Publishing Corporation. Bio Med Research International, Article ID 179867, 7 pages
Cascorbi I, Brockmoller J, Roots I (1996) A C4887A polymorphism in exon 7 of human CYP1A1: population frequency, mutation linkages, and impact on lung cancer susceptibility. Cancer Res 56:4965–4969
Chen YL, Tseng HS, Kuo WH, Yang SF, Chen DR, Tsai HT (2010) Glutathione S-transferase P1 (GSTP1) gene polymorphism increases age-related susceptibility to hepatocellular carcinoma. BMC Med Genet 11:46
Cholongitas E, Papatheodoridis GV, Vangeli M, Terreni N, Patch D, Burroughs AK (2005) Systematic review: the model for end-stage liver disease–should it replace Child-Pugh’s classification for assessing prognosis in cirrhosis? Aliment Pharmacol Ther 22(11–12):1079–1089
Ding S, Gong BD, Yu J, Gu J, Zhang HY, Shang ZB, Fei Q, Wang P, Zhu JD (2004) Methylation profile of the promoter CpG islands of 14 “drug-resistance” genes in hepatocellular carcinoma. World J Gastroenterol 10:3433–3440
Ding G, Xu W, Liu H, Zhang M, Huang Q, Liao Z (2013) CYP1A1 MspI polymorphism is associated with prostate cancer susceptibility: evidence from a meta-analysis. Mol Biol Rep 40:3483–3491
El-Serag HB, Rudolph KL (2007) Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology 132:2557–2576
El-Zayadi AR, Badran HM, Barakat EM, Mel-D A, Shawky S, Mohamed MK, Selim O, Saeid A (2005) Hepatocellular carcinoma in Egypt: a single center study over a decade. World J Gastroenterol 11:5193–5198
Forner A, Gilabert M, Bruix J, Raoul JL (2014) Treatment of intermediate-stage hepatocellular carcinoma. Nat Rev Clin Oncol 11(9):525–535
Gong FF, Lu SS, Hu CY, Qian ZZ, Feng F, Wu YL, Yang HY, Sun YH (2014) Cytochrome P450 1A1 (CYP1A1) polymorphism and susceptibility to esophageal cancer: an updated meta-analysis of 27 studies. Tumor Biol 35:10351–10361
Gross-Steinmeyer K, Eaton DL (2012) Dietary modulation of the biotransformation and genotoxicity of aflatoxin B (1). Toxicology 299:69–79
Hengstler JG, Arand M, Herrero ME, Oesch F (1998) Polymorphisms of N-acetyltransferases, glutathione S-transferases, microsomal epoxide hydrolase and sulfotransferases: influence on cancer susceptibility. Eur Pub Med Central 154:47–85
Huang S, Wu F, Luo M, Ma L, Gao K, Li J, Wu W, Huang S, Yang Q, Liu K, Zhao Y, Li L (2013) The glutathione S-transferase P1 341C > T polymorphism and cancer risk: a meta-analysis of 28 case-control studies. PLOS One 8(2):e56722
Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D (2011) Global cancer statistics. CA Cancer J Clin 61:69–90
Katoh T, Yamano Y, Tsuji M, Watanabe M (2008) Genetic polymorphisms of human cytosol glutathione S-transferases and prostate cancer. Pharmacogenomics 9:93–104
Kweekel DM, Koopman M, Antonini NF, Van der Straaten T, Nortier JW, Gelderblom H, Punt CJ, Guchelaar HJ (2008) GSTP1 Ile105Val poly-morphism correlates with progression-free survival in MCRC patients treated with or without irinotecan: a study of the Dutch colorectal cancer group. Br J Cancer 99:1316–1321
Li R, Shugart YY, Zhou W, An Y, Yang Y, Zhou Y, Zhang B, Lu D, Wang H, Qian J, Jin L (2009) Common genetic variations of the cytochrome P450 1A1 gene and risk of hepatocellular carcinoma in a Chinese population. Eur J Cancer 45:1239–1247
Li D, Dandara C, Parker MI (2010) Ther341C/T polymorphism in the GSTP1 gene is associated with increased risk of oesophageal cancer. BMC Genet 11:47
Li CG, Zhao ZM, Hu MG, Liu R (2012) Predictive role of glutathione-S-transferase gene polymorphisms in risk and prognosis of hepatocellular carcinoma. Asian Pacific J Cancer Prev 13:3247–3252
Li T, Zhao X, Wang L, Gao S, Zhao J, Fan Y, Wang K (2013) Glutathione S-transferase P1 correlated with oxida-tive stress in hepatocellular carcinoma. Int J Med Sci 10(6):683–690
Liu L, Wu G, Xue F, Li Y, Shi J, Han J, Zhang X, Na Y, Zhang H, Tang X, Pu H, Yuan Q, Zhang L, Yang M (2013) Functional CYP1A1 genetic variants, alone and in combination with smoking, contribute to development of head and neck cancers. Eur J Cancer 49:2143–2151
López-Cima MF, Álvarez-Avellón SM, Pascual T, Somoano AF, Tardón A (2012) Genetic polymorphisms in CYP1A1, GSTM1, GSTP1 and GSTT1 metabolic genes and risk of lung cancer in Asturias. BMC Cancer 12:433
National Research Council (1983) Polycyclic aromatic hydrocarbons: evaluation of sources and effects. National Academy Press, Washington, pp 197–262
Nerurkar PV, Okinaka L, Aoki C, Seifried A, Lum-Jones A, Wilkens LR, Le Marchand L (2000) CYP1A1, GSTM1, and GSTP1 genetic polymorphisms and urinary 1-hydroxypyrene excretion in non-occupationally exposed individuals. Cancer Epidemiol Biomark Prev 9:1119–1122
Qu K, Liu S, Wang Z, Huang Z, Liu S, Chang H, Xu X, Lin T, Dong Y, Liu C (2015) Polymorphisms of glutathione S-transferase genes and survival of resected hepatocellular carcinoma patients. World J Gastroenterol 21(14):4310–4322
Ryder SD (2003) Guidelines for the diagnosis and treatment of hepatocellular carcinoma (HCC) in adults. Gut 52(suppl 3):1111–1118
Salaspuro V, Salaspuro M (2004) Synergistic effect of alcohol drinking and smoking on in vivo acetaldehyde concentration in saliva. Int J Cancer 111:480–483
Silvestri L, Sonzogni L, De Silvestri A, Gritti C, Foti L, Zavaglia C, Leveri M, Cividini A, Mondelli MU, Civardi E, Silini EM (2003) CYP enzyme polymorphisms and susceptibility to HCV-related chronic liver disease and liver cancer. Int J Cancer 104(3):310–317
Smith G, Stanley LA, Sim E, Strange RC, Wolf CR (1995) Metabolic polymorphisms and cancer susceptibility. Cancer Surv 25:27–65
Yang G, Shu XO, Ruan ZX, Cai QY, Jin F, Gao YT, Zheng W (2005) Genetic polymorphisms in glutathione-S-transferase genes (GSTM1, GSTT1, GSTP1) and survival after chemotherapy for invasive breast carcinoma. Cancer 103:52–58
Yu MW, Chiu YH, Yang SY, Santella RM, Chern HD, Liaw YF, Chen CJ (1999) Cytochrome P450 1A1 genetic polymorphisms and risk of hepatocellular carcinoma among chronic hepatitis B carriers. Br J Cancer 80(3–4):598–603
Yu BW, Zhang LQ, Teng XL, Zhang Y, Zou LB, Ying HY (2015) Association between the CYP1A1 polymorphisms and hepatocellular carcinoma: a meta-analysis. Genet Mol Res 14(1):1076–1084
Zheng Y, Wang JJ, Sun L, Li HL (2012) Association between CYP1A1 polymorphism and colorectal cancer risk: a meta-analysis. Mol Biol Rep 39:3533–3540
Zhong S, Tang MW, Yeo W, Liu C, Lo YM, Johnson PJ (2002) Silencing of GSTP1 gene by CpG island DNA hypermethylation in HBV associated hepatocellular carcinomas. Clin Cancer Res 8:1087–1092
Acknowledgments
The authors thank Dr. Iman Fawzy (MD) for performing the statistical analysis.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Abo-Hashem, E.M., El-Emshaty, W.M., Farag, R.E.S. et al. Genetic Polymorphisms of Cytochrome P4501A1 (CYP1A1) and Glutathione S-Transferase P1 (GSTP1) and Risk of Hepatocellular Carcinoma Among Chronic Hepatitis C Patients in Egypt. Biochem Genet 54, 696–713 (2016). https://doi.org/10.1007/s10528-016-9749-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10528-016-9749-6