Abstract
Red tilapia is thought to be the result of mutant-colored female Mozambique tilapias mating with male Nile, blue, or Zanzibar tilapias and cultured widely in Asia and central and South America. However, there is limited information about its genetic diversity and stock structure. In this study, we investigated the genetic variability of red tilapia stocks to provide fundamental knowledge for genetic improvement by molecular-marker-assisted selective breeding programs. Individuals (n = 180) from three stocks (Chinese Taiwan, Israel, and Malaysia) of red tilapia were genotyped based on 14 microsatellite markers. The results showed that all microsatellite loci were detected with high levels of polymorphism, with a mean number of 14.87 ± 3.85 alleles per locus in all stocks. Taiwan and Israel stocks showed higher heterozygosity than did the Malaysia stock. The F-statistic analysis showed that there was no significant genetic differentiation between the Taiwan and Israel stocks (P > 0.05), whereas there was highly significant genetic divergence in the other pairwise stocks (P < 0.01), suggesting that Taiwan and Israel stocks could be regarded as a single genetic group distinct from Malaysia stock. This result was in accordance with the UPGMA dendrogram based on Nei’s genetic distances of three stocks. The analysis of molecular variances (AMOVAs) revealed highly significant genetic variation among three stocks (P < 0.01) and accounted for 8.68% of the total variance. The results reported above were confirmed by Bayesian analysis in genetic structure simulation, which indicated a distinct genetic difference between Taiwan and Israel stocks compared with Malaysia stock.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Tilapia (Oreochromis spp.), which is the common name of a group of cichlid fish that are native to tropical Africa, consists of Oreochromis, the mouthbrooding genera Sarotherodon, and the substrate-spawning Tilapia (Trewavas 1983). According to data reported by the Food and Agriculture Organization of the United Nations (FAO 2013), the annual aquaculture production of tilapia has continued to show strong growth, increasing almost three times from 1.33 million tons in 2003 to 3.90 million tons in 2013. Currently, tilapia is the second most important fish species in global aquaculture after carp, with China being the largest supplier of tilapia in the world. Red tilapia hybrids were produced from local crossbreeding between rare mutant-colored (reddish-orange) female Mozambique tilapia (O. mossambicus) and male Nile (O. niloticus), blue (O. aureus), or Zanzibar tilapia (O. urolepis hornorum) (Galman and Avtalion 1983; Wohlfarth et al. 1990; Sandeep et al. 2012). Red tilapia has many positive characteristics for commercial culture including pure red body color, porgy shape, no black coelarium and off-flavor, fast growth, high feed conversion rate (FCR), high dressing-out percentage, euryhalinity, and high adaptability to different environments (Romana-Eguia and Eguia 1999; Chen and Dai 2003).
Microsatellite DNA markers are distributed evenly throughout the genome at high abundance, exhibiting features of high levels of polymorphism, co-dominance inheritance, and ease for PCR analysis, which were widespread used in studies on genetic diversity and population structure of fish species and even improving stocks through marker-assisted selection (Liu and Cordes 2004; Zhao et al. 2011). Moreover, microsatellites have been used to differentiate between closely related populations of the same Oreochromis species and numerous primers corresponding to microsatellites have been developed for this purpose in tilapia (Kocher et al. 1998). Thus, microsatellites represent effective tools for the genetic study of tilapia. Zhang et al. (2010) analyzed the genetic diversity of six tilapia populations and the genetic relationship between Nile tilapia and Blue tilapia based on 20 microsatellites. In an analysis of GIFT (strain Nile tilapia), Li et al. (2009) showed high levels of genetic diversity in the GIFT stock and identified several microsatellite loci associated with body weight in females and one locus associated with body shape in males. However, basic information on the genetic diversity of red tilapia is scarce and the genetic ancestries are not well documented. In this study, we investigated the genetic diversity and genetic structure of three red tilapia stocks (Chinese Taiwan, Israel, and Malaysia) to provide general information that may facilitate genetic improvement by molecular-marker-assisted selective breeding programs.
Materials and methods
Sampling and DNA extraction
In this study, the stock of Malaysia red tilapia (designated MY) was introduced from Malaysia in 2009, and the stocks of Taiwan (China) red tilapia (designated TW) and Israel (designated IL) red tilapia were transferred from Fujian Province, China, in 2014 by the Freshwater Fisheries Research Center (FFRC) of the Chinese Academy of Fishery Sciences (Yang et al. 2015). All stocks were domesticated and bred at a local experimental aquaculture farm in Wuxi City (Jiangsu Province, China).
Fin clips (approximately 10 × 10 mm2) from 180 red tilapia individuals (60 individuals from each stock) were collected in 2014 and preserved in 75% ethanol. Genomic DNA was extracted using a standard phenol-chloroform method. The concentration of DNA in each sample was adjusted to 20 ng/μl and arrayed into 96-well PCR plates, which were stored at −20 °C for later PCR amplification.
Microsatellite selection and genotyping
Fourteen primer sets were selected from the National Center for Biotechnical Information (NCBI) database (Table 1) for amplification and identification of polymorphic products in tilapia based on the success of PCR amplification and heterozygosity levels. Primers were synthesized by Sangon Biotech (Shanghai) Co., Ltd., and the 5′-ends of the forward primers were labeled with a fluorescent dye (HEX or 6FAM). The PCR amplification was performed on an Eppendorf Mastercycler Pro 384 PCR thermocycler (Eppendorf, Germany). Each PCR sample (reaction volume, 25 μl) contained 10 μl Taq PCR MasterMix (2×) (TianGen Biotech Co., Ltd., Beijing) with 1 U Taq Polymerase, 0.5 mM of each dNTP, 20 mM Tris–HCl (pH 8.3), 100 mM KCl, 3 mM MgCl2, 2 μl primer mixture (10 μM), 1 μl genomic DNA (20 ng/μl), and 12 μl DNase/RNase-free deionized water. PCR amplification was conducted under the following conditions: 5-min pre-denaturation at 94 °C followed by 30 cycles of denaturing 30 s at 94 °C, annealing 30 s at specific annealing temperatures (Table 1), and prolonging 50 s at 72 °C, with a final prolonging at 72 °C for 10 min; subsequently, the reaction products were held at 4 °C for further detection. All individual genotypes were scored after the PCR products were resolved on Applied Biosystems 3730XL Genetic Analyzer (Applied Biosystems, USA) and sized relative to an internal size standard (GeneScan-500 ROX) using GeneMapper Version 3.5 software (Applied Biosystems, USA) (Fig. 1).
Statistical analysis
The number of alleles (N a ), effective number of alleles (N e ), expected and observed heterozygosities (H e and H o , respectively), and polymorphic information content (PIC) were calculated using Cervus v3.0 software (Kalinowski et al. 2007). The Hardy–Weinberg equilibrium (HWE) and linkage disequilibrium were analyzed by Genepop v4.4 software (Rousset 2008) using a Markov chain of 1,000,000 steps and 100,000 dememorization steps. The Shannon diversity index of alleles and Nei’s genetic distances (Nei 1978) were compiled using the Popgene v1.32 software (Yeh et al. 1997). F-statistics (F ST ) calculation and analysis of molecular variances (AMOVAs) were performed by using Arlequin v3.1 software (Excoffier et al. 2005). Phylogenetic trees were constructed using Nei’s genetic distance based on unweighted pair group methods with arithmetic (UPGMA) averages using MEGA v5.1 software (Tamura et al. 2011). Analysis of the genetic structure of populations was performed using STRUCTURE v2.3.3 software (Pritchard et al. 2000) with Bayesian methods. The parameters used in the structure analysis were assumed by an admixture model, with a burn-in of 50,000, with 100,000 Markov chain-Monte Carlo (MCMC) repetitions and five iterations per K (K = 2∼3).
Results
Polymorphisms of microsatellite loci
The characteristics of the microsatellite loci identified in this study are listed in Table 2. All 14 microsatellite loci were found to be polymorphic based on the genotypes of 180 individuals. Only one locus (UNH106) showed a highly significant deviation from Hardy–Weinberg equilibrium (P < 0.01). The number of alleles (N a ) ranged from nine (UNH178) to 22 (UNH880), with an average of 14.87 per locus, and the effective number of alleles (N e ) ranged from 1.95 (UNH176) to 6.10 (UNH985), with an average of 4.43 per locus. The observed/expected heterozygosity (H o /H e ) ranged from 0.43/0.49 (UNH176) to 0.89/0.86 (UNH999), with an average of 0.72/0.75. PIC ranged from 0.46 (UNH176) to 0.85 (UNH999), with an average of 0.72, indicating a high level of polymorphism in all the microsatellite loci (PIC > 0.5).
Genetic diversity among stocks
Data for all parameters of genetic diversity for the three red tilapia stocks are shown in Table 3. Three loci (UNH880, UNH997, and UNH106) in Malaysia stock showed significant divergence from Hardy–Weinberg equilibrium (P < 0.05), whereas the other two stocks did not. Israel stock presented the highest genetic diversity parameters followed by the Taiwan and Malaysia stocks. However, Taiwan stock showed the highest H o followed by Israel and Malaysia stocks, while Israel stock exhibited the highest PIC followed by Taiwan and Malaysia stocks.
Genetic divergence and genetic distance
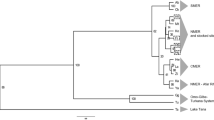
The pairwise F ST estimates presented in Table 4 showed a significant difference between MY and the other two stocks, indicating a moderate genetic differentiation (0.05 < F ST < 0.15) (Balloux and Lugon-Moulin 2002), whereas there was no genetic divergence between TW and IL. Nei’s genetic distance (D A ) between the stocks ranged from 0.023 to 0.396. The UPGMA dendrogram constructed according to Nei’s genetic distance is shown in Fig. 2. TW and IL formed a cluster that did not include MY. The AMOVA revealed genetic variation within and among stocks of 91.32 and 8.68%, respectively, with the variation found to be significant among stocks (P < 0.01) (Table 5).
Genetic structure
The logarithm probabilities Ln P (X/K) associated with different numbers of genetic clusters K, calculated from structure analysis of 180 individuals of red tilapia showed the highest value at K = 2 and the lowest value at K = 3. As shown in Fig. 3, the individuals in Taiwan and Israel stocks were mixed at some levels and were clearly distinct from the individuals in Malaysia stock.
Discussion
Analysis of microsatellite polymorphisms
Most of the microsatellite loci analyzed in this study were highly polymorphic (PIC > 0.5, Botsein et al. 1980) and did not exhibit divergence from Hardy–Weinberg equilibrium, which indicates that these microsatellites can be used efficiently to estimate the genetic variations of red tilapia stocks. All 14 microsatellite markers were detected with a higher degree of polymorphism than that reported by previous studies of the genetic diversity of red tilapia stocks based on microsatellite analysis, such as N a = 10 in a farmed red hybrid tilapia stock (Romana-Egui et al. 2004) and N a = 4.90 in the Florida red tilapia stock (Sandeep et al. 2012). These differences might be due to differences in the study sample (e.g., population size and genetic background), the microsatellite loci analyzed, and detection techniques. Most of the previous studies were conducted using small sample sizes (e.g., 8 individuals, Sandeep et al. 2012), which may lead to bias in the estimation of genetic diversity. Furthermore, the detecting technology with automatic DNA sequencers used in this study might provide higher resolution for genotyping, compared to that achieved by traditional silver-staining method, such as electrophoresed through 8% polyacrylamide gels (Romana-Egui et al. 2004).
Genetic diversity within stocks
The mean expected heterozygosity in three red tilapia stocks (0.62–0.75) was similar to that reported in farmed red hybrid tilapia stocks (0.70) by Romana-Egui et al. (2004) and in Florida red tilapia (0.74) by Sandeep et al. (2012) but much higher than that in the red tilapia (0.35) reported by Zhang et al. (2009). As mentioned previously, these discrepancies may be caused by differences in microsatellite markers and samples used in the studies. However, previous studies have shown significant positive correlations between genetic diversity and population size (Frankham 1996; Ha et al. 2009). Moreover, domestication is often accompanied by a decline in genetic variation due to genetic drift, selection, and inbreeding (Ha et al. 2009). The lower genetic diversity of Malaysia stock might be due to the small effective population size and possible inbreeding over several generations. Therefore, the level of inbreeding, genetic diversity, and broodfish population size should be considered in red tilapia breeding programs, especially in the selective breeding of independent Malaysia stock. Furthermore, our results indicate that appropriate breeding methods, such as cross-breeding, can be used to maintain genetic diversity.
Relationships among red tilapia stocks
The highly polymorphic microsatellite loci used in this study were selected to provide a reliable genetic evaluation of the stocks under investigation (Kalinowski 2002, 2005). The negative F ST value for Taiwan and Israel stocks indicates there is no significant genetic differentiation between these two stocks, which confirms their common origin. In contrast, the moderate differences in the F ST values for the other pairs of stocks indicate differences in the origin of parental populations and/or the occurrence of genetic drift during breeding. Similar results were found in three paddlefish (Polyodon spathula) stocks (Kaczmarczyk et al. 2012). Thorp (1982) reported genetic distances ranging between 0.03 and 0.20 in a single stock. Based on this observation, we infer that the genetic distance between Taiwan and Israel stocks is very small and typical for closely related populations, while the distance between one of these two stocks and Malaysia stock is large and typical for genetically disparate populations. This hypothesis is consistent with the UPGMA dendrogram constructed based on Nei’s genetic distance. The scale of the genetic distances between the stocks suggests that Taiwan stock is most similar to Israel stock, while the genetic differences between Taiwan and Malaysia stock reflect the substantial genetic distance between these two stocks.
AMOVA revealed a highly significant genetic variation among the three stocks (P < 0.01). Based on the correlation of genetic differentiation and gene flow (Nei 1987), it can be speculated that the divergence between Malaysia stock and the other two stocks is due to long-term geographic separation and limited natural gene flow. However, it is also possible that artificial gene flow might have contributed to the lack of divergence between Taiwan and Israel stocks because the two stocks had been farmed together in Fujian Province for several years before introduction into the FFRC. These results were confirmed by Bayesian analysis in genetic structure simulations, which indicate that Taiwan and Israel stocks are obviously different from Malaysia stock.
Conclusions
The three red tilapia stocks contained a high level of genetic diversity revealed by microsatellite marker analysis. However, the genetic variation in Taiwan and Israel stock was significantly higher than that in Malaysia stock. UPGMA dendrogram and genetic structure analysis showed that Taiwan and Israel stock can be considered as a single genetic group, which is separated from Malaysia stock. The high genetic variation in red tilapia stocks provides the basis of genetic improvement by molecular-marker-assisted selective breeding.
TW Taiwanese red tilapia; IL Israel red tilapia; MY Malaysia red tilapia; N a Number of alleles; N e Effective number of alleles; H o Observed heterozygosity; H e Expected heterozygosity; PIC Polymorphic information content; HWE Hardy–Weinberg Equilibrium; F ST F-statistics; AMOVA Analysis of molecular variances; UPGMA Un-weighted pair group methods with arithmetic; MCMC Markov Chain-Monte-Carlo
References
Balloux F, Lugon-Moulin N (2002) The estimation of population differentiation with microsatellite markers. Mol Ecol 11(22):155–165
Botsein D, White RL, Skolnick M, Davis RW (1980) Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet 32:314–331
Chen YS, Dai CJ (2003) Evaluation onnutritional components of the muscle in the Oreochromis niloticus×O.mossambicus. Journal of Fujian Normal University (natural science) 19(4): 62-66Cornuet JM, Luikart G (1996) description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics 144:2001–2014
Excoffier L, Laval G, Schneider S (2005) Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol. Bioinform. Online 1:47–50
FAO (2013) Food and Agriculture Organization of the United Nations (FAO). Fisheries and aquaculture department, Global Aquaculture Production 1950–2013
Frankham R (1996) Relationship of genetic variation to population size in wildlife. Conserv Biol 10:1500–1508
Galman OR, Avtalion RR (1983) A preliminary investigation of the characteristics of red tilapias from the Philippines and Taiwan. In: Fishelson, L, Yaron, Z (comps), proceedings of the international symposium on tilapia in aquaculture 8-13
Ha HP, Nguyen TTT, Poompuang S, Na-Nakorn U (2009) Microsatellites revealed no genetic differentiation between hatchery and contemporary wildpopulations of striped catfish, Pangasianodon hypophthalmus (Sauvage 1878) in Vietnam. Aquaculture 291:154–160
Kaczmarczyk D, Luczynski M, Brzuzan P (2012) Genetic variation in three paddlefish (Polyodon spathula Walbaum) stocks based on microsatellite DNA analysis. Czech J Anim Sci 57(8):345–352
Kalinowski ST (2002) How many alleles per locus should be used to estimate genetic distance? Heredity 88(1):62–65
Kalinowski ST (2005) Dopolymorphic loci require large sample sizes to estimate genetic distance? Heredity 94(1):33–36
Kalinowski ST, Taper ML, Marshall TC (2007) Revising how the computer program cervus accommodates genotyping error increases success in paternity assignment. Mol Ecol 16:1099–1106
Kocher TD, Lee WJ, Sobolewaka H, Penman D, Mcandrew B (1998) A genetic linkage map of a cichlid fish, the tilapia (Oreochromis niloticus). Genetics 148:1225–1232
Li JL, Tang YK, Chen WH, Yu JH, Dong ZJ, Xu P (2009) Association analysis of microsatellite DNA markers with body weight and body shape in GIFT. J Fish Sci China 16(6):824–832
Liu ZJ, Cordes JF (2004) DNA marker technologies and their applications in aquaculture genetics. Aquaculture 238(1–4):1–37
Nei M (1978) Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89(3):583–590
Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York, pp 159–164
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multi-locus data. Genetics 155:945–959
Romana-Eguia MRR, Eguia RV (1999) Growth of five Asian red tilapia strains in saline environments. Aquaculture 173:161–170
Romana-Egui MRR, Ikeda M, Basiao ZU, Taniguchi N (2004) Genetic diversity in farmed Asian Nile and red hybrid tilapia stocks evaluated from microsatellite and mitochondrial DNA analysis. Aquaculture 236:131–150
Rousset F (2008) GENEPOP’007: a complete re-implementation of the GENEPOP software for Windows and Linux. Mol Ecol Resour 8:103–106
Sandeep M, Sun F, Liu F, Li J, David PB, Yue GH (2012) Novel polymorphic microsatellites from Florida red tilapia and cross-species amplification in Mozambique and Nile tilapia. J Genet 91:e97–e99
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetic analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28(10):2731–2739
Thorp JP (1982) The molecular dock hypothesis: biochemical evolution, genetic differentiation, and systematic. Annu Rev Ecol Syst 13:139–168
Trewavas E (1983) Tilapiine fishes of the genera Sarotherodon, Oreochromis and Danakilia. British Museum (Natural History), London, England
Wohlfarth GW, Rothbard S, Hulata G, Szweigman D (1990) Inheritance of red body coloration in Taiwanese tilapias and in Oreochromis mossambicus. Aquaculture 84:219–234
Yeh FC, Yang RC, Boyle T, Ye ZH, Mao JX (1997) POPGENE, the user-friendly shareware for population genetic analysis. Molecular biology and biotechnology centre. University of Alberta, Canada
Yang H, Zhu WB, Dong ZJ, L F, Gong CP, Liu N, Yuan XH (2015) Morphological variation analysis of three populations of red tilapia. J Shanghai Ocean Univ 24(5):678–684
Zhao MZ, Li Q, Qin Q, Cai YX, Xu ZQ, Wang MH, Chen XH, Ge JC, Bian WJ (2011) Genetic diversity based on microsatellite markers in five channel catfish (Ictalurus punctatus) populations. J Aquac 32(1):24–30
Zhang T, Lu MX, Ye X, Quan YC, Gao FY, Huang ZH, Bai JJ (2009) The genetic structure of four populations of Oreochromis aureus by microsatellite DNA analysis. Acta Hydrobiol Sin (in Chinese) 33:498–508
Zhang YD, Gan X, Tang ZS, Chen Z, Su XH, Tang ZY, Li LP, Lin Y (2010) Analysis of genetic diversity in six tilapia populations. J Northwest A & F Univ (Nat Sci Ed) 38(10):58–66
Acknowledgements
This work was supported by the National Fund for Basic R&D of Non-Profit Research Institutes of China [grant number 2015JBFM07].
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Zhu, W., Yang, H., Yuan, X. et al. High genetic diversity and differentiation in three red tilapia stocks revealed by microsatellite DNA marker analysis. Aquacult Int 25, 1997–2006 (2017). https://doi.org/10.1007/s10499-017-0150-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10499-017-0150-1