Abstract
Vibrio parahaemolyticus is a gram-negative motile bacterium inhabiting marine, estuarine, and coastal environments. It is a critical aquatic pathogen in shrimp farming, and strains possessing pir A and pir B toxins can cause acute hepatopancreatic necrosis disease (AHPND) in shrimps. The study aimed to isolate V. parahaemolyticus from the gut and hepatopancreas of shrimps from the selected districts of Tamil Nadu for a duration of 6 months from December 2022 to May 2023 by random sampling, and thirty-two strains were confirmed as V. parahaemolyticus from 110 presumptive bacterial isolates. All 32 isolates used in the AP4 PCR protocol failed to detect any isolates carrying AHPND pir toxins (pir A and pir B). Forty-four percent of isolates have shown β-haemolytic activity on blood agar. Of the 32 isolates, two, eleven, and twenty-nine harboured tdh, trh, and T3SS1 genes, respectively, and none of them possessed T3SS2 gene. Isolates of V. parahaemolyticus were resistant to gentamycin, vancomycin, and erythromycin and highly susceptible to ciprofloxacin, ofloxacin, chloramphenicol, amoxyclav, trimethoprim, streptomycin, cefoxitin, and ceftazidime. AMR genes encoding qnrA, tet A, blaSHV, and aac-3-IIa were present in 6%, 16%, 16%, and 22% of the isolates. AMR genes catA1 and str B were negative for all 32 isolates. It is concluded that the prevalence and incidence of V. parahaemolyticus were 29% in the studied coastal districts of Tamil Nadu. All isolates were found to be negative for AHPND. Hence, shrimp farms in the studied area were free from infection of AHPND.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In aquaculture, crustaceans are economically significant and monumental in demand for products with high unit value. Crustacean production has risen continuously from 2000 onwards with a growth rate of 9.9%/year and reached 8.4 MT with a value of USD 61.06 billion (FAO 2022). In crustaceans, Pacific white-leg shrimp (Penaeus vannamei) is a more popular and demanding product with a unit value of USD 26.7 billion, always making first place on the table in both quality and quantity (Kumar et al. 2021). India contributes significantly to global shrimp aquaculture output, and just one product accounts for more than 70% of the nation’s whole value of seafood exports (Navaneeth et al. 2020). In Indian shrimp culture production, P. vannamei is a consequential attribute as a noteworthy contributor. With rising demand, the shrimp culture industry intensified, and high scaling growth increased the risk of disease exposure to aquaculture enterprises. It resulted in socioeconomic loss and hindered the shrimp culture industry’s growth. Over the past few years, the emergence of acute hepatopancreatic necrosis disease (AHPND) has inflicted severe economic losses on the global shrimp farming industry. AHPND is caused by Vibrio parahaemolyticus with a particular strain containing two Photorhabdus insect–related (Pir) toxins, pir A and pir B. A bacterial disease known as AHPND has washed out farmed populations of Pacific white-leg shrimp and black tiger shrimp (P. monodon) (Tang and Bondad-Reantaso 2019). The first outbreak of AHPND originated in China in 2009, and it gradually spread among other Asian countries, including Vietnam (2010), Malaysia in 2011, Thailand in 2012, the Philippines in 2015, Mexico in 2013, and South America (2016) (Ananda Raja et al. 2017b; Muthukrishnan et al. 2019). Within 25–30 days of stocking PL, mortality started, and occasionally, mass mortality was also observed 10 days after PL stocking. As a result of these characteristics, this disease was previously called “early mortality syndrome” (EMS). Approximately 60% of shrimp production declined in the AHPND-affected areas compared with the 2012 production data and an annual economic loss of USD 1 billion globally (FAO 2013).
V. parahaemolyticus is a gram-negative halophilic, tiny rod and motile bacterium belonging to the Vibrionaceae family inhabiting marine, estuarine, and coastal environments (Su and Liu 2007; Wang et al. 2011; Zhang and Orth 2013). It has two major virulence systems: haemolysin and type III secretion system. Haemolysin comprises tdh, trh, and tlh, while T3SS is T3SS1, T3SS2α, and T3SS2β. The virulence factors responsible for V. parahaemolyticus’ haemolysis and cytotoxicity in host cells are tdh and trh (Broberg et al. 2011). V. parahaemolyticus possesses two distinct sets of type III secretion systems (T3SSs) situated on two separate chromosomes: T3SS1 on chromosome 1 and T3SS2 on chromosome 2 (Calder et al. 2014). T3SS1 primarily plays a role in cytotoxicity, whereas T3SS2 causes enterotoxicity (Hiyoshi et al. 2011). Such virulence genes are responsible for the pathogenicity of V. parahaemolyticus.
The study aims to isolate V. parahaemolyticus from the gut and hepatopancreas of shrimps from the selected districts of Tamil Nadu and screen the isolates for the occurrence of virulence genes with special reference to APHND pir toxin and the antimicrobial resistance genes.
Materials and methods
Sample collection and isolation of bacteria
Samples were collected from shrimp farms in four coastal districts of Tamil Nadu (Nagapattinam, Pudukkottai, Cuddalore, and Ramanathapuram) from December 2022 to May 2023 (Fig. 1). P. vannamei was specifically selected for this study due to its present share as highly cultured shrimp in India. All samples were aseptically collected and transported in chilled conditions using a thermally insulated box within 24 h to the bacteriology laboratory, Department of Fish Pathology and Health Management, Fisheries College and Research Institute Thoothukudi for further analysis.
The hepatopancreas and gut of shrimps were dissected aseptically for all the samples and streaked on thiosulfate citrate bile salt sucrose (TCBS) agar (HiMedia, Mumbai) plates using a sterile loop. Plates were incubated at 32 °C overnight. Presumptive green colonies on TCBS agar plates were again streaked and purified on TSAS (trypticase soy agar with 2% salt) (HiMedia, Mumbai) plates (Canizalez-Roman et al. 2011). Pure bacterial isolates were preserved in TSBS (tryptic soy broth with 2% salt) (HiMedia, Mumbai) containing 20% glycerol at − 80 °C (Fadanka et al. 2022).
Phenotypic characterization of bacteria
Phenotypic characterization and identification of bacterial isolates were performed using the standard protocol (Brenner et al. 2005; Ananda Raja et al. 2017a, c). A series of tests were carried out, such as gram stain, motility, catalase, oxidase, citrate, urease, Voges and Proskauer (VP), methyl red (MR), sugar fermentation (glucose, arabinose, sucrose, lactose, raffinose, galactose, rhamnose), decarboxylase (arginine, ornithine, lysine), and growth at 0, 3, 6, and 8% NaCl concentrations. To identify V. parahaemolyticus, all test media were enriched with 2% NaCl, and the experiments were duplicated and then incubated at 32 °C overnight.
DNA extraction from bacteria
Pure cultures of bacterial isolates were inoculated into 5 ml of TSBS and incubated for 24 h at 32 °C. After 24 h, 1 ml of culture was taken aseptically and centrifuged at 8000 × g for 10 min, and the bacterial pellet was obtained by discarding the supernatant. Two hundred microliters of DNA-XPress Reagent (HiGenoMB, HiMedia, Mumbai) was added to the bacterial pellet. The mixture was thoroughly mixed and incubated at 60 °C for 1 h. It was then centrifuged at 10,000 × g for 10 min, and the supernatant was carefully transferred to a fresh sterile tube. An equivalent volume of absolute alcohol (200 µl) was added to the supernatant, followed by centrifugation at 10,000 × g for 5 min. The supernatant was discarded, and the DNA pellet was washed twice with 200 µl of 95% alcohol, followed by centrifugation at 8000 × g for 5 min each time. After removing the supernatant, the DNA pellet was allowed to air dry for 5 min, and 75 µl of nuclease-free water was added and stored at 4 °C for further use.
Detection of virulence and AMR genes by polymerase chain reaction (PCR)
The PCR amplification of all reactions was carried out using a thermal cycler (Bio-Rad, USA) in a 25 µl reaction mixture with 2 µl each forward and reverse primer, 6.5 µl PCR grade water, 12.5 µl master mix, and 2 µl template DNA. The bacterial isolates were confirmed as V. parahaemolyticus following standard protocol and previously published primer VpM, toxR, and GyrB (Bauer and Rørvik 2007; Luan et al. 2007), as in Table 1. The PCR amplification of putative virulence genes of V. parahaemolyticus such as thermostable direct haemolysin (tdh), tdh-related haemolysin (trh), thermolabile haemolysin (tlh), type III secretion system one (T3SS1), type III secretion system two alpha (T3SS2α), and type III secretion system two beta (T3SS2β) was carried out using appropriate primers and protocols as mentioned in Table 1. Detection of antimicrobial resistance genes encoding resistance to β-lactams (blaSHV), aminoglycosides (aac-3-IIa), chloramphenicol (catA1), streptomycins (str B), quinolones (qnrA), and tetracyclines (tet A) was done as per standard primers listed in Table 1. All 32 confirmed V. parahaemolyticus isolates were screened for the presence of virulence and AMR genes enlisted above. In a similar way, the confirmed V. parahaemolyticus isolates were tested for AHPND using the standard AP4 protocol (Dangtip et al. 2015), cited in Table 1. Shrimps were screened for the presence of WSSV, IHHNV, TSV, IMNV, YHV, and DIV 1 as per the WOAH diagnostic manual to get a specific study on AHPND and V. parahaemolyticus without any association of any other bacterial/viral disease.
Determination of haemolytic activity
Blood agar plates (HiMedia, Mumbai) were used for the haemolytic activity. Plates were inoculated with a sterile loop from 16 h young bacterial cultures. After inoculation, plates were incubated for 24 h at 32 °C.
Antibiotic susceptibility test
The standard disc diffusion method (Bauer et al. 1966) was used to study the antibiotic susceptibility of V. parahaemolyticus isolates. According to the Kirby-Bauer method, plates were prepared with Mueller Hinton agar (MHA) (HiMedia, Mumbai). The purified bacterial isolates were inoculated in TSBS and incubated at 32 °C for 8 h until light to moderate turbidity developed. Inoculum turbidity was compared with standard 0.5 McFarland (0.5 ml 1.175% barium chloride and 99.5 ml 0.36 N sulphuric acid) and adjusted to 1 Optical Density (Ananda Raja et al. 2017a). A sterile, non-toxic cotton swab on a wooden applicator was dipped into an OD-adjusted inoculum containing a young bacterial culture. The entire agar surface of the MHA plate was streaked with a swab three times, and the plate was turned at a 60° angle between each streaking to obtain complete lawn culture. The plates with inoculum were allowed to dry for 10 min with the lid in place. Antibiotic discs (HiMedia, Mumbai) were placed aseptically with at least 30 mm apart centres. The plates were incubated at 32 °C overnight and examined for the size of the inhibition zone (Ananda Raja et al. 2017a). Fourteen antibiotics of 11 different classes were used for this study such as β lactam—amoxyclav (25 μg); second-generation cephalosporin—cefoxitin (30 μg); third-generation cephalosporin—ceftazidime (30 μg); fourth-generation cephalosporins—cefepime (30 μg); aminoglycosides—gentamycin (30 μg), streptomycin (10 μg), and kanamycin (5 μg); quinolones and fluoroquinolones—ciprofloxacin (30 μg) and ofloxacin (5 μg); tetracyclines—oxytetracycline (30 μg); sulphonamides—trimethoprim-sulphamethoxazole (25 μg); phenicol—chloramphenicol (30 μg); glycopeptides—vancomycin (30 μg); and macrolides—erythromycin (10 μg). The inhibition zone diameter (millimeter) was recorded and interpreted according to the Clinical and Laboratory Standard Procedure (CLSI) 2020 guidelines. The multiple antibiotic or drug resistance (MAR) was also calculated based on the procedure (Krumperman 1983). A heatmap showing an antibiogram was prepared using OriginPro 2023b.
Results
Prevalence, identification, and characterization of V. parahaemolyticus
Over a period of 6 months from December 2022 to May 2023, a total of 67 shrimp samples were collected from four coastal districts of Tamil Nadu (Nagapattinam, Pudukkottai, Cuddalore, and Ramanathapuram) and processed for the isolation of V. parahaemolyticus. One hundred ten bacterial isolates were obtained from 67 shrimp samples using the conventional method (TCBS agar) and subsequently confirmed for V. parahaemolyticus using biochemical and molecular methods.
Phenotypic tests demonstrated that all V. parahaemolyticus isolates were gram-negative, rod-shaped motile, catalase, oxidase, urease positive and glucose, arabinose, galactose fermentative whereas non-fermentative for sucrose, lactose, and raffinose.
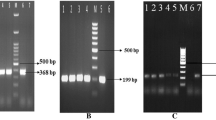
Following biochemical and PCR tests, 32 of 110 isolates were found to be positive for V. parahaemolyticus by primer VpM, toxR, and GyrB (Fig. 2). Of the confirmed 32 V. parahaemolyticus strains, 9 were isolated from the gut, and the remaining were from the hepatopancreas.
V. parahaemolyticus confirmation by amplification of different gene-specific primers such as GyrB, VpM, and toxR. a GyrB lanes—M, molecular weight marker; 1–5, representative isolates; P, positive control; N: negative control. b toxR lanes—M, molecular weight marker, 1–4: representative isolates; P, positive control; N, negative control. c VpM lanes—M, molecular weight marker, 1–13: representative isolates; P, positive control; N, negative control
PCR detection of virulence, AMR genes, and AHPND
Results of haemolysin virulence genes showed that 2 (6%) and 11 (34%) of V. parahaemolyticus isolates possessed tdh and trh genes, while all the 32 (100%) isolates harboured tlh gene (Fig. 3). Regarding the type III secretion system, the T3SS1 gene was present in 29 (91%) isolates (Fig. 3); none of the 32 possessed the T3SS2 gene (both T3SS2α and T3SS2β). AMR genes such as tetracycline (tet A), beta-lactam (blaSHV), chloramphenicol (catA1), streptomycin (str B), aminoglycosides (aac-3-IIa), and quinolinones (qnrA) were studied and 2 (6%), 5 (16%), 5 (16%), and 7 (22%) isolates carried qnrA, tet A, blaSHV, and aac-3-IIa genes, respectively. However, genes catA1 and str B were negative for all 32 isolates.
Amplification of virulence genes of V. parahaemolyticus such as tdh, tlh, trh, and T3SS1. A tdh lanes—M, molecular weight marker; 1–5, representative isolates; P, positive control; N, negative control. B tlh lanes—M, molecular weight marker; 1–9, representative isolates; P, positive control; N, negative control. C trh lanes—M, molecular weight marker, 1–12, representative isolates; P, positive control; N, negative control. D T3SS1 lanes—M, molecular weight marker; 1–17, representative isolates; P, positive control; N, negative control
All the confirmed V. parahaemolyticus isolates were screened for AHPND pir toxins to check the presence of acute hepatopancreatic necrosis disease (AHPND) using AP4 protocol. AHPND pir toxin was absent in all the 32 V. parahaemolyticus studied. WOAH-listed diseases such as WSSV, IHHNV, TSV, IMNV, YHV, and DIV 1 were screened for their presence, and all the samples were tested negative for enlisted diseases by PCR.
Determination of haemolytic activity
The haemolytic activity on blood agar was studied according to the protocol of Twedt et al. (1970), and isolates were recorded as β-haemolytic positive with a clear zone around the colony. β-Haemolysis was observed in 14 (44%) isolates of V. parahaemolyticus in the present study.
Antimicrobial susceptibility of V. parahaemolyticus
An antibiotic susceptibility test was done for all 32 V. parahaemolyticus isolates, for 14 antibiotics belonging to 11 classes such as β-lactams, 2nd-, 3rd-, and 4th-generation cephalosporins, aminoglycosides, quinolones, fluoroquinolones, tetracyclines, sulphonamides, phenicol, glycopeptides, and macrolides Table 2. The resistance pattern of V. parahaemolyticus toward antibiotics given in Table 3. Figure 4 shows a heatmap of the antibiogram depicting resistance of antibiotics in a colour scale from dark blue to dark red and categorized into resistant, susceptible, and intermediate.
MAR index
Multiple antibiotic resistance (MAR) index was estimated for all isolates (Table 3), and the findings revealed that the MAR of 32% of the isolates was less than 0.2. In comparison, a significant portion, 68% of the isolates, had a MAR index range from 0.21 to 0.40. The MAR index was as high as 0.36 in one isolate that was resistant to five antibiotics (VA-CPM-CAZ-E-GEN).
Discussion
V. parahaemolyticus is a gram-negative motile bacterium inhabiting marine, estuarine, and coastal environments (Baffone et al. 2006; Wang et al. 2011). The study results of Caburlotto et al. (2016) and Coly et al. (2013) reported that the prevalence of V. parahaemolyticus was 28% and 30% from crustaceans in Italy and seafood in Senegal, respectively. However, other studies have found wide variations in prevalence values, ranging from 2.8% in a survey on crabs and prawns sold in Abidjan, Ivory Coast (Traorã et al. 2012) to 50.5% in frozen shrimp, prawns, and crabs bought in local markets or from import–export businesses in Boulogne-sur-Mer, France (Robert-Pillot et al. 2014). Pathogenic V. parahaemolyticus was found in Thailand, the world’s leading producer and exporter of farmed shrimps (Yano et al. 2014). In our study, 32 V. parahaemolyticus isolates were confirmed with a 29% prevalence from P. vannamei shrimp farms of studied coastal districts in Tamil Nadu, which showed relatively same prevalence rate reported by Coly et al. (2013) and Caburlotto et al. (2016).
Because members of the family Vibrionaceae belong to the monophyletic group (clades) that are closely related (Sawabe et al. 2013), it becomes challenging to differentiate different species among Vibrio spp. The genes (VpM, toxR, and GyrB) targeting V. parahaemolyticus were chosen and tested for all isolates, and those that tested positive were confirmed to be V. parahaemolyticus.
The virulence factors linked to V. parahaemolyticus haemolysis and cytotoxicity activity in the host cell include tdh and tdh-related haemolysin (trh) (Broberg et al. 2011). The presence of thermostable direct haemolysin (tdh) and/or tdh-related haemolysin (trh) indicate the human pathogenic nature of V. parahaemolyticus isolates (Tada et al. 1992). The pathogenic genes tdh and trh, which cause diseases in human and marine animals, are often absent from the strains obtained from environmental samples (Deepanjali et al. 2005). However, a previous study showed that environmental sources had a smaller percentage (1–2%) of human pathogenic strains (Sakazaki et al. 1968). In the present research, out of a total of 32 isolates, 2 (6%) and 11 (34%) isolates harboured the tdh gene and trh genes, respectively, which showed the pathogenic nature of the isolates obtained from shrimp samples and is a supreme concern for aquatic animals and human safety.
Another haemolysin of V. parahaemolyticus, thermolabile haemolysin (tlh), is likewise responsible for the lysis of red blood cells and is encoded by the tlh gene (Shinoda et al. 1991). All clinical and environmental strains of V. parahaemolyticus express tlh (Bej et al. 1999). The gene is markedly increased when the intestinal infection is reproduced (Gotoh et al. 2010). All isolates harboured the tlh gene in our study, indicating that the tlh gene could be considered a biomarker for identifying V. parahaemolyticus.
Isolates were further screened for other virulence genes of V. parahaemolyticus, such as T3SS genes. Two T3SS genes such as T3SS1 and T3SS2 are present in V. parahaemolyticus, on chromosomes 1 and 2, respectively (Calder et al. 2014). T3SS1 primarily causes cytotoxicity, and T3SS2 causes enterotoxicity (Hiyoshi et al. 2011). Ninety-one percent of the total isolates in our study harboured the T3SS1 gene; however, all isolates were negative for T3SS2 genes (both T3SS2α and T3SS2β). Therefore, it could be concluded that V. parahaemolyticus isolates were pathogenic but would cause cytotoxicity rather than enterotoxicity.
The bottom line of this study was to conduct a surveillance of AHPND along the southeast coast of India, specifically on the Tamil Nadu coast. All 32 confirmed V. parahaemolyticus isolates were investigated for AHPND pir toxins. However, the 32 isolates studied using AP4 PCR failed to detect any isolates carrying pir toxins. Recent studies in India regarding AHPND showed the absence of AHPND in India (Ananda Raja et al. 2017b, 2023; Kumar et al. 2020; Navaneeth et al. 2020). Hence, the current research confirmed that AHPND was not present in the studied locations during the sampling period.
The haemolytic activity is one of the essential virulence factors exhibited by V. parahaemolyticus phenotypically (Twedt et al. 1970). Forty-four percent of V. parahaemolyticus isolates in our study were β-haemolytic. It pinpointed the virulence nature and potency of the isolates in terms of haemolytic action.
The V. parahaemolyticus isolates in the present study were highly resistant to erythromycin and vancomycin, which was consistent with results obtained in the earlier studies (Hamdan et al. 2017; Hu et al. 2020; Elsherif et al. 2023). In our research, isolates showed 72% resistance towards gentamycin among the aminoglycosides group, while previous studies (Lee et al. 2018; Kim et al. 2021; Zhou et al. 2022; Elsherif et al. 2023) showed contrasting results against gentamycin, and were found to be susceptible. The resistance of many antibiotics has been transferred from sensitive to resistant mode with time and uncountable usage. Hence, there are many chances that antibiotics could be sensitive and become resistant one day with continuous use over time.
All the isolates were susceptible to ciprofloxacin, ofloxacin, and chloramphenicol, and it could be understood that these antibiotics are highly effective against V. parahaemolyticus. Similarly, previous studies have shown that the susceptibility of V. parahaemolyticus isolates against chloramphenicol, ofloxacin, and ciprofloxacin is natural (Xu et al. 2016; Lee et al. 2018; Ryu et al. 2019). Since these are the most frequently used antibiotics to treat infections caused by Vibrio spp. (Letchumanan et al. 2014), this finding was satisfactory. Streptomycin resistance was commonly observed in the isolates of V. parahaemolyticus, and the resistance which could be due to the everyday use of antibiotics (Kim et al. 2021). This could be the reason why most of the studies showed resistance towards streptomycin (Jiang et al. 2014; Xu et al. 2016; Ryu et al. 2019; Xie et al. 2020; Elsherif et al. 2023). However, in the present study, the results were contrasting regarding streptomycin, and 84% were sensitive. As discussed earlier, resistance to particular antibiotics is variable as the usage and choice of antibiotics vary from place to place. Some investigations had reported that resistance to trimethoprim-sulphamethoxazole was 75%, 38.2%, and 64.7% in V. parahaemolyticus (Ahmed et al. 2018; Zhou et al. 2022; Elsherif et al. 2023). In contrast, some studies showed that V. parahaemolyticus was 80%, 78.22%, 97.7%, and 100% susceptible to trimethoprim-sulphamethoxazole (Jiang et al. 2014; Lee et al. 2018; Ryu et al. 2019; Li et al. 2020). In the present study, V. parahaemolyticus isolates were sensitive (94%) to trimethoprim-sulphamethoxazole. Hence, our study’s findings align with those of earlier investigations. This observed variation might be due to variations in contamination of source water from place to place, since usage of any antibiotics in shrimp aquaculture is strictly prohibited in India with good awareness among the farmers (Ananda Raja et al. 2012). V. parahaemolyticus isolates from this study showed 75% and 82% susceptibility to second-generation cephalosporin (cefoxitin) and third-generation cephalosporin (ceftazidime) as well as 59% intermediate levels of resistance and 38% sensitive to a fourth-generation cephalosporin (cefepime). The earlier investigations had reported the same results (Jiang et al. 2014; Lee et al. 2018; Elhadi et al. 2022; Zhou et al. 2022). At the same time, some researchers have shown contradictory findings, such as pathogenic V. parahaemolyticus isolated from AHPND-affected P. vannamei farmed in the Mekong Delta exhibited resistance against the ceftazidime 100% (Ha et al. 2023). A total of 97.2% resistance of ceftazidime against V. parahaemolyticus isolated from shrimp, crab, and gastroenteritis patients was reported in Egypt (Ahmed et al. 2018). In the present research, 75% of V. parahaemolyticus showed intermediate resistance towards kanamycin. A similar finding was reported in which 92% and 74.5% of intermediate resistance of V. parahaemolyticus to kanamycin were observed, isolates from sea cucumber and aquatic products, respectively (Jiang et al. 2014; Xu et al. 2016). Antibiogram revealed that isolates were 50% sensitive and 50% intermediate resistant to oxytetracycline. Seventy-two percent susceptibility of V. parahaemolyticus was reported to oxytetracycline from fish samples (Lee et al. 2018). Isolates of V. parahaemolyticus in our study were 91% susceptible to amoxyclav. As per earlier studies, V. parahaemolyticus isolates were also demonstrated to be 75%, 95.4%, 95.7%, and 75.83% sensitive against amoxyclav (Jiang et al. 2014; Ryu et al. 2019; Tan et al. 2020; Elhadi et al. 2022).
Multiple antibiotic or drug resistance (MAR) was calculated according to Krumperman (1983). The present study showed that 32% of the isolates had less than 0.2 MAR index value. In the study, 70% and 76.59% of V. parahaemolyticus isolates had MAR index values lower than 0.2 (Ryu et al. 2019; Elhadi et al. 2022). A significant section of isolates (65%) in the present study had a MAR index within the range of 0.21 to 0.30. The MAR index was as high as 0.36 in one isolate that was resistant to five antibiotics. The MAR index ranges from 0.07 to 0.50 in V. parahaemolyticus isolated in marine and freshwater fish in Selangor (Lee et al. 2018). High values of the MAR index, such as 0.77 and 0.80, were observed in V. parahaemolyticus strains isolated from crustaceans and humans in Egypt and retail aquatic products in Nanjing, China, respectively (Ahmed et al. 2018; Zhou et al. 2022). The fluctuation in the MAR index may be due to variations in sample sources, geographic dispersion, and testing procedures.
Conclusion
Determining the virulence potency to cause severe diseases in humans and shrimps and the antimicrobial resistance of V. parahaemolyticus isolates such study is foremost essential. The present study highlighted the characterization and prevalence of V. parahaemolyticus, associated virulence genes, AMR genes, and anti-microbial resistance with AHPND surveillance in four coastal districts of Tamil Nadu. The most important aspect of this study proved that India is free from the infection of AHPND in addition to earlier studies.
Data availability
Data will be made available on request.
References
Ahmed HA, El Bayomi RM, Hussein MA et al (2018) Molecular characterization, antibiotic resistance pattern and biofilm formation of Vibrio parahaemolyticus and V. cholerae isolated from crustaceans and humans. Int J Food Microbiol 274:31–37
Ananda Raja R, Panigrahi A, Kumar S (2012) Epidemiological investigation of brackish water culture systems in West Bengal. India Journal of Applied Aquaculture 24(1):49–59
Ananda Raja R, Panigrahi A, Debasis D, Sujeet K (2017a) Investigations on white spot disease outbreak in Penaeus monodon (Fabricius, 1798) in association with Vibrio mimicus infection in the Sunderbans, West Bengal. India Indian Journal of Fisheries 64(1):56–60
Ananda Raja R, Sridhar R, Balachandran C, Palanisammi A, Ramesh S, Nagarajan K (2017b) Pathogenicity profile of Vibrio parahaemolyticus in farmed Pacific white shrimp, Penaeus vannamei. Fish Shellfish Immunol 67:368–381. https://doi.org/10.1016/j.fsi.2017.06.020
Ananda Raja R, Sridhar R, Balachandran C, Palanisammi A, Ramesh S, Nagarajan K (2017c) Prevalence of Vibrio spp. with special reference to Vibrio parahaemolyticus in farmed penaeid shrimp Penaeus vannamei (Boone, 1931) from selected districts of Tamil Nadu, India India. Indian Journal of Fisheries 64(3):122–128
Ananda Raja R, Sridhar R, Balachandran C, Palanisammi A, Ramesh S, Nagarajan K (2023) Susceptibility of farmed Pacific white shrimp, Penaeus vannamei to non-AHPND strain of Vibrio parahaemolyticus. Indian Journal of Veterinary and Animal Sciences Research 50(6):53–68
Baffone W, Tarsi R, Pane L et al (2006) Detection of free-living and plankton-bound vibrios in coastal waters of the Adriatic Sea (Italy) and study of their pathogenicity-associated properties. Environ Microbiol 8:1299–1305. https://doi.org/10.1111/j.1462-2920.2006.01011.x
Bauer A, Rørvik LM (2007) A novel multiplex PCR for the identification of Vibrio parahaemolyticus, Vibrio cholerae and Vibrio vulnificus. Lett Appl Microbiol 45:371–375. https://doi.org/10.1111/j.1472-765X.2007.02195.x
Bauer AW, Kirby WMM, Sherris JC, Turck M (1966) Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol 45:493–496. https://doi.org/10.1093/ajcp/45.4_ts.493
Bej AK, Patterson DP, Brasher CW et al (1999) Detection of total and hemolysin-producing Vibrio parahaemolyticus in shellfish using multiplex PCR amplification of tl, tdh and trh. J Microbiol Methods 36:215–225. https://doi.org/10.1016/S0167-7012(99)00037-8
Brenner DJ, Krieg NR, Staley JT, Garrity GM (2005) Bergey’s manual® of systematic bacteriology: volume two: the proteobacteria, part A introductory essays. Springer
Broberg CA, Calder TJ, Orth K (2011) Vibrio parahaemolyticus cell biology and pathogenicity determinants. Microbes Infect 13:992–1001. https://doi.org/10.1016/j.micinf.2011.06.013
Caburlotto G, Suffredini E, Toson M et al (2016) Occurrence and molecular characterisation of Vibrio parahaemolyticus in crustaceans commercialised in Venice area, Italy. Int J Food Microbiol 220:39–49. https://doi.org/10.1016/j.ijfoodmicro.2015.12.007
Calder T, de Souza SM, Attah V et al (2014) Structural and regulatory mutations in Vibrio parahaemolyticus type III secretion systems display variable effects on virulence. FEMS Microbiol Lett 361:107–114. https://doi.org/10.1111/1574-6968.12619
Canizalez-Roman A, Flores-Villasenor H, Zazueta-Beltran J et al (2011) Comparative evaluation of a chromogenic agar medium–PCR protocol with a conventional method for isolation of Vibrio parahaemolyticus strains from environmental and clinical samples. Can J Microbiol 57:136–142
Coly I, Gassama Sow A, Seydi M, Martinez-Urtaza J (2013) Vibrio cholerae and Vibrio parahaemolyticus detected in seafood products from Senegal. Foodborne Pathog Dis 10:1050–1058. https://doi.org/10.1089/fpd.2013.1523
Dangtip S, Sirikharin R, Sanguanrut P et al (2015) AP4 method for two-tube nested PCR detection of AHPND isolates of Vibrio parahaemolyticus. Aquaculture Reports 2:158–162. https://doi.org/10.1016/j.aqrep.2015.10.002
Deepanjali A, Kumar HS, Karunasagar I, Karunasagar I (2005) Seasonal variation in abundance of total and pathogenic Vibrio parahaemolyticus bacteria in oysters along the southwest coast of India. Appl Environ Microbiol 71:3575–3580. https://doi.org/10.1128/AEM.71.7.3575-3580.2005
Elhadi N, Yamani LZ, Aljeldah M et al (2022) Serological and antibiotic resistance patterns as well as molecular characterization of Vibrio parahaemolyticus isolated from coastal waters in the eastern province of Saudi Arabia. Journal of Epidemiology and Global Health 12:524–540. https://doi.org/10.1007/s44197-022-00071-3
Elsherif MF, Saad SM, Amin RA (2023) Incidence and antibiotic resistance patterns of Vibrio parahaemolyticus recovered from different types of fish and shrimp. Benha Veterinary Medical Journal 44:1–4. https://doi.org/10.21608/BVMJ.2023.184043.1623
Fadanka S, Minette S, Mowoh N (2022) Preparation of bacteria glycerol stocks
FAO (2013) Report of the FAO/MARD technical workshop on early mortality syndrome (EMS) or acute hepatopancreatic necrosis syndrome (AHPNS) of cultured shrimp (under TCP/VIE/3304). FAO Rome; 2013. FAO Rome
FAO (2022) The state of world fisheries and aquaculture 2022. Towards blue transformation. Food and Agriculture Organization of the United Nations Rome, Italy
Gotoh K, Kodama T, Hiyoshi H et al (2010) Bile acid-induced virulence gene expression of Vibrio parahaemolyticus reveals a novel therapeutic potential for bile acid sequestrants. PLoS ONE 5:e13365. https://doi.org/10.1371/journal.pone.0013365
Gow SP, Waldner CL, Harel J, Boerlin P (2008) Associations between antimicrobial resistance genes in fecal generic Escherichia coli isolates from cow-calf herds in western Canada. Appl Environ Microbiol 74:3658–3666. https://doi.org/10.1128/AEM.02505-07
Ha PTH, Thi QVC, Thuy NP, Luan NT (2023) Multi-antibiotics resistance phenotype of pathogenic Vibrio parahaemolyticus isolated from acute hepatopancreatic necrosis disease in Litopenaeus vannamei farmed in the Mekong Delta. J World Aquaculture Soc. https://doi.org/10.1111/jwas.12945
Hamdan RH, Peng TL, Ong BL, et al (2017) Antibiotics resistance of Vibrio spp. isolated from diseased seabass and tilapia in cage culture. In: International Seminar on Livestock Production and Veterinary Technology. 554–560
Hiyoshi H, Kodama T, Saito K et al (2011) VopV, an F-actin-binding type III secretion effector, is required for Vibrio parahaemolyticus-induced enterotoxicity. Cell Host Microbe 10:401–409. https://doi.org/10.1016/j.chom.2011.08.014
Hu Y, Li F, Zheng Y et al (2020) Isolation, molecular characterization and antibiotic susceptibility pattern of Vibrio parahaemolyticus from aquatic products in the southern Fujian Coast. China Journal of Microbiology and Biotechnology 30:856. https://doi.org/10.4014/jmb.2001.01005
Jiang Y, Yao L, Li F et al (2014) Characterization of antimicrobial resistance of Vibrio parahaemolyticus from cultured sea cucumbers (Apostichopus japonicas). Lett Appl Microbiol 59:147–154. https://doi.org/10.1111/lam.12258
Kim D-H, Rajapaksha L, Gunasekara CWR et al (2021) Phylogenetic relationships and antibiotic resistance of Vibrio parahaemolyticus isolates related to acute hepatopancreatic necrosis disease in Korea. Aquaculture 545:737253. https://doi.org/10.1016/j.aquaculture.2021.737253
Krumperman PH (1983) Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Appl Environ Microbiol 46:165–170. https://doi.org/10.1128/aem.46.1.165-170.1983
Kumar R, Ng TH, Wang H-C (2020) Acute hepatopancreatic necrosis disease in penaeid shrimp. Rev Aquac 12:1867–1880. https://doi.org/10.1111/raq.12414
Kumar V, Roy S, Behera BK et al (2021) Acute hepatopancreatic necrosis disease (AHPND): virulence, pathogenesis and mitigation strategies in shrimp aquaculture. Toxins 13:524
Lee C-T, Chen I-T, Yang Y-T et al (2015) The opportunistic marine pathogen Vibrio parahaemolyticus becomes virulent by acquiring a plasmid that expresses a deadly toxin. Proc Natl Acad Sci 112:10798–10803. https://doi.org/10.1073/pnas.1503129112
Lee L-H, Ab Mutalib N-S, Law JW-F et al (2018) Discovery on antibiotic resistance patterns of Vibrio parahaemolyticus in Selangor reveals carbapenemase producing Vibrio parahaemolyticus in marine and freshwater fish. Front Microbiol 9:2513. https://doi.org/10.3389/fmicb.2018.02513
Letchumanan V, Chan K-G, Lee L-H (2014) Vibrio parahaemolyticus: a review on the pathogenesis, prevalence, and advance molecular identification techniques. Front Microbiol 5:705. https://doi.org/10.3389/fmicb.2014.00705
Li Y, Xie T, Pang R et al (2020) Food-borne Vibrio parahaemolyticus in China: prevalence, antibiotic susceptibility, and genetic characterization. Front Microbiol 11:1670
Luan X-Y, Chen J-X, Zhang X-H et al (2007) Comparison of different primers for rapid detection of Vibrio parahaemolyticus using the polymerase chain reaction. Lett Appl Microbiol 44:242–247. https://doi.org/10.1111/j.1472-765X.2006.02074.x
Mammeri H, Van De Loo M, Poirel L et al (2005) Emergence of plasmid-mediated quinolone resistance in Escherichia coli in Europe. Antimicrob Agents Chemother 49:71–76. https://doi.org/10.1128/aac.49.1.71-76.2005
Muthukrishnan S, Defoirdt T, Ina-Salwany MY et al (2019) Vibrio parahaemolyticus and Vibrio harveyi causing acute hepatopancreatic necrosis disease (AHPND) in Penaeus vannamei (Boone, 1931) isolated from Malaysian shrimp ponds. Aquaculture 511:734227. https://doi.org/10.1016/j.aquaculture.2019.734227
Navaneeth KA, Bhuvaneswari T, Rajan JJS et al (2020) Characterization of Vibrio parahaemolyticus isolates from shrimp farms of southeast coast of India with special reference to acute hepatopancreatic necrosis disease (AHPND) status. Aquaculture 518:734813. https://doi.org/10.1016/j.aquaculture.2019.734813
Noriea NF III, Johnson CN, Griffitt KJ, Grimes DJ (2010) Distribution of type III secretion systems in Vibrio parahaemolyticus from the northern Gulf of Mexico. J Appl Microbiol 109:953–962. https://doi.org/10.1111/j.1365-2672.2010.04722.x
Robert-Pillot A, Copin S, Himber C et al (2014) Occurrence of the three major Vibrio species pathogenic for human in seafood products consumed in France using real-time PCR. Int J Food Microbiol 189:75–81. https://doi.org/10.1016/j.ijfoodmicro.2014.07.014
Ryu AR, Mok JS, Lee DE et al (2019) Occurrence, virulence, and antimicrobial resistance of Vibrio parahaemolyticus isolated from bivalve shellfish farms along the southern coast of Korea. Environ Sci Pollut Res 26:21034–21043. https://doi.org/10.1007/s11356-019-05426-1
Sakazaki R, Tamura K, Kato T et al (1968) Studies on the enteropathogenic, facultatively halophilic bacteria, Vibrio parahaemolyticus III. Enteropathogenicity. Jpn J Med Sci Biol 21:325–331. https://doi.org/10.7883/yoken1952.21.325
Sawabe T, Ogura Y, Matsumura Y et al (2013) Updating the Vibrio clades defined by multilocus sequence phylogeny: proposal of eight new clades, and the description of Vibrio tritonius sp. nov. Frontiers in microbiology 4:414. https://doi.org/10.3389/fmicb.2013.00414
Shinoda S, Matsuoka H, Tsuchie T et al (1991) Purification and characterization of a lecithin-dependent haemolysin from Escherichia coli transformed by a Vibrio parahaemolyticus gene. Microbiology 137:2705–2711. https://doi.org/10.1099/00221287-137-12-2705
Su Y-C, Liu C (2007) Vibrio parahaemolyticus: a concern of seafood safety. Food Microbiol 24:549–558. https://doi.org/10.1016/j.fm.2007.01.005
Sunde M, Norström M (2005) The genetic background for streptomycin resistance in Escherichia coli influences the distribution of MICs. J Antimicrob Chemother 56:87–90. https://doi.org/10.1093/jac/dki150
Tada J, Ohashi T, Nishimura N et al (1992) Detection of the thermostable direct hemolysin gene (tdh) and the thermostable direct hemolysin-related hemolysin gene (trh) of Vibrio parahaemolyticus by polymerase chain reaction. Mol Cell Probes 6:477–487. https://doi.org/10.1016/0890-8508(92)90044-X
Tan CW, Rukayadi Y, Hasan H et al (2020) Prevalence and antibiotic resistance patterns of Vibrio parahaemolyticus isolated from different types of seafood in Selangor, Malaysia. Saudi Journal of Biological Sciences 27:1602–1608. https://doi.org/10.1016/j.sjbs.2020.01.002
Tang KFJ, Bondad-Reantaso MG (2019) Impacts of acute hepatopancreatic necrosis disease on commercial shrimp aquaculture. Rev Sci Tech 38:477–490. https://doi.org/10.20506/rst.38.2.2999
Traorã SG, Bonfoh B, Krabi R et al (2012) Risk of Vibrio transmission linked to the consumption of crustaceans in coastal towns of Côte d’Ivoire. J Food Prot 75:1004–1011. https://doi.org/10.4315/0362-028X.JFP-11-472
Twedt RM, Novelli RE, Spaulding PL, Hall HE (1970) Comparative hemolytic activity of Vibrio parahaemolyticus and related vibrios. Infect Immun 1:394–399. https://doi.org/10.1128/iai.1.4.394-399.1970
Van TTH, Chin J, Chapman T et al (2008) Safety of raw meat and shellfish in Vietnam: an analysis of Escherichia coli isolations for antibiotic resistance and virulence genes. Int J Food Microbiol 124:217–223. https://doi.org/10.1016/j.ijfoodmicro.2008.03.029
Wang R, Huang J, Zhang W et al (2011) Detection and identification of Vibrio parahaemolyticus by multiplex PCR and DNA–DNA hybridization on a microarray. J Genet Genomics 38:129–135. https://doi.org/10.1016/j.jgg.2011.02.002
Xie T, Yu Q, Tang X et al (2020) Prevalence, antibiotic susceptibility and characterization of Vibrio parahaemolyticus isolates in China. FEMS microbiology letters 367:136. https://doi.org/10.1093/femsle/fnaa136
Xu X, Cheng J, Wu Q et al (2016) Prevalence, characterization, and antibiotic susceptibility of Vibrio parahaemolyticus isolated from retail aquatic products in North China. BMC Microbiol 16:1–9. https://doi.org/10.1186/s12866-016-0650-6
Yano Y, Hamano K, Satomi M et al (2014) Prevalence and antimicrobial susceptibility of Vibrio species related to food safety isolated from shrimp cultured at inland ponds in Thailand. Food Control 38:30–36. https://doi.org/10.1016/j.foodcont.2013.09.019
Zhang L, Orth K (2013) Virulence determinants for Vibrio parahaemolyticus infection. Curr Opin Microbiol 16:70–77. https://doi.org/10.1016/j.mib.2013.02.002
Zhou H, Liu X, Hu W et al (2022) Prevalence, antimicrobial resistance and genetic characterization of Vibrio parahaemolyticus isolated from retail aquatic products in Nanjing. China Food Research International 162:112026. https://doi.org/10.1016/j.foodres.2022.112026
Acknowledgements
The authors sincerely thank PMMSY-ICAR, NSPAAD Phase-II for the funding support and also grateful to Tamil Nadu Dr. J. Jayalalithaa Fisheries University for the facilities and support to carry out this research.
Author information
Authors and Affiliations
Contributions
Swapnil Ananda Narsale: Investigation, Validation, Visualization, Writing – original draft. Bagthasingh Chrisolite: Conceptualization, Resources, Supervision, Writing – review & editing. Panchavarnam Sivasankar: Conceptualization, Writing – review & editing. Palaniappan Subash: Investigation, Validation, Visualization Mohamed Mansoor: Conceptualization, Formal analysis Muthumariappan Selvamagheswaran: Formal analysis Sourabh Debbarma: Investigation, Validation, Visualization Magesh Kumar P: Formal analysis Sampa Baidya: Formal analysis Rishikesh Kadam: Visualization, Formal analysis
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Handling Editor: Amany Abbass
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Highlights
• Isolates of V. parahaemolyticus possess virulence genes such as tdh, trh, and T3SS1.
• All isolates were found to be negative for AHPND.
• Most isolates were resistant to gentamycin, vancomycin, and erythromycin, with AMR genes such as tet A, blaSHV, aac-3-IIa, and qnrA.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Narsale, S.A., Chrisolite, B., Sivasankar, P. et al. Isolation, characterization, virulence genes, antimicrobial resistant genes, and antibiotic susceptibility pattern of Vibrio parahaemolyticus in relation to AHPND from shrimp farms in coastal districts of Tamil Nadu. Aquacult Int 32, 3835–3851 (2024). https://doi.org/10.1007/s10499-023-01353-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10499-023-01353-8