Abstract
Integration of biological activities of the algal and bacterial communities enhances the bioremediation potency of aquaculture systems. The effects of nitrogen removal mediated by Ulva lactuca on the bacterial community structure and the abundance of nitrogen cycle functional genes were investigated using an integrated multi-trophic aquaculture (IMTA) system mainly composed of sea cucumber (Stichopus japonicus), shrimp (Penaeus japonicus), and crab (Portunus trituberculatus). The experimental treatments were separated into two groups: control group (C: without U. lactuca) and algae treatment (A: with U. lactuca). Microbial diversity and abundance indexes, including the Sobs, Shannon, Ace, and Chao1 indexes, were higher in the U. lactuca treatment group in both water and sediment. Flavobacteriaceae and Rhodobacteraceae were the dominant families in both the U. lactuca and control treatment groups in October and November, respectively. In sediment, Bacillaceae was the dominant family in the U. lactuca treatment group throughout the experimental period, whereas Desulfocapsaceae was the dominant family in the control group in October and November. Moreover, the nitrogen cycle functional genes nifH, amoA, nxrB, norB, and nrfA were more abundant in the U. lactuca treatment group than in the control group. Results of water quality and its correlation with bacterial community were comprehensively investigated, revealing that U. lactuca influenced the bacterial community structure and nitrogen cycle by increasing DO in the IMTA system. In conclusion, U. lactuca co-cultured in an IMTA system could represent a novel approach for enhancing nitrogen removal, based on the interaction between the algal and bacterial communities.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Coastal eutrophication caused by shrimp and fish aquaculture has been an increasingly serious concern in recent years. The integrated multi-trophic aquaculture (IMTA) is an eco-friendly and sustainable system (Browdy et al. 2012; Troell et al. 2009) which capitalizes on the synergistic interactions among aquatic species to cultivate some species that occupy different trophic levels, provides complementary ecosystem functions in a way (Abreu et al. 2009; Neori et al. 2004), and reduces the amount of required nutrients and organic waste outputs (Neori et al. 2004; Samocha et al. 2015). For instance, sea cucumbers with market economic value need not require special diet, which can utilize suspended organic particles to support their growth, such as shrimp uneaten feed, feces, and plankton (Lander et al. 2013), and limit the proliferation of anaerobic bacteria by bioturbation in shrimp polyculture systems (Martínez-Porchas et al. 2010; Uthicke 1999). Additionally, algae can be used for water remediation (Agarwal et al. 2020; Bonanno et al. 2020) through assimilating dissolved inorganic nutrients including ammonia and phosphate and converting these into valuable biomass (Neori et al. 2004; Yang et al. 2015). The genus Ulva as green macroalgae with fast growth rate and high carbohydrate content are well known for their excellent capacity to absorb, utilize, and remove nutrients (nitrogen and phosphorus) and productivity (Cunha et al. 2019; Gao et al. 2020; Shahar and Guttman 2021), commonly used for IMTA system and aquaculture wastewater treatment, playing an important ecological role in marine ecosystems (Massocato et al. 2022). Ulva can absorb both ammonia and nitrate nitrogen; however, its absorption of ammonia nitrogen is much higher than nitrate nitrogen (Hadley et al. 2014; Naldi and Wheeler 2002), for example, in IMTA systems, the efficiency of ammonia removal by Ulva lactuca may exceed 80% (Al-Hafedh et al. 2015; Macchiavello and Bulboa 2014). In addition, in the IMTA system, Ulva sp. as primary producers due to providing a food source for other organisms (Guerreiro et al. 2018; Santizo-Taan et al. 2019) are usually co-cultured with other aquatic organisms (e.g., shrimp, shellfish, and fish), not only reducing the effluent nutrient loads released into the marine environment but also reducing the demand for commercial feed, thus mitigating the detrimental effects of aquaculture, increasing the economic value of aquatic products, and enhancing aquaculture development sustainability (Bolton et al. 2008; Cruz-Suárez et al. 2010; Shpigel et al. 2018).
In aquatic ecosystems, algae can provide grazing and habitat for a variety of species. In addition, algae transform dissolved inorganic carbon into organic matter that can be directly utilized by heterotrophic bacteria via photosynthesis, and release oxygen, in addition to releasing metabolites to surrounding environment, which affect the reproduction and respiratory metabolism of heterotrophic bacteria (Hollants et al. 2013; Zhang et al. 2015). In contrast, heterotrophic bacteria can also degrade organic compounds to produce carbon dioxide, nutrients, vitamins, and growth-promoting factors for supporting algal growth (Hollants et al. 2013). Thus, the interaction between heterotrophic bacteria and algae is mutualistic (Ramanan et al. 2016); in other words, algal blooms are associated with increasing activity of heterotrophic bacteria (Ramanan et al. 2016; Sigman and Hain 2012). However, algae can absorb inorganic nutrients (nitrogen, phosphorus, etc.) that are competitively utilized by bacteria (D’Silva and Kyndt 2020; Liu et al. 2019; Urakawa et al. 2019).
Higher dissolved oxygen (DO) concentrations and higher pH value result from algae’s photosynthesis (Areco et al. 2021; Li et al. 2021, 2019). DO plays an important role in the bioremediation of aquaculture wastewater in IMTA systems, especially in nitrogen transformation, because it is involved in many biological processes such as photosynthesis, nitrification, and respiration (Devi et al. 2012; Fang et al. 2018; Lananan et al. 2014). Generally, pH can substantially affect aquatic organism and bacterial communities (Giordani et al. 2019) but may also be altered by bacteria, algae, or both (García-de-la-Fuente et al. 2011; Giordani et al. 2019). Most bacteria participate in the nitrogen cycle by stimulating nitrification and denitrification (Banks et al. 2013), in both water and sediment (Ma et al. 2015). In aquaculture ecosystems, microorganisms utilize nitrogen and phosphorus as energy sources (Jasmina et al. 2020), assimilating these elements as proteins and polyphosphates used for cell growth and metabolism (Lananan et al. 2014; Rawat et al. 2011). Growth performance of bacteria reflects their adaptation, assimilation, and survival in the surrounding environment, while the regulation of external factors, such as pH, temperature, and DO, closely related to microorganism growth performance (Lananan et al. 2014).
Algal cultivation in IMTA system is a promising approach to enhance water quality. Some studies have demonstrated that algal–bacterial interactions, such as those occurring in algae-bacteria-based aquaponics systems, can improve productivity, nitrogen utilization efficiency, and water quality. This study aimed to investigate the effect of U. lactuca algae cultivation on nitrogen removal and microbiota composition in water and sediment of IMTA system, to provide a novel practical approach of co-cultured U. lactuca algae which presented a high nitrogen bioremediation ability in IMTA system.
Materials and methods
Experimental design
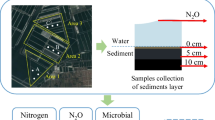
This experiment samples were collected from an integrated multi-trophic aquaculture (IMTA) system that included Stichopus japonicus (sea cucumber), Penaeus japonicus (shrimp), Portunus trituberculatus (crab), and U. lactuca (algae) in Qingdao Ruizi Co., Ltd. which is located (35°64′74″N; 119°84′22″E) in Langya town, Qingdao city, Shandong province, along the Yellow Sea. The aquaculture pond had a surface area of 40,255 m2 and a depth of more than 2 m. The experimental treatments were separated into two groups: control group (C: without U. lactuca) and algae treatment (A: with U. lactuca), which are shown in Fig. 1b. There were three parallel ponds in each treatment. A total of 300,000 S. japonicus (mean weight ≈ 1.7 g, stocking density ≈ 7.5 m2−1) were added in the pond on April 11. A total of 120,000 Penaeus japonicus (mean length ≈ 1 cm, stocking density ≈ 3 m2−1) were cultured in the pond on May 1. A total of 240,000 U. lactuca (mean length ≈ 1 cm, mean weight ≈ 17 g, stocking density ≈ 6 m2−1) were added in the pond on May 4, cultured in cages, as shown in Fig. 1b (Aalgae treatment). On May 15, 1500 seeds of P. trituberculatus (0.05 g.crab−1, stocking density ≈ 0.04 m2−1) were added in pond. During the experimental period, we feed 200-kg ice fresh fish bait (0.019-g organic nitrogen.g−1) per pond every day. The oxygen was supplied by an aeration system through nanotubules at the pond’s bottom. According to the water quality, the exchange of water was carried out through the sea tide and water level difference for IMTA system.
Sample collection
The experimental samples were collected from the control group (C) and the algae treatment (A). Control group and algae treatment consist of three ponds, respectively. Three random samples were obtained monthly from the area without algae of each pond in September, October, and November, completely mixed as a repeat sample, where the samples were acquired from water and sediment. A 1000-mL volume of water was collected using a plexiglass water collector. A total of 200 mL of water was filtered through a 0.22-μm acetate fiber membrane, and the residues on the membranes were used to analyze the microbial community. The remaining 800 mL of water was filtered through a 0.45-μm microporous membrane and utilized to determine the chemical indexes of the water. Sediment samples were gathered at 0–8 cm below the sediment’s surface with a plexiglass mud picker. The sediment sample size for DNA extraction was 5 g. The filter membranes and sediment samples were flash-frozen and kept at − 80 °C until they were analyzed.
Water quality
Water quality indicators were measured each time water samples were collected. The YSI incorporated device (Yellow Springs, OH, USA) was used to detect water temperature, dissolved oxygen (DO), salinity, and pH value. The ammonia nitrogen content was measured using indophenol blue spectrophotometry (Pai et al. 2001). Spectrophotometry was used to measure nitrite nitrogen (Aydın et al. 2005). The UV spectrophotometric approach was used to detect nitrate nitrogen (Miles et al. 1998). The potassium persulfate oxidation was used to detect total nitrogen and total phosphorus (Zhou et al. 2007).
High-throughput sequencing of bacteria and bioinformatic analysis
The FastDNA® Spin Kit for Soil (MP Biomedicals, USA) was used to extract the total DNA from all water and sediment samples. Agarose gel electrophoresis was used to confirm the integrity of the DNA. We used a NanoDrop spectrophotometer (Thermo Scientific, USA) to determine the concentration of bacterial DNA. The V3–V4 region of the 16SrRNA gene was conserved as a bacterial DNA-specific sequence area using the primers 338F (5′-ACTCCTACGGGAGGCAGCAG-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) (Xu et al. 2016). The 16SrRNA gene was then amplified via polymerase chain reaction (PCR) on a MyCycler™ thermal cycler (Bio-Rad, USA). Majorbio then purified and sequenced the bacterial DNA using Illumina MiSeq. The raw reads were submitted to the NCBI Sequence Read Archive (SRA) database (GenBank accession: PRJNA948567).
Quantitative PCR of functional genes of nitrogen cycle
PCR was performed with primers for the nifH (nitrogenase), amoA (ammonia monooxygenase), Amx (anaerobic ammonia oxidase), nxrB (nitrite oxidase), nirK (nitrite reductase), norB (nitric oxide reductase), nosZ (nitrous oxide reductase), and nrfA (nitrite reductase, dissimilatory nitrite reduction to ammonium [DNRA]) genes as depicted in Table 1 using 2 × Phanta® Max Master Mix (Vazyme, Nanjing, China). Table S1 (supplementary information) shows the specifics of the PCR conditions. The products were purified with a FastPure® Gel DNA Extraction Mini Kit (Vazyme), subcloned into a 5-min TA/Blunt-Zero Cloning Vector (Vazyme), propagated in Fast-T1 DH5α (Vazyme), and sequenced using an ABI3700 sequencer (USA). FastPure® Plasmid Mini Kit (Vazyme) was used to extract the corrected plasmids for subsequent qPCR. Standards were created by serial dilution of plasmids containing the target gene and measured with an Agilent 220 TapeStation System (Agilent Technologies, Santa Clara, CA, USA). We conducted qPCR assays for the nitrogen cycling genes nifH, amoA, Amx, nxrB, nirK, norB, nosZ, and nrfA (Fig. 1), using ChamQ Universal SYBR qPCR Master Mix (Vazyme) and an Applied Biosystems 7500 Fast Real-Time PCR System (Thermo Fisher Scientific, Waltham, MA, USA). The reaction system is shown in Table S2(supplementary information). The reaction conditions for qPCR are listed in Table S3 (supplementary information).
Data analysis
FLASH program spliced paired-end (PE) reads based on the overlap relationship (Magoč and Salzberg 2011). For quality inspection and filtration of original sequencing sequences, Fastp software was employed (Chen et al. 2018). After data optimization, we utilized UPARSE software (Edgar 2013) for operational taxonomic units (OTU) clustering and statistical analysis of biological information for the sequence, which had a 97% similarity (Edgar 2013; Stackebrandt, Goebel 1994). Before the data analysis, gene sequences from each sample have been normalized according to the minimum number of sample sequences, removing chloroplasts and mitochondria. For each sequence, the RDP classifier software (Wang et al. 2007) was utilized for species classification analysis. Statistical analysis was used to determine the community structure of the samples at various classification levels based on the results of taxonomic analysis. MOTHUR (Schloss et al. 2009) was used to calculate alpha diversity. LEfSe (https://huttenhower.sph.harvard.edu/lefse/) was used to perform linear discriminant analysis (LDA) on samples with varying treatments. QIIME was used to compute weighted UniFrac distances for principal coordinate analysis (PCoA) (Lawley and Tannock 2017) and ANOSIM analysis. The vegan package in R (http://www.r-project.org) was used for redundancy analysis (RDA). The SPSS Statistics 22 software was utilized for statistical analysis of water quality differences and bacterial community differences through T-test, with value of p < 0.05 deemed significant and p < 0.01 regarded extremely significant (Liu et al. 2018). Function prediction analysis of bacterial community under control group and algae treatment were examined with the Prokaryotic Taxa Annotation Database (FAPROTAX) using python software (Louca et al. 2016).
Results
Organisms’ production
Table 2 shows the final average weight and total production of all organisms at the end of the aquaculture period. The final average weight and total production of S. japonicus (sea cucumber), P. japonicus (shrimp), P. trituberculatus (crab) in the algae treatment were more than those in the control group. Simultaneously, the algae treatment significantly increased the final average weight of crab.
Water quality
The results of water quality indexes with each treatment in the IMTA system (C: control group; A: with U. lactuca) are shown in Table 3. During the experimental period, the salinity, DO, and pH value showed an increasing trend, whereas the temperature showed a decreasing trend. At the same sampling time, the temperature and salinity in the algae treatment group were slightly lower compared to those in the control group (p > 0.05). The DO and pH value were higher in the algae treatment group than in the control group. Moreover, the DO at the three sampling times were significantly different between the algae treatment and control groups (p < 0.05).
The results of nutrient contents in water in the control and algae treatment groups over time (September, October, and November) are shown in Fig. 2. During the experiment, the concentrations of ammonia-N (Fig. 2a), nitrite-N (Fig. 2b), nitrate–N (Fig. 2c), total nitrogen (Fig. 2d), and total phosphorus (Fig. 2e) were decreased with an extended sampling time in both the algae treatment and control groups. The ammonia-N in the algae treatment group was lower than that in the control group, with significant differences in September and October (p < 0.05). In the algae treatment groups, nitrite-N and nitrate–N concentrations were lower than those in the control group. During the experiment, total nitrogen in the algae treatment group was lower than that in the control group, with a significant difference in October and November (p < 0.05). Total phosphorus in the algae treatment group was higher than that in the control group, with a significant difference in September (p < 0.05). All concentrations of nutrients in the aquaculture system conform to the fishery water quality standard.
Nutrients including ammonia nitrogen, nitrite nitrogen, nitrate nitrogen, total nitrogen, and total phosphorus in water in three experimental groups (C: control group; A: with U. lactuca) at September, October, and November. Samples from different locations were analyzed for significant differences only among the same time. *The asterisk means that the value is significantly different (p < 0.05)
Composition and diversity of bacteria in water and sediment
In this study, 45,763–219,748 effective sequences, with an average of 131,195, were detected in water samples, and these clustered into 645–1437 OTUs, with an average of 1041. The microbial compositions of the control and algae treatment groups were similar, but the proportions differed with the sampling time. The dominant bacteria on family level in the water samples included Cyanobiaceae (0.17–32.78%), Rhodobacteraceae (12.93–20.72%), Flavobacteriaceae (10.82–20.09%), Microbacteriaceae (2.33–11.82%), SAR116_clade (1.64–5.93%), and Cryomorphaceae (2.71–5.50%), which accounted for approximately 66.09% of the total bacteria (Fig. 3a). The dominant bacteria on genus level in the water samples included HIMB11 (9.11–13.27%), NS5_marine_group (6.16–12.15%), Synechococcus_CC9902 (0.12–12.01%), Cyanobium_PCC-6307 (0.05–23.11%), Candidatus_Aquiluna (2.19–11.48%), and norank_f__Cryomorphaceae (2.55–5.29%), which accounted for approximately 57.77% of the total bacteria (Fig. 3b). Alpha-diversity indexes are shown in Table 4. The Sobs, Shannon, Ace, and Chao1 indexes of the algae treatment group were higher than those of the control group. According to the Sobs, Ace, and Chao1 indexes, there were significant differences between the algae treatment and control groups in September and October (p < 0.05). The alpha-diversity results showed that algae culture could increase the diversity and abundance of the bacterial community in water, especially significantly increasing abundances.
Moreover, 49,090–204,192 effective sequences, with an average of 127,681, were detected in sediment samples, which clustered into 2457–3638 OTUs, with an average of 2952. The dominant bacteria on family level in the sediment samples included Bacillaceae (6.05–16.26%), Desulfocapsaceae (4.38–14.72%), Rhodobacteraceae (1.21–9.40%), Flavobacteriaceae (3.42–7.61%), and Woeseiaceae (1.83–5.63%), which accounted for approximately 38.2% of the total bacteria (Fig. 4a). The dominant bacteria on genus level in the sediment samples included Bacillus (5.66–15.72%), norank_f__Desulfocapsaceae (3.82–12.93%), Woeseia (1.83–5.63%), norank_f__unclassified (2.29–4.76%), Actibacter (1.70–4.17%), norank_f__Desulfobulbaceae (0.51–3.74%), Ilumatobacter (1.43–3.54%), Halioglobus (1.42–2.32%), and Filomicrobium (1.05–1.84%), which accounted for approximately 39.89% of the total bacteria (Fig. 4b). In the algae treatment group, Bacillaceae and Bacillus were the dominant bacteria at the three sampling times (September, October, and November). These results indicated that U. lactuca cultivation had a marked influence on the dominant bacteria in the sediment samples. Alpha-diversity indexes of the sediment samples are shown in Table 5. During all sampling times, the Sobs, Shannon, Ace, and Chao1 indexes of the algae treatment group were higher than those of the control group. In October and November, the Shannon, Ace, and Chao1 indexes in the algae treatment group were significantly higher than those in the control group (p < 0.05). The results of alpha diversity showed that U. lactuca culture could increase the diversity and abundance of the bacterial community in sediment, especially significantly increasing abundances.
PCoA (Fig. 5), utilizing weighted UniFrac distances, was used to analyze the bacterial community compositions of all water (Fig. 5a) and sediment (Fig. 5b) samples in the control and algae treatment groups. The first axis of Fig. 5b shows that the bacterial communities of the sediment samples differed between the control and algae treatment groups. Meanwhile, the analysis of similarity (ANOSIM) statistical dissimilarities revealed a significant difference in bacterial communities in sediment between the control and algae treatment groups (p = 0.04) (Fig. 5b).
Correlation analysis between environmental factor levels and bacterial community abundances
As shown in Fig. 6, correlations between environmental parameters and the bacterial community structure of water (Fig. 6a) and sediment (Fig. 6b) samples were determined using redundancy analysis. Temperature (r2 = 0.8491, p = 0.001), DO (r2 = 0.6192, p = 0.001), pH (r2 = 0.7081, p = 0.001), salinity (r2 = 0.5189, p = 0.007), nitrate–N (r2 = 0.4071, p = 0.015), and nitrite-N (r2 = 0.4281, p = 0.036) were found to be the significant environmental determinants of the bacterial community in water samples. Moreover, temperature (r2 = 0.524, p = 0.002), total nitrogen (r2 = 0.623, p = 0.002), pH (r2 = 0.4574, p = 0.007), ammonia-N (r2 = 0.4986, p = 0.009), DO (r2 = 0.4063, p = 0.011), and nitrate–N (r2 = 0.4089, p = 0.013) were the significant environmental determinants of the bacterial community in sediment samples. Considering the effect of sampling time on the results, correlations between environmental parameters and the bacterial community structure of each water and sediment samples collected from September to November were determined using redundancy analysis (Fig. S2 are given in supplementary information). DO (r2 = 0.9591, p = 0.0194) (Fig. S2b1) was the significant environmental determinants of the bacterial community in sediment in September, and total nitrogen (r2 = 0.8767, p = 0.0389) (Fig. S2b3) was the significant environmental determinants of the bacterial community in sediment in November.
Redundancy analysis (RDA) to show correlation between environmental parameters and bacterial community structure of water and sediment samples in control group and algae treatment. a Water samples. b Sediment samples. The quantitative environmental factors were represented by the red arrow, whose length can represent the degree of impact of environmental factors on samples. Positive and negative correlation are represented by the angle between the arrows of environmental factors (acute angle, positive correlation; obtuse angle, negative correlation; right angle, no connection)
Microbial biomarkers in water and sediment between control and algae treatment group
The LEfSe algorithm detected 7 and 13 differentially abundant taxonomic clades as active biomarkers and identified divergence between the control and algae treatment group, respectively, in water (Fig. S1a1) and sediment (Fig. S1b1) samples (Fig. S1 is given in supplementary information). The representative differentially abundant bacterial taxa included f__Moraxellaceae (p = 0.002), g__C1-B045 (p = 0.03), f__Planococcaceae (p = 0.007), g__Planococcus (p = 0.037), o__Bacillales (p = 0.024), and g__Portibacter (p = 0.04), which were enriched in the water samples of the algae treatment group. Further, f__Woeseiaceae (p = 0.031), g__Woeseia (p = 0.031), o__Steroidobacterales (p = 0.031), p__Campylobacterota (p = 0.004), o__Campylobacterales (p = 0.004), c__Campylobacteria (p = 0.004), g__Sulfurovum (p = 0.004), and f__Sulfurovaceae (p = 0.004) were enriched in the sediment samples of the algae treatment group. Biomarkers had high LDA scores in water (Fig. S1a2; LDA > 2) and sediment (Fig. S1b2; LDA > 3.6), indicating statistically and biologically significant variations in abundance among the observed microbial communities.
Functional predictions in water and sediment under control and algae treatments
The results of functional predictions of bacterial communities in water (Fig. 7a) showed that high abundance of nitrate_reduction was significantly higher in the algae treatment group than in the control group in November. Low abundances of nitrite_ammonification, nitrate_ammonification, nitrate_denitrification, denitrification, and nitrous_oxide_denitrification in water were higher in the algae treatment group than in the control group in September and October. The results of functional prediction of bacterial communities in sediment (Fig. 7b) showed that high abundances of nitrate_reduction, nitrite_ammonification, and nitrate_ammonification were higher in the algae treatment group than in the control group at all sample times. Low abundance of nitrogen_fixation in sediment was higher in the algae treatment group than in the control group in October.
Abundances of nitrogen cycle functional genes
Data presented in Fig. 8 pertain to the nitrogen cycle functional genes that were detected via real-time PCR (q-PCR), including nifH, amoA, Amx, nxrB, nirK, norB, nosZ, and nrfA. The nifH gene (1.4 × 102–1.6 × 108 copies/mL) is a biological nitrogen fixation-associated gene. The amoA (1.9 × 103– 2.4 × 108 copies/mL) and nxrB (1.9 × 104–1.1 × 108 copies/mL) genes are the key genes in nitrification. Further, the Amx gene (6.1 × 105–3.7 × 108 copies/mL) is an anammox-related gene, and nirK (3.6 × 102–1.3 × 107 copies/mL), norB (2.4 × 103–1.7 × 107 copies/mL), and nosZ (6.5 × 104–2.5 × 109 copies/mL) are the main genes involved in the denitrification process; meanwhile, the nrfA gene (7.5 × 104–1.1 × 109 copies/mL) is the key gene involved in the DNRA process.
Abundances of nitrogen cycling functional genes in water (/ml) and sediment (/g) in control group and algae treatment (C: control group; A: with U. lactuca) at different sampling times (September, October, November). a, b nifH (nitrogenase). c, d amoA (ammonia monooxygenase). e, f Amx (anaerobic ammonia oxidase). g, h nxrB (nitrite oxidase). i, j nirK (nitrite reductase). k, l norB (nitric oxide reductase). m, n nosZ (nitrous oxide reductase). o, p nrfA (nitrite reductase, dissimilatory nitrite reduction to ammonium [DNRA])
Many Amx, nosZ, nxrB, and nrfA genes were simultaneously detected in water and sediment. The nifH, amoA, nxrB, and nirK genes in water and sediment were determined to be more prevalent in the algae treatment group than in the control group. In both water and sediment, the abundances of the amoA gene in September to November, the nxrB gene in September and October, and the nirK gene in September, were significantly higher in the algae treatment group (p < 0.05) than in the control group. The abundance of the nifH gene in September to November in sediment and that in October in water was significantly higher in the algae treatment group (p < 0.05) than in the control group. Further, the abundances of the nrfA gene in September to November and the norB gene in September and October were significantly higher in the algae treatment group (p < 0.05) than in the control group but only in sediment. However, the abundances of the Amx gene in September, the norB gene in November, and the nosZ gene in October and November were significantly lower in the algae treatment (p < 0.05) than in the control group, in water and sediment.
Discussion
High ammonia level is harmful to the cultured shrimp and can lead to death by including a sharp decline in DO in water, leading to the production of harmful gases, destruction of the stability of the aquatic environment, and promotion of the invasion of various pathogens, which they are not conducive to shrimp culture. In this study, cultivation of U. lactuca significantly increased the concentration of dissolved oxygen and significantly decreased the concentrations of ammonia nitrogen and total nitrogen in aquaculture water environment. Studies have shown that macroalgae (e.g., U. lactuca) in IMTA systems can absorb dissolved inorganic nutrients for their own growth (Neori et al. 2004) and release oxygen through photosynthesis process, resulting in an increase in the DO concentration (Areco et al. 2021; Li et al. 2021), and promoting the growth of ammonia-oxidizing bacteria. Additionally, algae can increase the relative abundance of denitrifying bacteria and significantly improve the removal efficiency of total nitrogen (Zhou et al. 2022), which is consistent with the results of this study. In addition, it is worth noting that U. lactuca cultivation increased the final average weight and total production of all organisms; particularly, a significant increase of the final average weight of P. trituberculatus (crab) was observed in this study. The reason may be that macroalgae can contribute to the primary production of global habitat formation, increasing bioactive substances that improve the health status and production performance of aquaculture organisms (Michalak et al. 2022).
Aside from absorbing nutrients, algae can improve water quality collaboratively with surrounding microorganisms for bioremediation in IMTA systems. Macroalgae (Ulva lactuca) surfaces harbor various epiphytic bacterial communities with functions related to algal life and water quality, mainly including Rhodobacteraceae (Proteobacteria) and Flavobacteriaceae (Bacteroidetes) (Comba González et al. 2021; Hmani et al. 2023). In this study, Rhodobacteraceae and Flavobacteriaceae as prominent bacteria in water samples were identical to the epiphytic bacteria on U. lactuca’s surface; however, both have no significant difference in proportion between the control and algae treatments in either water and sediment. Therefore, epiphytic bacteria on U. lactuca’s surface have little effect on the bacterial communities in water and sediment environments. In this study, ammonia-N levels were substantially lower in the algae treatment group than in the control group, indicating that algae treatment can stimulate the proliferation of ammonia-oxidizing bacteria and expedite ammonia oxidation. Most of the major families in the algae treatment group were belonged to Proteobacteria and Bacteroidetes, which comprise important microorganisms in the nitrogen cycle (Rurangwa and Verdegem 2014) and contain ammonia-oxidizing bacteria with a highly stable deamination capacity (Zhao et al. 2013). In sediments, U. lactuca can increase the abundance of Bacillaceae, including ammonia oxidizers under low DO conditions, as well as heterotrophic nitrobacteria, aerobic-denitrifying bacteria, and non-isolated anammox strains (Dos Santos et al. 2021). Among these, members have heterotrophic nitrification and aerobic denitrification capacity (Zhang et al. 2012), removing dissolved ammonia and nitrite and restoring water quality.
In our IMTA system, temperature, DO, pH value, and nitrate–N were found to be significantly important environmental factors influencing bacterial communities in water and sediments in our study. In particular, the effects of DO and total nitrogen on sediment bacterial communities were significant. However, the cultivation of U. lactuca significantly increased the concentration of DO, which is required for various biological processes, including photosynthesis, nitrification, and respiration (Devi et al. 2012; Fang et al. 2018; Lananan et al. 2014), playing important roles in the bioremediation of aquaculture wastewater, especially in promoting nitrogen conversion. In addition to increasing dissolved oxygen content, algae also absorb nutrients including nitrogen and phosphorus required for bacterial growth, thereby affecting bacterial communities. Moreover, compared to the control group, U. lactuca cultivation can decrease the water temperature, but there is no significant difference, due to providing increased shading, which reduces the amount of sunlight that can penetrate deeper into the water (Ahonen et al. 2023). Additionally, algal photosynthesis consumes CO2 (Zhang et al. 2022), decreasing its availability in the surrounding area, which can suppress the absorption of infrared radiation by CO2 molecules (Elahi et al. 2020) and lead to a decrease in the water temperature.
Modern molecular biological techniques for functional gene detection have enabled us to explore the relationship between bacterial communities and nitrogen cycles in aquaculture systems and study the characteristics and regulation of the nitrogen cycle. In this study, the abundances of nifH, amoA, nxrB, norB, and nrfA genes in the algae treatment group were higher than those in control group, in both water and sediment environments. As a highly conserved gene for biological nitrogen fixation, nifH is the most commonly used molecular marker in studies on nitrogen-fixing microorganisms (Zehr et al. 2003). Generally, nifH gene abundance is negatively correlated with DO and positively correlated with temperature because nitrogen fixation is an anaerobic process. However, our results showed that nifH gene abundance was higher in the algae treatment group, which showed a lower temperature and higher DO content compared to control group. Some reports have shown that in deep-sea samples, the nifH gene originates from two sources, the anaerobic high-temperature seabed and cold oxygen-rich deep waters (Mehta et al. 2003). This could explain the high nifH gene abundance in the algae treatment group under low-temperature and oxygen-rich conditions. Some members of Bacillaceae, the dominant bacteria in the algae treatment group, are nitrogen-fixing bacteria (Han et al. 2019). Additionally, more Vibrionaceae species were detected in the algae treatment group (6.82%) in water in November; some members of this family are pathogenic and cause severe economic losses in aquaculture (Banchi et al. 2022; Costa et al. 2022), whereas others are marine nitrogen-fixing bacteria that show nitrogenase activity (Rubio-Portillo et al. 2016). These findings validate the increased nifH gene abundance in the algae treatment group.
The increased abundance of amoA and nxrB genes in sediment and water after algal treatment also validated the conclusion, and that algae treatment can stimulate the proliferation of ammonia-oxidizing bacteria and expedite ammonia oxidation. The amoA gene, which is highly conserved, encodes ammonia monooxygenase, a specific enzyme harbored by ammoxidation bacteria that can catalyze the oxidation of ammonia to hydroxylamine (McTavish et al. 1993). Oxygen is a key environmental factor involved in ammoxidation. In this study, the increased abundance of amoA in the algae treatment group was likely due to the higher DO content. The abundance of amoA in sediment was highest in October and lowest in November, which could be due to the produced ammonia by microorganisms as a result of accumulation of organic matter during the aquaculture process, thereby promoting the peak growth of ammonia-oxidizing bacteria in October, whereas harvesting occurred in November, resulting in a decrease in organic matter and the availability of ammonia for ammonia-oxidizing bacteria. The nxrB gene, encoding the nitrite oxidase subunit, is a specific functional marker for the oxidation of nitrite to nitrate (Lücker et al. 2010). Nitrite-oxidizing bacteria were mainly detected in the sediment and water of the algae treatment group, and these included Nitrospina and Nitrospira. Typically, the entire nitrification process needs to be carried out under aerobic conditions; however, nitrite-oxidizing bacteria require more oxygen than ammonia-oxidizing bacteria (Blackburne et al. 2007). Therefore, the cultivation of U. lactuca increased the abundance of nitrite-oxidizing bacteria by increasing the DO content.
Lower levels of ammonia nitrogen, nitrite, nitrate, and total nitrogen were observed in the algae treatment group compared to control group. From September to November, nitrite-N and nitrate–N concentrations decreased, with ammonia-N decreasing first and then remaining stable, indicating the activation of nitrogen cycle pathways related to both denitrification and DNRA gene expression. This inference could be confirmed by function predictions, including the terms nitrate_reduction and nitrite/nitrate_ammonification of bacterial communities in water and sediment, as well as functional genes related to the nitrogen cycle, including nirK, norB, and nrfA, in sediment. The norB gene, encoding NO reductase (Andrea et al. 2002), was more abundant in the algae treatment group than in the control group. Expression of this gene is associated with the rapidly catalyzed reduction of NO (which is highly toxic to cells) to N2O, and it has been detected in some aerobic-denitrifying bacteria (Yang et al. 2020). Common denitrification processes facilitate the gradual conversion of nitrate into N2, which eventually escapes from the aquaculture system, resulting in nitrogen loss. However, the DNRA process can ultimately result in the reduction of nitrate to ammonia under conditions of sufficient carbon, which is conducive to nitrogen retention (Li et al. 2022). The nrfA gene, which encodes an enzyme catalyzing the DNRA process, was more abundant in the algae treatment group than in the control group, especially in sediment. Many environmental factors affect the abundance of the nrfA gene, including nitrate, ammonia, nitrite, organic carbon, and sulfide contents (Robertson et al. 2016; Yin et al. 2017). However, according to the reaction kinetic principles, ammonia, which is the final product of the DNRA process, is an important driving factor. In this study, U. lactuca promoted nitrification process which was in line with literature (Wang et al. 2019) to further provide a substrate for the DNRA process, which could explain the increase in nrfA gene abundance.
Nutrient availability, light intensity, temperature, dietary composition (Pereira et al. 2015), and aquatic organisms determine the bacterial community structure in IMTA systems. The cultivation of U. lactuca can increase the diversity and abundance of bacteria in water and sediment. Temperature, DO, pH value, and nitrate nitrogen were discovered to be environmental elements closely associated with the abundance of the bacterial community in this study, with DO being the decisive factor in how U. lactuca cultivation affected the bacterial community. U. lactuca can produce oxygen, through photosynthesis, and promote microbial metabolism (Dame 1996), ultimately affecting the bacterial community structure, especially increasing the abundance of aerobic bacteria in water and sediment owing to increases in the DO concentrations. Moreover, metabolites released by U. lactuca are related to bacterial community (especially heterotrophic bacteria), contributing to the change in bacterial diversity and community structure; however, the exact reason for this is yet to be studied. Additionally, we should consider the limitations of predicting bacterial functions based on representative gene sequencing and common nitrogen cycling function genes in research results, while more precise analysis of the function of bacterial communities through metagenomes is further work for our study.
Conclusion
A comprehensive analysis of nitrogen cycle gene abundance and bacterial composition has shown that U. lactuca cultivation is beneficial for a productive aquaculture environment. The cultivation of U. lactuca influenced the bacterial community structure by increasing DO in the IMTA system. Further, U. lactuca increased the abundances of functional genes of the nitrogen cycle, including the nifH gene, required for nitrogen fixation; amoA and nxrB genes, for nitrification; and nirK, norB, and nrfA genes, for denitrification, to promote nitrogen conversion. This study has elucidated the interactions between U. lactuca and bacterial communities and their effect with respect to maximizing nitrogen cycle efficiency and sustainability in IMTA systems.
Data availability
All supporting data generated during this study are included in this published article. Sequence data that support the findings of this study have been deposited in the National Center for Biotechnology Information with the primary accession code PRJNA948567.
References
Abreu MH, Varela DA, Henríquez L, Villarroel A, Yarish C, Sousa-Pinto I, Buschmann AH (2009) Traditional vs. integrated multi-trophic aquaculture of Gracilaria chilensis C. J. Bird, J. McLachlan & E. C. Oliveira: Productivity and physiological performance. Aquaculture 293:211–220. https://doi.org/10.1016/j.aquaculture.2009.03.043
Agarwal A, Mhatre A, Pandit R, Lali AM (2020) Synergistic biorefinery of Scenedesmus obliquus and Ulva lactuca in poultry manure towards sustainable bioproduct generation. Bioresour Technol 297:122462. https://doi.org/10.1016/j.biortech.2019.122462
Ahonen SA, Seppälä J, Karjalainen JS, Kuha J, Vähätalo AV (2023) Increasing air temperature relative to water temperature makes the mixed layer shallower, reducing phytoplankton biomass in a stratified lake. Freshw Biol 68:577–587. https://doi.org/10.1111/fwb.14048
Al-Hafedh YS, Alam A, Buschmann AH (2015) Bioremediation potential, growth and biomass yield of the green seaweed, Ulva lactuca in an integrated marine aquaculture system at the Red Sea coast of Saudi Arabia at different stocking densities and effluent flow rates. Rev Aquacult 7:161–171. https://doi.org/10.1111/raq.12060
Andrea B, Bärbel F, Rainer C (2002) Characterization of the norB gene, encoding nitric oxide reductase, in the nondenitrifying cyanobacterium Synechocystis sp. strain PCC6803. Appl Environ Microbiol 68:668–672. https://doi.org/10.1128/AEM.68.2.668-672.2002
Angnes G, Nicoloso RS, da Silva MLB, de Oliveira PAV, Higarashi MM, Mezzari MP, Miller PRM (2013) Correlating denitrifying catabolic genes with N2O and N2 emissions from swine slurry composting. Bioresour Technol 140:368–375. https://doi.org/10.1016/j.biortech.2013.04.112
Areco MM, Salomone VN, Afonso MDS (2021) Ulva lactuca: a bioindicator for anthropogenic contamination and its environmental remediation capacity. Mar Environ Res 171:105468. https://doi.org/10.1016/j.marenvres.2021.105468
Aydın A, Ercan Ö, Taşcıoğlu S (2005) A novel method for the spectrophotometric determination of nitrite in water. Talanta 66:1181–1186. https://doi.org/10.1016/j.talanta.2005.01.024
Banchi E, Manna V, Fonti V, Fabbro C, Celussi M (2022) Improving environmental monitoring of Vibrionaceae in coastal ecosystems through 16S rRNA gene amplicon sequencing. Environ Sci Pollut Res 29:67466–67482. https://doi.org/10.1007/s11356-022-22752-z
Banks JL, Ross DJ, Keough MJ, Macleod CK, Keane J, Eyre BD (2013) Influence of a burrowing, metal-tolerant polychaete on benthic metabolism, denitrification and nitrogen regeneration in contaminated estuarine sediments. Mar Pollut Bull 68:30–37. https://doi.org/10.1016/j.marpolbul.2013.01.002
Bian R, Sun Y, Li W, Ma Q, Chai X (2017) Co-composting of municipal solid waste mixed with matured sewage sludge: the relationship between N2O emissions and denitrifying gene abundance. Chemosphere 189:581–589. https://doi.org/10.1016/j.chemosphere.2017.09.070
Blackburne R, Yuan Z, Keller J (2007) Partial nitrification to nitrite using low dissolved oxygen concentration as the main selection factor. Biodegradation 19:303–312. https://doi.org/10.1007/s10532-007-9136-4
Bolton JJ, Robertson-Andersson DV, Shuuluka D, Kandjengo L (2008) Growing Ulva (Chlorophyta) in integrated systems as a commercial crop for abalone feed in South Africa: a SWOT analysis. J Appl Phycol 21:575–583. https://doi.org/10.1007/s10811-008-9385-6
Bonanno G, Veneziano V, Piccione V (2020) The alga Ulva lactuca (Ulvaceae, Chlorophyta) as a bioindicator of trace element contamination along the coast of Sicily. Italy. Sci Total Environ 699:134329. https://doi.org/10.1016/j.scitotenv.2019.134329
Browdy CL, Hulata G, Liu Z, Allan GL, Sommerville C, Andrade TP, Pereira R, Yarish C, Shpigel M, Chopin T, Robinson S, Avnimelech Y, Lovatelli A (2012) Novel and emerging technologies: can they contribute to improving aquaculture sustainability? in: R.P. Subasinghe, J.R.A., D.M. Bartley, S.S. De Silva, M. Halwart, N. Hishamunda, C.V. Mohan & P. Sorgeloos, eds (Ed.), Proceedings of the Global Conference on Aquaculture 2010. FAO, Phuket, Thailand, pp 149–191
Chen S, Zhou Y, Chen Y, Gu J (2018) fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34:i884–i890. https://doi.org/10.1093/bioinformatics/bty560
Comba González NB, Niño Corredor AN, López Kleine L, Montoya Castaño D (2021) Temporal changes of the epiphytic bacteria community from the marine macroalga Ulva lactuca (Santa Marta, Colombian-Caribbean). Curr Microbiol 78:534–543. https://doi.org/10.1007/s00284-020-02302-x
Costa C, Ferreira GD, Simoes M, Silva JL, Campos MJ (2022) Real-time PCR protocol for detection and quantification of three pathogenic members of the Vibrionaceae family. Microorganisms 10:2060. https://doi.org/10.3390/microorganisms10102060
Cruz-Suárez LE, León A, Peña-Rodríguez A, Rodríguez-Peña G, Moll B, Ricque-Marie D (2010) Shrimp/Ulva co-culture: a sustainable alternative to diminish the need for artificial feed and improve shrimp quality. Aquaculture 301:64–68. https://doi.org/10.1016/j.aquaculture.2010.01.021
Cunha ME, Quental-Ferreira H, Parejo A, Gamito S, Ribeiro L, Moreira M, Monteiro I, Soares F, Pousão-Ferreira P (2019) Understanding the individual role of fish, oyster, phytoplankton and macroalgae in the ecology of integrated production in earthen ponds. Aquaculture 512:734297. https://doi.org/10.1016/j.aquaculture.2019.734297
D’Silva A, Kyndt JA (2020) Bacterial diversity greatly affects ammonia and overall nitrogen levels in aquabioponics bioflocs systems, based on 16S rRNA gene amplicon metagenomics. Appl Microbiol 6:169. https://doi.org/10.35248/2471-9315.20.6.169
Dame R (1996) Ecology of marine bivalves: an ecosystem approach. CRC Mar Sci Ser 43(7):272. https://doi.org/10.4319/lo.1998.43.7.1764
Devi MP, Subhash GV, Mohan SV (2012) Heterotrophic cultivation of mixed microalgae for lipid accumulation and wastewater treatment during sequential growth and starvation phases: effect of nutrient supplementation. Renew Energy 43:276–283. https://doi.org/10.1016/j.renene.2011.11.021
Ding Y, Wang J, Liu Y, Chen S (2005) Isolation and identification of nitrogen-fixing bacilli from plant rhizospheres in Beijing region. J Appl Microbiol 99:1271–1281. https://doi.org/10.1111/j.1365-2672.2005.02738.x
Dos Santos CED, Costa RB, Rabelo C, Ferraz Junior ADN, Persinoti GF, Pozzi E, Foresti E, Damianovic M (2021) Hacking biofilm developed in a structured-bed reactor (SBRRIA) with integrated processes of nitrogen and organic matter removal. Bioprocess Biosyst Eng 44:1841–1851. https://doi.org/10.1007/s00449-021-02564-0
Edgar RC (2013) uParse: highly accurate otu sequences from microbial amplicon reads. Nat Methods 10:996–998. https://doi.org/10.1038/nmeth.2604
Elahi R, Nagassou D, Mohsenian S, Trelles JP (2020) Enhanced solar radiation absorption by carbon dioxide in thermodynamic nonequilibrium: a computational study. Sol Energy 195:369–381. https://doi.org/10.1016/j.solener.2019.11.015
Fang Y, Chen X, Hu Z, Liu D, Gao H, Nie L (2018) Effects of hydraulic retention time on the performance of algal-bacterial-based aquaponics (AA): focusing on nitrogen and oxygen distribution. Appl Microbiol Biotechnol 102:9843–9855. https://doi.org/10.1007/s00253-018-9338-1
Gao G, Burgess JG, Wu M, Wang S, Gao K (2020) Using macroalgae as biofuel: current opportunities and challenges. Bot Mar 63:355–370. https://doi.org/10.1515/bot-2019-0065
García-de-la-Fuente R, Cuesta G, Sanchís-Jiménez E, Botella S, Abad M, Fornes F (2011) Bacteria involved in sulfur amendment oxidation and acidification processes of alkaline ‘alperujo’ compost. Bioresour Technol 102:1481–1488. https://doi.org/10.1016/j.biortech.2010.09.103
Giordani A, Rodriguez RP, Sancinetti GP, Hayashi EA, Beli E, Brucha G (2019) Effect of low pH and metal content on microbial community structure in an anaerobic sequencing batch reactor treating acid mine drainage. Miner Eng 141:105860. https://doi.org/10.1016/j.mineng.2019.105860
Guerreiro I, Magalhães R, Coutinho F, Couto A, Sousa S, Delerue-Matos C, Domingues VF, Oliva-Teles A, Peres H (2018) Evaluation of the seaweeds Chondrus crispus and Ulva lactuca as functional ingredients in gilthead seabream (Sparus aurata). J Appl Phycol 31:2115–2124. https://doi.org/10.1007/s10811-018-1708-7
Hadley S, Wild-Allen K, Johnson C, Macleod C (2014) Modeling macroalgae growth and nutrient dynamics for integrated multi-trophic aquaculture. J Appl Phycol 27:901–916. https://doi.org/10.1007/s10811-014-0370-y
Han S, Li J, Zhou Q, Liu G, Wang T (2019) Harmless disposal and resource utilization of wastes from the lake in China: dewatering, composting and safety evaluation of fertilizer. Algal Res 43:101623. https://doi.org/10.1016/j.algal.2019.101623
Henrya S, Baudoinb E, López-Gutiérreza JC, Martin-Laurenta F, Braumanb A, Philippot L (2004) Quantification of denitrifying bacteria in soils by nirK gene targeted real-time PCR. J Microbiol Methods 59:327–335. https://doi.org/10.1016/j.mimet.2004.07.002
Hmani I, Ktari L, Ismail A, El Bour M (2023) Biotechnological potential of Ulva ohnoi epiphytic bacteria: enzyme production and antimicrobial activities. Front Mar Sci 10:1042527. https://doi.org/10.3389/fmars.2023.1042527
Hollants J, Leliaert F, Verbruggen H, Willems A, De Clerck O (2013) Permanent residents or temporary lodgers: characterizing intracellular bacterial communities in the siphonous green alga Bryopsis. Proc R Soc Ser B Biol Sci 280:20122659. https://doi.org/10.1098/rspb.2012.2659
Jasmina MY, Syukria F, Kamarudina MS, Karim M (2020) Potential of bioremediation in treating aquaculture sludge: review article. Aquaculture 519:734905. https://doi.org/10.1016/j.aquaculture.2019.734905
Lananan F, Hamid SHA, Din WNS, Ali N, Khatoon H, Jusoh A, Endut A (2014) Symbiotic bioremediation of aquaculture wastewater in reducing ammonia and phosphorus utilizing effective microorganism (EM-1) and microalgae (Chlorella sp.). Int Biodeterior Biodegrad 95:127–134. https://doi.org/10.1016/j.ibiod.2014.06.013
Lander TR, Robinson SMC, MacDonald BA, Martin JD (2013) Characterization of the suspended organic particles released from salmon farms and their potential as a food supply for the suspension feeder, Mytilus edulis in integrated multi-trophic aquaculture (IMTA) systems. Aquaculture 406–407:160–171. https://doi.org/10.1016/j.aquaculture.2013.05.001
Lawley B, Tannock GW (2017) Analysis of 16S rRNA gene amplicon sequences using the QIIME Software Package. Methods Mol Biol 1537:153–163. https://doi.org/10.1007/978-1-4939-6685-1_9
Li X, Zhang Z, Xie Q, Yang R, Guan T, Wu D (2019) Immobilization and release behavior of phosphorus on phoslock-inactivated sediment under conditions simulating the photic zone in eutrophic shallow lakes. Environ Sci Technol 53:12449–12457. https://doi.org/10.1021/acs.est.9b04093
Li X, Zhang Z, Xie Q, Wu D (2021) Effect of algae on phosphorus immobilization by lanthanum-modified zeolite. Environ Pollut 276:116713. https://doi.org/10.1016/j.envpol.2021.116713
Li S, Liao Y, Pang Y, Dong X, Strous M, Ji G (2022) Denitrification and dissimilatory nitrate reduction to ammonia in long-term lake sediment microcosms with iron(II). Sci Total Environ 807:150835. https://doi.org/10.1016/j.scitotenv.2021.150835
Liu T, Zhang AN, Wang J, Liu S, Jiang X, Dang C, Ma T, Liu S, Chen Q, Xie S, Zhang T, Ni J (2018) Integrated biogeography of planktonic and sedimentary bacterial communities in the Yangtze River. Microbiome 6:16. https://doi.org/10.1186/s40168-017-0388-x
Liu W, Tan H, Li S, Fu X, Luo G, Yu Y, Zhang N, Yao M (2019) Effects of C/N ratio on nitrogen removal with denitrification phase after a nitrification-based biofloc aquaculture cycle. Aquacult Eng 86:101994. https://doi.org/10.1016/j.aquaeng.2019.101994
Louca S, Parfrey LW, Doebeli M (2016) Decoupling function and taxonomy in the global ocean microbiome. Science 353:1272–1277. https://doi.org/10.1126/science.aaf4507
Lücker S, Wagner M, Maixner F, Pelletier E, Koch H, Vacherie B, Rattei T, Damsté JS, Spieck E, Paslier DL, Daims H (2010) A Nitrospira metagenome illuminates the physiology and evolution of globally important nitrite-oxidizing bacteria. PNAS 107:13479–13484. https://doi.org/10.1073/pnas.1003860107
Ma Y, Hu A, Yu CP, Yan Q, Yan X, Wang Y, Deng F, Xiong H (2015) Response of microbial communities to bioturbation by artificially introducing macrobenthos to mudflat sediments for in situ bioremediation in a typical semi-enclosed bay, southeast China. Mar Pollut Bull 94:114–122. https://doi.org/10.1016/j.marpolbul.2015.03.003
Macchiavello J, Bulboa C (2014) Nutrient uptake efficiency of Gracilaria chilensis and Ulva lactuca in an IMTA system with the red abalone Haliotis rufescens. Lat Am J Aquat Res 42:523–533. https://doi.org/10.3856/vol42-issue3-fulltext-12
Magoč T, Salzberg SL (2011) FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27:2957–2963. https://doi.org/10.1093/bioinformatics/btr507
Martínez-Porchas M, Martínez-Córdova LR, Porchas-Cornejo MA, López-Elías JA (2010) Shrimp polyculture: a potentially profitable, sustainable, but uncommon aquacultural practice. Rev Aquacult 2:73–85. https://doi.org/10.1111/j.1753-5131.2010.01023.x
Massocato TF, Robles-Carnero V, Moreira BR, Castro-Varela P, Pinheiro-Silva L, Oliveira WdS, Vega J, Avilés A, Bonomi-Barufi J, Rörig LR, Figueroa FL (2022) Growth, biofiltration and photosynthetic performance of Ulva spp. cultivated in fishpond effluents: an outdoor study. Front Mar Sci 9:981468. https://doi.org/10.3389/fmars.2022.981468
McTavish H, Fuchs JA, Hooper AB (1993) Sequence of the gene coding for ammonia monooxygenase in Nitrosomonas europaea. J Bacteriol 175:2436–2444. https://doi.org/10.1128/jb.175.8.2436-2444.1993
Mehta MP, Butterfield DA, Baross JA (2003) Phylogenetic diversity of nitrogenase (nifH) genes in deep-sea and hydrothermal vent environments of the Juan de Fuca Ridge. Appl Environ Microbiol 69:960–970. https://doi.org/10.1128/AEM.69.2.960-970.2003
Michalak I, Tiwari R, Dhawan M, Alagawany M, Farag MR, Sharun K, Emran TB, Dhama K (2022) Antioxidant effects of seaweeds and their active compounds on animal health and production – a review. Veterinary Quarterly 42:48–67. https://doi.org/10.1080/01652176.2022.2061744
Miles SF, David JH, Charles HC, Bernhard W, John D, Pat G (1998) A low power ultra violet spectrophotometer for measurement of nitrate in seawater: introduction, calibration and initial sea trials. Anal Chim Acta 377:167–177. https://doi.org/10.1016/s0003-2670(98)00616-3
Naldi M, Wheeler PA (2002) 15N measurement of ammonium and nitrate uptake by Ulva fenestrata (Chlorophyta) and Gracilaria pacifica (Rhodophyta): comparison of net nutrient disappearance, release of ammonium and nitrate, and 15N accumulationin algal tissue. J Phycol 38:135–144. https://doi.org/10.1046/j.1529-8817.2002.01070.x
Neori A, Chopin T, Troell M, Buschmann AH, Kraemer GP, Halling C, Shpigel M, Yarish C (2004) Integrated aquaculture: rationale, evolution and state of the art emphasizing seaweed biofiltration in modern mariculture. Aquaculture 231:361–391. https://doi.org/10.1016/j.aquaculture.2003.11.015
Oyarzúa P, Bovio-Winkler P, Etchebehere C, Suárez-Ojeda ME (2021) Microbial communities in an anammox reactor treating municipal wastewater at mainstream conditions: Practical implications of different molecular approaches. J Environ Chem Eng 9:106622. https://doi.org/10.1016/j.jece.2021.106622
Pai SC, Tsau YJ, Yang TI (2001) pH and buffering capacity problems involved in the determination of ammonia in saline water using the indophenol blue spectrophotometric method. Anal Chim Acta 434:209–216. https://doi.org/10.1016/s0003-2670(01)00851-0
Pereira C, Santos L, Silva AP, Silva YJ, Cunha A, Romalde JL, Nunes ML, Almeida A (2015) Seasonal variation of bacterial communities in shellfish harvesting waters: preliminary study before applying phage therapy. Mar Pollut Bull 90:68–77. https://doi.org/10.1016/j.marpolbul.2014.11.019
Pester M, Maixner F, Berry D, Rattei T, Lücker S, Nowka B, Daims H (2014) NxrB encoding the beta subunit of nitrite oxidoreductase as functional and phylogenetic marker for nitrite-oxidizing Nitrospira. Environ Microbiol 16:3055–3071. https://doi.org/10.1111/1462-2920.12300
Ramanan R, Kim BH, Cho DH, Oh HM, Kim HS (2016) Algae-bacteria interactions: evolution, ecology and emerging applications. Biotechnol Adv 34:14–29. https://doi.org/10.1016/j.biotechadv.2015.12.003
Rawat I, Ranjith Kumar R, Mutanda T, Bux F (2011) Dual role of microalgae: phycoremediation of domestic wastewater and biomass production for sustainable biofuels production. Appl Energy 88:3411–3424. https://doi.org/10.1016/j.apenergy.2010.11.025
Robertson EK, Roberts KL, Burdorf LDW, Cook P, Thamdrup B (2016) Dissimilatory nitrate reduction to ammonium coupled to Fe(II) oxidation in sediments of a periodically hypoxic estuary. Limnol Oceanogr 61:365–381. https://doi.org/10.1002/lno.10220
Rubio-Portillo E, Santos F, Martinez-Garcia M, de Los Rios A, Ascaso C, Souza-Egipsy V, Ramos-Espla AA, Anton J (2016) Structure and temporal dynamics of the bacterial communities associated to microhabitats of the coral Oculina patagonica. Environ Microbiol 18:4564–4578. https://doi.org/10.1111/1462-2920.13548
Rurangwa E, Verdegem MCJ (2014) Microorganisms in recirculating aquaculture systems and their management. Rev Aquacult 5:1–14. https://doi.org/10.1111/raq.12057
Samocha TM, Fricker J, Ali AM, Shpigel M, Neori A (2015) Growth and nutrient uptake of the macroalga Gracilaria tikvahiae cultured with the shrimp Litopenaeus vannamei in an integrated multi-trophic aquaculture (IMTA) system. Aquaculture 446:263–271. https://doi.org/10.1016/j.aquaculture.2015.05.008
Santizo-Taan R, Bautista-Teruel M, Maquirang JRH (2019) Enriched Ulva pertusa as partial replacement of the combined fish and soybean meals in juvenile abalone Haliotis asinina (Linnaeus) diet. J Appl Phycol 32:741–749. https://doi.org/10.1007/s10811-019-01977-5
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. https://doi.org/10.1128/AEM.01541-09
Shahar B, Guttman L (2021) Integrated biofilters with Ulva and periphyton to improve nitrogen removal from mariculture effluent. Aquaculture 532:736011. https://doi.org/10.1016/j.aquaculture.2020.736011
Shpigel M, Shauli L, Odintsov V, Ashkenazi N, Ben-Ezra D (2018) Ulva lactuca biofilter from a land-based integrated multi trophic aquaculture (IMTA) system as a sole food source for the tropical sea urchin Tripneustes gratilla elatensis. Aquaculture 496:221–231. https://doi.org/10.1016/j.aquaculture.2018.06.038
Sigman DM, Hain MP (2012) The biological productivity of the ocean. Nat Educ 3:21
Stackebrandt E, Goebel BM (1994) Taxonomic note: a place for DNA-DNA reassociation and 16s rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol 44:846–849. https://doi.org/10.1099/00207713-44-4-846
Troell M, Joyce A, Chopin T, Neori A, Buschmann AH, Fang JG (2009) Ecological engineering in aquaculture — potential for integrated multi-trophic aquaculture (IMTA) in marine offshore systems. Aquaculture 297:1–9. https://doi.org/10.1016/j.aquaculture.2009.09.010
Urakawa H, Rajan S, Feeney ME, Sobecky PA, Mortazavi B (2019) Ecological response of nitrification to oil spills and its impact on the nitrogen cycle. Environ Microbiol 21:18–33. https://doi.org/10.1111/1462-2920.14391
Uthicke S (1999) Sediment bioturbation and impact of feeding activity of Holothuria (Halodeima) atra and Stichopus chloronotus, two sediment feeding holothurians, at Lizard Island, Great Barrier Reef. Bull Mar Sci 64:129–141
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267. https://doi.org/10.1128/AEM.00062-07
Wang Q, Zhu R, Zheng Y, Bao T, Hou L (2019) Effects of sea animal colonization on the coupling between dynamics and activity of soil ammonia-oxidizing bacteria and archaea in maritime Antarctica. Biogeosciences 16:4113–4128. https://doi.org/10.5194/bg-16-4113-2019
Welsh A, Chee-Sanford JC, Connor LM, Löffler FE, Sanford RA (2014) Refined NrfA phylogeny improves PCR-based nrfA gene detection. Appl Environ Microbiol 80:2110–2119. https://doi.org/10.1128/AEM.03443-13
Xu N, Tan G, Wang H, Gai X (2016) Effect of biochar additions to soil on nitrogen leaching, microbial biomass and bacterial community structure. Eur J Soil Biol 74:1–8. https://doi.org/10.1016/j.ejsobi.2016.02.004
Yang Y, Chai Z, Wang Q, Chen W, He Z, Jiang S (2015) Cultivation of seaweed Gracilaria in Chinese coastal waters and its contribution to environmental improvements. Algal Res 9:236–244. https://doi.org/10.1016/j.algal.2015.03.017
Yang T, Xin Y, Zhang L, Gu Z, Li Y, Ding Z, Shi G (2020) Characterization on the aerobic denitrification process of Bacillus strains. Biomass and Bioenergy 140:105677. https://doi.org/10.1016/j.biombioe.2020.105677
Yao R, Li H, Yang J, Zhu W, Yin C, Wang X, Xie W, Zhang X (2022) Combined application of biochar and N fertilizer shifted nitrification rate and amoA gene abundance of ammonia-oxidizing microorganisms in salt-affected anthropogenic-alluvial soil. Appl Soil Ecol 171:104348. https://doi.org/10.1016/j.apsoil.2021.104348
Yin G, Hou L, Jiang X, Liu M, Wang R, Li X, Zheng Y, Yu C, Lin X (2017) DNRA in intertidal sediments of the Yangtze Estuary. J Geophys Res 122:1–11. https://doi.org/10.1002/2017JG003766
Zehr JP, Short SM, Jenkins BD, Steward GF (2003) Nitrogenase gene diversity and microbial community structure: a cross-system comparison. Environ Microbiol 5:539–554. https://doi.org/10.1046/j.1462-2920.2003.00451.x
Zhang Q-L, Liu Y, Ai G-M, Miao L-L, Zheng H-Y, Liu Z-P (2012) The characteristics of a novel heterotrophic nitrification-aerobic denitrification bacterium, Bacillus methylotrophicus strain L7. Bioresour Technol 108:35–44. https://doi.org/10.1016/j.biortech.2011.12.139
Zhang X, Song Y, Liu D, Keesing JK, Gong J (2015) Macroalgal blooms favor heterotrophic diazotrophic bacteria in nitrogen-rich and phosphorus-limited coastal surface waters in the Yellow Sea. Estuarine Coastal Shelf Sci 163:75–81. https://doi.org/10.1016/j.ecss.2014.12.015
Zhang L, He K, Wang T, Liu C, An Y, Zhong J (2022) Frequent algal blooms dramatically increase methane while decrease carbon dioxide in a shallow lake bay. Environ Pollut 312:120061. https://doi.org/10.1016/j.envpol.2022.120061
Zhao Y, Huang J, Zhao H, Yang H (2013) Microbial community and N removal of aerobic granular sludge at high COD and N loading rates. Bioresour Technol 143:439–446. https://doi.org/10.1016/j.biortech.2013.06.020
Zhou J, Chen Z, Li S (2007) Oxidation efficiency of different oxidants of persulfate method used to determine total nitrogen and phosphorus in solutions. Commun Soil Sci Plant Anal 34:725–734. https://doi.org/10.1081/css-120018971
Zhou Y, Zhou Y, Chen S, Guo N, Xiang P, Lin S, Bai Y, Hu X, Zhang Z (2022) Evaluating the role of algae in algal-bacterial granular sludge: nutrient removal, microbial community and granular characteristics. Bioresource Technology 365:128165. https://doi.org/10.1016/j.biortech.2022.128165
Acknowledgements
Thank Qingdao Ruizi Co., Ltd. for providing the research site and services. We acknowledge the sequencing service from the Majorbio Company (Shanghai, China).
Funding
This study was supported by the National Key Research and Development Program of China (No. 2023YFD2401704), China Agriculture Research System (No. CARS-48), and Central Public-interest Scientific Institution Basal Research Fund, CAFS (No. 2023TD50).
Author information
Authors and Affiliations
Contributions
Shuo Kong wrote the main manuscript text. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
Husbandry and experimental procedures were performed in accordance with research protocols which approved by the Institutional Animal Care and Use Committee, Yellow Sea Fisheries Research Institute, China.
Conflict of interest
The authors declare no competing interests.
Additional information
Handling Editor: Ronan Sulpice
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kong, S., Ghonimy, A., Chen, Z. et al. Ulva lactuca changed bacteria community structure and enhanced nitrogen removal capability in a shrimp-sea cucumber-crab-algae integrated multi-trophic aquaculture (IMTA) system. Aquacult Int (2024). https://doi.org/10.1007/s10499-024-01598-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10499-024-01598-x