Abstract
Treatment of advanced BRAFV600-mutant melanoma using BRAF inhibitors (BRAFi) eventually leads to drug resistance and selects for highly metastatic tumor cells. We compared the most differentially dysregulated miRNA expression profiles of vemurafenib-resistant and highly-metastatic melanoma cell lines obtained from GEO DataSets. We discovered miR-152-5p was a potential regulator mediating melanoma drug resistance and metastasis. Functionally, knockdown of miR-152-5p significantly compromised the metastatic ability of BRAFi-resistant melanoma cells and overexpression of miR-152-5p promoted the formation of slow-cycling phenotype. Furthermore, we explored the cause of how and why miR-152-5p affected metastasis in depth. Mechanistically, miR-152-5p targeted TXNIP which affected metastasis and BRAFi altered the methylation status of MIR152 promoter. Our study highlights the crucial role of miR-152-5p on melanoma metastasis after BRAFi treatment and holds significant implying that discontinuous dosing strategy may improve the benefit of advanced BRAFV600-mutant melanoma patients.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Melanoma is one of the most aggressive cancers with high degree of heterogeneity and plasticity [1]. Genetically, melanoma harbors the highest mutation load of any cancer, of which BRAF, NF1, CDKN2A, NRAS and TP53 are the most prevalent significantly mutated genes [2, 3]. Phenotypically, melanoma cells could switch back and forth between states of proliferation and invasion to drive disease progression [4]. In addition, accumulating data indicate that epigenomes alterations resulting from histone modifications and DNA methylome trigger metastatic dissemination of melanoma cells [5, 6]. The complex entanglement of genetic mutation, phenotypic switching and epigenetic alteration brings about the extremely difficult to treatment and fast deterioration of advanced melanoma with a median survival less than 9 months.
Hot-spot mutation of BRAFV600 is one of the most prevalent genetic alterations in human melanoma (35–50% of melanomas), which hyperactivates the mitogen-activated protein kinase (MAPK) pathway and results in uncontrolled proliferation, inhibition of apoptosis, immune evasion and disease progression of melanoma [7]. Hence, selective inhibitors of mutated BRAF (BRAFi), including vemurafenib and dabrafenib, have been developed and have improved the progression-free and overall survival of melanoma patients with BRAFV600 mutations [8]. Despite achieving observably clinical responses, acquired drug resistance frequently emerges after 2 to 18 months [9]. To a more severity of consequence, study has found that BRAFi induced resistance selected for highly malignant brain and lung-metastasizing melanoma cells [10]. O'Connell et al. found that melanoma patients treated with the BRAFi vemurafenib could give rise to a subpopulation of highly invasive cells by activating Wnt5A/ROR2 axis [11]. Caramel et al. confirmed that development of BRAFi induced resistance was associated with an EMT phenotype [12]. All of these studies highlighted the fact that metastatic progression may be linked to BRAFi induced resistance.
MicroRNAs (miRNA) are a class of small, non-protein-coding RNAs that play important roles in virtually all biological pathways in mammals and other multicellular organisms [13]. On one hand, some miRNAs contribute to BRAFi induced resistance in melanoma [14,15,16]. On the other hand, specific miRNAs have been identified as promoters or suppressors of metastatic progression through regulation of cancer phenotype or metastatic gene pathways [17,18,19,20,21,22]. Here we explored the potential miRNAs that have both dysregulated in BRAFi induced resistance and metastasis in advanced melanoma. We provide strong evidence that miR-152-5p is a critical link between BRAFi induced resistance and metastasis. These findings together have important therapeutic and diagnostic implications.
Materials and methods
Primary tissue samples
Tissue samples from four patients with advanced cutaneous melanoma were acquired through the department of pathology with patients’ consents and the approval of the Research Ethics Committee of the First Affiliated Hospital of China Medical University. All tumor tissue samples were surgically removed and stored in liquid nitrogen until use. The clinical information of the four patients was listed in Table 1.
Cell culture
Human melanoma cell lines (A375 and SK-MEL-28) were purchased from the Committee on Type Culture Collection of Chinese Academy of Sciences (Shanghai, China). A375 cell line was grown in DMEM medium (Gibco) supplemented with 10% heat-inactivated fetal bovine serum (FBS, Gibco) and 1 mM sodium pyruvate (Invitrogen). SK-MEL-28 cell line was cultured in EMEM medium (Gibco) supplemented with 10% heat-inactivated FBS. Cells were maintained in a 37 ℃ and 5% CO2 culture environment. BRAFi resistant melanoma cell lines (A375R and SK-MEL-28R) were established from their parental cell lines by gradually increasing concentration vemurafenib (Selleck) and maintained in a final concentration of 1 µM vemurafenib.
RNA isolation and real-time quantitative PCR
Total RNA was extracted using Trizol reagent (Invitrogen) and treated with DNase I (Invitrogen). The miRNAs expression was examined by All-in-One™ miRNA qRT-PCR Detection Kit (GeneCopoeia™) according to manufacturer’s instructions. The mRNAs expression was reverse-transcribed and quantified by PrimeScript™ RT reagent Kit (Takara) and SYBR™ Green PCR Master Mix (Applied Biosystems) following the instructions of manufacturer. U6 or GAPDH was used as an endogenous control. Expression of miRNA and protein were normalized to U6 and GAPDH, respectively. Primers were listed in Supplementary Table S1.
Wound healing assay
3 × 105 cells were seeded into 6-well plates. When the cells reached 100% confluency, cells were transfected with 100 nM miR-152-5p antagomir/agomir (GenePharma) or/and 1 µg pcDNA/pcDNA-TXNIP and scratch was made in the plate using a pipette tip. Images were collected at 0 h and 24 h for A375R/SK-MEL-28R and at 0 h and 8 h for A375/SK-MEL-28 under inverted microscope.
Cell invasion assay
Matrigel assay was performed using Transwell chambers (8 µm pore size, Millipore) that were pre-coated with 200 ng/mL Matrigel (BD Biosciences). Cells were pre-treated with 100 nM miR-152-5p antagomir/agomir or/and 1 µg pcDNA/pcDNA-TXNIP and then 5 × 104 cells were seeded into the upper chamber serum-free medium and the bottom chamber contained medium with 10% FBS. Following 24 h incubation, the invasive cells invading into the bottom chamber were fixed cells in methanol and stained with crystal violet.
Cell cycle analysis
To synchronize the cell cycle, cells were serum-starved for 3 days and subsequently stimulated with 10% heat-inactivated FBS. After 24 h, cells were collected and treated with propidium staining (Cell Cycle and Apoptosis Analysis Kit, Beyotime) according to manufacturer’s instructions.
Western blot
Cell lines were lysed using RIPA protein extraction reagent (Beyotime) supplemented with a protease inhibitor cocktail and PMSF (Roche). After quantification (BCA protein assay, Beyotime), protein extract was electrophoresed on a 10% SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred onto 0.22 µm nitrocellulose membrane (Sigma). The membranes were subsequently blocked with 5% skim milk for 30 min and incubated overnight at 4 ℃ with primary antibodies against JARID1B (3273, CST), SOX10 (ab155279, Abcam), Vimentin (5741, CST), E-cadherin (3195, CST), Fibronectin (ab2413, Abcam), TXNIP (ab188865, Abcam) and GAPDH (2118, CST). Then membranes were washed and incubated with secondary antibody goat anti-rabbit IgG H&L (HRP) (ab6721). After washing, membranes were stained by ECL chromogenic substrate and visualized Bio-Rad ChemiDoc XRS. All the WB gel bands were listed in Supplementary Material. The western blot bands were quantified by Quantity One software (Bio-Rad ChemiDoc XRS). GAPDH was used as a control.
Immunohistochemistry staining
4 µm-thick sections were deparaffinized, rehydrated in graded alcohols and antigens were retrieved in citrate buffer. After inhibition of endogenous peroxidase activity with 3% H2O2 for 30 min, Ultra-Vision Protein Block (BD) was applied to block non-specific background staining. The tissue sections were incubated with an anti-TXNIP antibody (ab188865, Abcam) overnight at 4 ℃. After washing thrice with PBS, the sections were incubated with horseradish peroxidase-conjugated (HRP) goat anti-rabbit IgG for 30 min, followed by reaction with diaminobenzidine (DAB) and counterstaining with Mayer’s hematoxylin.
Luciferase reporter assay
A375 cells were transfected with 100 nM miR-152-5p agomir and 40 ng of pMIR-Reporter vector (Promega) containing the wild-type and mutated 3′-UTR of TXNIP. Luciferase activity was detected by Dual-Luciferase Reporter Assay System (Promega) after 48 h of transfection. Renilla luciferase activity was normalized against Firefly luciferase activity.
Methylation-specific PCR (MSP)
Genomic DNA was extracted from A375, A375R, SK-MEL-28 and SK-MEL-28R cell lines using the DNeasy Blood and Tissue Kit (Qiagen). Bisulfite conversion of DNA was performed using the EpiTect Bisulfite Kit (Qiagen). MSP was performed using the EpiTect MSP Kit (Qiagen) with the following cycling conditions: 95 °C for 10 min; 35 cycles of 94 °C for 15 s, 52 °C for 30 s, and 72 °C for 30 s; final extension at 72 °C for 10 min. EpiTect PCR Control DNA Set (Qiagen) was used as control. MSP primers are listed in Supplementary Table S1. The MSP products were electrophoresed in 1.5% agarose gel and photographed by BIO-RAD ChemiDoc XRS.
Statistical analysis
Statistical analysis was carried out with R version 3.6.0 or GraphPad Prism software 7.0. All results are shown as mean ± SD of three independent experiments. The significance of differences between two groups was performed with Student’s t-test. In case of multiple groups comparisons, one-way ANOVA followed by Dunnett’s test was applied. P ≤ 0.05 was considered statistically significant.
Results
The miRNA profiles of BRAFi induced resistance and metastasis in BRAF mutated melanoma cells
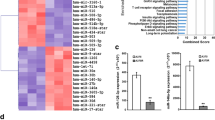
In an attempt to explore potential miRNAs that link BRAFi induced resistance and metastasis, we obtained 2 datasets from GEO DataSets, GSE107576 (miRNA profiles of parental and VMF-resistant melanoma cell lines, Table S2) and GSE77090 (microRNAs involved in malignant melanoma progression, Table S3) (Fig. 1a, b). Next, we selected out 7 common miRNAs (Fig. 1c), which were notably dysregulated both in BRAFi-resistant and highly metastatic cell lines with LogFC ≥ 0.5 and P value ≤ 0.05. In Fig. 1d, we identified 5 miRNAs were positively correlated, while 2 miRNAs were negatively correlated in BRAFi resistant and metastatic cell lines.
miRNA profiles of BRAFi induced resistance and metastasis in BRAF mutated melanoma cells. a Heatmap of the most differentially expressed miRNAs in vemurafenib resistant A375 (A375R) cells compared to parental A375 cells (GSE107576), P value ≤ 0.05 and Log(Fold Change) ≥ 0.5. b Heatmap of the most differentially expressed miRNAs in highly metastatic MA-1, MC-1, MA-2 and MC-2 cells compared to parental A375 cells (GSE77090), P value ≤ 0.05 and Log(Fold Change) ≥ 0.5. c 7 miRNAs were significantly differentially expressed both in GSE107576 and GSE77090. d Presentation of the 7 differentially expressed miRNAs. e Relative expression of the 7 miRNAs in primary tumor sites before and after BRAFi treatment by qRT-PCR. f Expression of the 7 miRNAs in metastatic tumor sites before and after BRAFi treatment by qRT-PCR. g qRT-PCR assays for miR-152-5p expression in vemurafenib resistant A375/SK-MEL-28 (A375R/SK-MEL-28R) cells and their parental cell lines. Error bars represent the SD, ***P < 0.001
To confirm whether BRAFi treatment modulated these miRNAs to impact on metastasis, we used qRT-PCR to detect the relative expression of these miRNAs from four paired advanced melanoma patients before- and post-BRAFi treatment. As shown in Fig. 1e and f, miR-204-5p was upregulated in primary tumor sites while downregulated in metastatic tumor sites upon BRAFi treatment. The expression of miR-199b-5p was downregulated in primary tumor sites while upregulated in metastatic tumor sites upon BRAFi treatment. The expression of miR-455-5p and miR-152-5p were all upregulated upon BRAFi treatment in primary tumor sites and metastatic tumor sites. After references searching, the expression of miR-204-5p was consistent with previous reports, which was increased upon BRAFi treatment and served as tumor suppressor in melanoma [14, 23]. miR-455-5p promoted melanoma metastasis through inhibition of the tumor suppressor gene CPEB1[24]. miR-199b-5p/VEGF axis was responsible for acquisition of a pro-angiogenic status in BRAFi resistant melanoma cells and associated with invasive and aggressive phenotype in cutaneous melanoma [16, 25]. Therefore, we focused on miR-152-5p that hasn’t been fully studied. Then we established BRAFi resistant cell lines and confirmed miR-152-5p was significantly increased in BRAFi resistant cell lines (Fig. 1g).
miR-152-5p modulates metastasis of BRAFi resistant melanoma cells and promotes the formation of slow-cycling phenotype
To determine the effect of miR-152-5p on metastasis of BRAFi resistant cells, BRAFi resistant cells were treated with miR-152-5p antagomir (antagomir-152) (Fig. 2a). The introduction of antagomir-152 into the BRAFi resistant cells led to a significantly increase of cell proliferation (Fig. 2b), inhibition of migration (Fig. 2c) and reduction of invading cells (Fig. 2d) compared with negative control group. Conversely, exogenous expression of miR-152-5p (agomir-152) (Fig. 2e) in BRAFi sensitive cells increased the proportion of G2/M cells (Fig. 2f) and promoted the expression of JARID1B, while decreased the expression of SOX10 (Fig. 2g, h). Moreover, miR-152-5p promoted the epithelial-mesenchymal transition (EMT) of BRAFi sensitive cells (Fig. 2h). In all, miR-152-5p promoted the formation of slow-cycling phenotype with highly invasive property in BRAFi-mutant melanoma cells.
miR-152-5p modulates metastasis of BRAFi resistant melanoma cells and promotes the formation of slow-cycling phenotype. a Knockdown of miR-152-5p by antagomir-152. b miR-152-5p antagomir significantly decreased the proliferation of BRAFi resistant melanoma cells. c, d Wound-healing assay and Transwell assay after the transfection with antagomir-152 or miR-NC in BRAFi resistant melanoma cells, scale bar = 20 µm. e Overexpression of miR-152-5p by agomir-152. f Cell cycle analysis of BRAFi sensitive melanoma cells after the transfection with agomir-152 or miR-NC. g agomir-152 affects the expression of JARID1B and MITF. h agomir-152 promotes the EMT-like transition of BRAFi sensitive melanoma cells. Error bars represent the SD, **P < 0.01, ***P < 0.001.
miR-152-5p targets metastasis suppressor TXNIP
As miRNAs function through inhibition of downstream target genes, we further predicated potential target genes of miR-152-5p from miRTarBase and selected those genes with relevance in cancer based on their associated Gene Ontology terms using DAVID bioinformatics tools. After GO filtering, five target genes were subjected for experimental validation (Fig. 3a). We then examined the expression of these five potential target genes in BRAFi sensitive and resistant cells. As shown in Fig. 3b and c, TXNIP was the most significantly downregulated gene in BRAFi resistant cells. Transfection of BRAFi sensitive cells with agomir-152 resulted in a significant downregulation of TXNIP, which suggested that miR-152-5p targeted TXNIP (Fig. 3d). Furthermore, luciferase activity of the pMIR-Reporter vector containing the wild-type 3′-UTR of TXNIP was vigorously reduced after agomir-152 co-transfection in A375 cells (Fig. 3e, f). In conclusion, these results indicated that TXNIP was a major target of miR-152-5p in BRAF-mutant melanoma.
TXNIP is the direct target of miR-152-5p. a Scheme of targets prediction for miR-152-5p. b TXNIP is significantly downregulated in A375R cells. c Relative expression of TXNIP in melanoma cells via qRT-PCR (up) and western blot (down). d agomir-152 significantly decreases the expression of TXNIP in melanoma cells via qRT-PCR (up) and western blot (down). e Schematic illustration of predicted binding sites of miR-152-5p with the wild-type and mutated 3′-UTR of TXNIP mRNA. f Luciferase activities were detected in luciferase reporters containing wild-type or mutated 3′-UTR of TXNIP mRNA when transfected with agomir-152 or miR-NC. Error bars represent the SD, *P < 0.05, ***P < 0.001
TXNIP regulates migration and invasion of BRAFi resistant melanoma cells
To further verify the functional role of TXNIP on BRAFi induced resistance and metastasis in advanced melanoma, we examined the expression of TXNIP from clinical specimens. TXNIP was significantly downregulated post-BRAFi treatment both in primary and metastatic tumor sites (Fig. 4a) and the metastatic tumors that newly formed post-BRAFi treatment showed a dramatic decrease of TXNIP compared with the metastatic tumors that formed before-BRAFi treatment (Fig. 4b). We next over-expressed TXNIP in BRAFi resistant cell lines (Fig. 4c). The results revealed that TXNIP expression was negatively correlated with cell migration and invasion in BRAFi resistant melanoma cells (Fig. 4d). In the rescue assays, TXNIP overexpression partially reversed the effects of miR-152-5p on cell migration and invasion (Fig. 4e, f). Together, these results above suggested that miR-152-5p promoted cell migration and invasion through suppressing TXNIP.
TXNIP regulates migration and invasion of BRAFi resistant melanoma cells. a TXNIP is downregulated post-BRAFi treatment in primary and metastatic tumor sites. b TXNIP expression was detected by immunohistochemistry staining of two-paired metastatic tumor tissues before- and post-BRAFi treatment. c, d Overexpression of TXNIP reduced the migration and invasion of BRAFi resistant melanoma cells. e, f Overexpression of TXNIP partially abolished the effects of miR-152-5p overexpression on cell migration and invasion. Scale bar = 20 µm. Error bars represent the SD, *P < 0.05, **P < 0.01 ***P < 0.001
miR-152-5p expression is regulated by BRAFi induced demethylation of its promoter
To find the cause of miR-152-5p overexpression in BRAFi resistant melanoma cells, we retrieved the putative promoter region sequences of miR-152-5p. Bioinformatics analysis of CpG island prediction by MethPrimer indicated that the 600–100 bp upstream of MIR152 gene was enriched with CpG islands (Fig. 5a). We assessed the methylation status of the miR-152-5p promoter by methylation-specific PCR (MSP) in BRAFi-resistant and BRAFi-sensitive melanoma cells. We observed complete methylation of the miR-152-5p promoter in BRAFi-sensitive melanoma cells and demethylation of the miR-152-5p promoter in BRAFi-resistant melanoma cells, which suggested the loss of methylation upon BRAFi treatment (Fig. 5b). To further validate the role of CpG methylation in miR-152-5p expression, BRAFi-sensitive melanoma cells were treated with 5′-Aza-2′-deoxycytidine (DAC) for 2 days. Treatment with DAC notably increased miR-152-5p expression (Fig. 5c), while reduced TXNIP expression in both mRNA and protein level (Fig. 5d, f). This further consolidated the negative regulation of miR-152-5p on TXNIP. Interestingly, we observed a gradual downregulation of miR-152-5p and simultaneous restoration of TXNIP when vemurafenib was removed from the maintaining medium of BRAFi-resistant A375 cells, which indicated the spontaneous re-methylation of miR-152-5p promoter without BRAFi (Fig. 5f). Taken together, these results suggested that BRAFi could induce demethylation of miR-152-5p promoter, which led to upregulation of miR-152-5p in BRAFi-mutant melanoma cells.
BRAFi induces demethylation of miR-152-5p promoter. a Schematic illustration of CpG islands in the upstream of miR-152-5p promoter. b Methylation of the miR-152-5p promoter was assessed by methylation-specific PCR in melanoma cells. c–e DAC treatment notably affected the expression of miR-152-5p and TXNIP. f Relative expression of miR-152-5p and TXNIP after vemurafenib removal in A375R cells. Error bars represent the SD, *P < 0.05, **P < 0.01 ***P < 0.001.
Discussion
In this study, we presented evidence that long term BRAFi treatment induced demethylation of miR-152-5p promoter. This epigenetic change led to enhanced expression of miR-152-5p in BRAFi-resistant cells. Functional experiments demonstrated that miR-152-5p was a critical regulator of metastasis in BRAFi-resistant cells and promoted phenotype switching, which resulted in the formation of a slow-growing and much more invasive cell subtype. In depth mechanism exploration revealed that miR-152-5p regulated metastasis of BRAFi-resistant cells by targeting TXNIP. Our results suggested that miR-152-5p closely linked BRAFi-resistance and metastasis in BRAFi-mutant melanoma cells.
Through bioinformatics analysis of miRNA expression profiles in BRAFi-resistant and highly-metastatic melanoma cells and validation of clinical paired specimens from advanced melanoma patients, we identified miR-152-5p as a critical mediator of BRAFi and metastasis. miR-152-5p is a member of miR-148/152 family that shares the same seed sequence of approximately 6–7 nucleotides [26]. miR-152-5p is a tumor suppressor that suppresses the progression of several types of cancer cells [27, 28]. MSP results have indicated that miR-152 expression was significantly correlated with its promoter methylation [29,30,31], suggesting that MIR152 gene could be modulated by epigenetic alterations. In consistent with previous report that BRAFi could induce the demethylation of EGFR in melanomas[12], we identified BRAFi was capable of inducing demethylation of MIR152 gene promoter. To our knowledge, BRAFi induced methylation alteration of miR-152-5p promoter has not previously been reported. Interestingly, miR-152-5p has also been identified to regulate DNA methylation by targeting DNA methyltransferase 1 (DNMT1) [30, 32, 33]. However, forced expression of miR-152-5p didn’t exhibit a negative correlation between miR-152-5p and DNMT1 in A375 cells (Fig. S1A). We tried to decipher which direct factor was responsible for demethylation of miR-152-5p promoter. As the most notable regulator of DNA methylation, the expression of DNMT1 didn’t significantly changed after BRAFi treatment (Fig. S1B). A major challenge in the clinical management of BRAFV600 melanoma was the restoration of glycolysis metabolism upon development of BRAFi induced resistance [34]. This metabolism switching may remarkably influence the cofactors mediating the activity of DNA demethylases, which may be the cause of BRAFi induced demethylation [35].
Unlike most cancer types that comprise a distinct population of cancer stem cells with property to self-renew and resistance to therapy, melanoma cells have the ability to switch phenotypes between proliferative and invasive that implies that most cells would have the potential to adopt a stem cell-like identity [36]. Invading and disseminating cancer cells can undergo EMT, which is associated with a gain of stem cell-like behavior, which implies phenotype switching involving transient EMT–MET (mesenchymal-epithelial transition) processes [37]. The phenotype switching model indicates that melanoma cells undergo transcriptional signature switching in vivo regulated by local microenvironmental conditions [38]. Although melanoma is a heterogeneous tumor of neuroectodermal origin, BRAFi-resistant cells displayed a distinct EMT-like phenotype with spindle-shaped morphology as well as upregulation of EMT markers [12]. The flexible and transient nature of EMT pathways determines reversible switches between proliferative and invasive phenotypes [39]. BRAFi-resistant cells expressed higher levels of genes coding for cancer stem cell markers (JARID1B and Fibronectin) [10].
Our results demonstrated that BRAFi induced phenotype switching was dependent on miR-152-5p that promoted EMT-like transition, which led to a SOX10low and JARID1Bhigh cell phenotypes with highly metastatic ability in advanced melanoma. The transcription factor SOX10 is a key regulator of pigment cell formation during embryonic development and is expressed throughout all stages of melanocyte differentiation [40]. Melanoma of the ‘proliferative’ type expresses high levels of the melanocyte-lineage-specific transcription factor SOX10 [1]. SOX10low melanoma cells displaying slow-cycling phenotype are notoriously resistant to vemurafenib in BRAFV600-mutant melanoma [41]. Depletion of SOX10 sensitized mutant BRAF melanoma cells to BRAFi [42]. JARID1B (KDM5B/PLU-1/RBP2-H1) belongs to a highly-conserved family of jumonji/ARID1 H3K4 demethylases, which are involved in tissue development, cancer, and stem cell biology[43]. JARID1Bhigh cells exhibit stem cell properties that cycle slowly, self-renew and can give rise to JARID1Bhigh progeny [44]. Treatment with vemurafenib led to enrichment of JARID1Bhigh subpopulation [43, 45]. Moreover, investigators identified that JARID1Bhigh subpopulation was of strong invasive properties [46]. Also of note was the slow-cycling phenotype heterogeneously comprising both oncogene-induced senescence (OIS) cells (SOX10low) and cells with stem cell properties (JARID1Bhigh) [41, 44, 47]. In a study of human hematological malignancies, researchers found that senescence-associated reprogramming could promote cancer stemness and tumor aggressiveness[48, 49]. Despite the discovery of pigment cells can overcome OIS by triggering the emergence of tumor-initiating mononucleated stem-like cells from senescent cells, the senescent-stem switching still need more studies[50].
TXNIP (thioredoxin-interacting protein) is a ubiquitous protein inhibitor of thioredoxin, an oxidoreductase that partners with thioredoxin reductase and thioredoxin peroxidase to reduce oxidized proteins and scavenge free radicals [51]. In particular, TXNIP was a proved metastasis suppressor in melanoma [52,53,54]. In the present study, TXNIP was predicted and then identified as a target of miR-152-5p. Moreover, we found overexpression of TXNIP partly suppressed miR-152-5p exerted metastasis-promoting role. Decline of TXNIP enriched GLUT1 at the plasma membrane leading to induction of glycolysis and concomitant acceleration of cell migration [55]. Upregulation of TXNIP shifted cell metabolism away from aerobic glycolysis, which was required for the therapeutic response of BRAFV600E melanoma cells to vemurafenib [56]. We hypothesized that during the process of acquiring BRAFi induced resistance, TXNIP was degraded by increasing miR-152-5p expression. Consequently, glycolysis took the dominant metabolic way and then the BRAFi induced resistance established.
In summary, our study shed light on the molecular link of BRAFi induced resistance and metastasis. miR-152-5p promoter was hypomethylated during the acquisition of BRAFi induced resistance in BRAFV600E melanoma cells. miR-152-5p wield its metastasis-promoting effects by directly targeting TXNIP. The spontaneous inactivation of miR-152-5p/TXNIP axis with BRAFi withdraw implying that discontinuous dosing strategy may not only forestall BRAFi induced resistance, but also inhibit the formation of highly invasive phenotype in advanced BRAFV600-mutant melanoma.
References
Verfaillie A, Imrichova H, Atak ZK, Dewaele M, Rambow F, Hulselmans G et al (2015) Decoding the regulatory landscape of melanoma reveals TEADS as regulators of the invasive cell state. Nat Commun 6:6683
Hayward NK, Wilmott JS, Waddell N, Johansson PA, Field MA, Nones K et al (2017) Whole-genome landscapes of major melanoma subtypes. Nature 545(7653):175–180
Cancer Genome Atlas N (2015) Genomic classification of cutaneous melanoma. Cell 161(7):1681–1696
Widmer DS, Cheng PF, Eichhoff OM, Belloni BC, Zipser MC, Schlegel NC et al (2012) Systematic classification of melanoma cells by phenotype-specific gene expression mapping. Pigment Cell Melanoma Res 25(3):343–353
Richards HW, Medrano EE (2009) Epigenetic marks in melanoma. Pigment Cell Melanoma Res 22(1):14–29
Wouters J, Vizoso M, Martinez-Cardus A, Carmona FJ, Govaere O, Laguna T et al (2017) Comprehensive DNA methylation study identifies novel progression-related and prognostic markers for cutaneous melanoma. BMC Med 15(1):101
Shi H, Hugo W, Kong X, Hong A, Koya RC, Moriceau G et al (2013) Acquired resistance and clonal evolution in melanoma during BRAF inhibitor therapy. Cancer Discov 4(1):80–93
Maxwell R, Garzon-Muvdi T, Lipson EJ, Sharfman WH, Bettegowda C, Redmond KJ et al (2017) BRAF-V600 mutational status affects recurrence patterns of melanoma brain metastasis. Int J Cancer 140(12):2716–2727
Paulitschke V, Berger W, Paulitschke P, Hofstatter E, Knapp B, Dingelmaier-Hovorka R et al (2015) Vemurafenib resistance signature by proteome analysis offers new strategies and rational therapeutic concepts. Mol Cancer Ther 14(3):757–768
Zubrilov I, Sagi-Assif O, Izraely S, Meshel T, Ben-Menahem S, Ginat R et al (2015) Vemurafenib resistance selects for highly malignant brain and lung-metastasizing melanoma cells. Cancer Lett 361(1):86–96
O'Connell MP, Marchbank K, Webster MR, Valiga AA, Kaur A, Vultur A et al (2013) Hypoxia induces phenotypic plasticity and therapy resistance in melanoma via the tyrosine kinase receptors ROR1 and ROR2. Cancer Discov 3(12):1378–1393
Wang J, Huang SK, Marzese DM, Hsu SC, Kawas NP, Chong KK et al (2015) Epigenetic changes of EGFR have an important role in BRAF inhibitor-resistant cutaneous melanomas. J Investig Dermatol 135(2):532–541
Jansson MD, Lund AH (2012) MicroRNA and cancer. Mol Oncol 6(6):590–610
Diaz-Martinez M, Benito-Jardon L, Alonso L, Koetz-Ploch L, Hernando E, Teixido J (2018) miR-204-5p and miR-211-5p contribute to BRAF inhibitor resistance in melanoma. Cancer Res 78(4):1017–1030
Sahoo A, Sahoo SK, Joshi P, Lee B, Perera RJ (2019) MicroRNA-211 loss promotes metabolic vulnerability and BRAF inhibitor sensitivity in melanoma. J Investig Dermatol 139(1):167–176
Fattore L, Ruggiero CF, Pisanu ME, Liguoro D, Cerri A, Costantini S et al (2019) Reprogramming miRNAs global expression orchestrates development of drug resistance in BRAF mutated melanoma. Cell Death Differ 26(7):1267–1282
Pencheva N, Tavazoie SF (2013) Control of metastatic progression by microRNA regulatory networks. Nat Cell Biol 15(6):546–554
Rambow F, Bechadergue A, Luciani F, Gros G, Domingues M, Bonaventure J et al (2016) Regulation of melanoma progression through the TCF4/miR-125b/NEDD9 cascade. J Investig Dermatol 136(6):1229–1237
Boyle GM, Woods SL, Bonazzi VF, Stark MS, Hacker E, Aoude LG et al (2011) Melanoma cell invasiveness is regulated by miR-211 suppression of the BRN2 transcription factor. Pigment Cell Melanoma Res 24(3):525–537
Kappelmann M, Kuphal S, Meister G, Vardimon L, Bosserhoff AK (2013) MicroRNA miR-125b controls melanoma progression by direct regulation of c-Jun protein expression. Oncogene 32(24):2984–2991
Bell RE, Khaled M, Netanely D, Schubert S, Golan T, Buxbaum A et al (2014) Transcription factor/microRNA axis blocks melanoma invasion program by miR-211 targeting NUAK1. J Invest Dermatol 134(2):441–451
Guo W, Wang H, Yang Y, Guo S, Zhang W, Liu Y et al (2017) Down-regulated miR-23a contributes to the metastasis of cutaneous melanoma by promoting autophagy. Theranostics 7(8):2231–2249
Luan W, Qian Y, Ni X, Bu X, Xia Y, Wang J et al (2017) miR-204-5p acts as a tumor suppressor by targeting matrix metalloproteinases-9 and B-cell lymphoma-2 in malignant melanoma. OncoTargets Ther 10:1237–1246
Shoshan E, Mobley AK, Braeuer RR, Kamiya T, Huang L, Vasquez ME et al (2015) Reduced adenosine-to-inosine miR-455-5p editing promotes melanoma growth and metastasis. Nat Cell Biol 17(3):311–321
Babapoor S, Wu R, Kozubek J, Auidi D, Grant-Kels JM, Dadras SS (2017) Identification of microRNAs associated with invasive and aggressive phenotype in cutaneous melanoma by next-generation sequencing. Lab Investig 97(6):636–648
Chen Y, Song YX, Wang ZN (2013) The microRNA-148/152 family: multi-faceted players. Mol Cancer 12(1):43
Ma J, Yao YL, Wang P, Liu YH, Zhao LN, Li ZQ et al (2014) MiR-152 functions as a tumor suppressor in glioblastoma stem cells by targeting Kruppel-like factor 4. Cancer Lett 355(1):85–95
Zheng X, Chopp M, Lu Y, Buller B, Jiang F (2013) MiR-15b and miR-152 reduce glioma cell invasion and angiogenesis via NRP-2 and MMP-3. Cancer Lett 329(2):146–154
Daniunaite K, Dubikaityte M, Gibas P, Bakavicius A, Rimantas Lazutka J, Ulys A et al (2017) Clinical significance of miRNA host gene promoter methylation in prostate cancer. Hum Mol Genet 26(13):2451–2461
Stumpel DJ, Schotte D, Lange-Turenhout EA, Schneider P, Seslija L, de Menezes RX et al (2011) Hypermethylation of specific microRNA genes in MLL-rearranged infant acute lymphoblastic leukemia: major matters at a micro scale. Leukemia 25(3):429–439
Tsuruta T, Kozaki K, Uesugi A, Furuta M, Hirasawa A, Imoto I et al (2011) miR-152 is a tumor suppressor microRNA that is silenced by DNA hypermethylation in endometrial cancer. Cancer Res 71(20):6450–6462
Braconi C, Huang N, Patel T (2010) MicroRNA-dependent regulation of DNA methyltransferase-1 and tumor suppressor gene expression by interleukin-6 in human malignant cholangiocytes. Hepatology 51(3):881–890
Huang J, Wang Y, Guo Y, Sun S (2010) Down-regulated microRNA-152 induces aberrant DNA methylation in hepatitis B virus-related hepatocellular carcinoma by targeting DNA methyltransferase 1. Hepatology 52(1):60–70
Parmenter TJ, Kleinschmidt M, Kinross KM, Bond ST, Li J, Kaadige MR et al (2014) Response of BRAF-mutant melanoma to BRAF inhibition is mediated by a network of transcriptional regulators of glycolysis. Cancer Discov 4(4):423–433
Raffel S, Falcone M, Kneisel N, Hansson J, Wang W, Lutz C et al (2017) BCAT1 restricts alphaKG levels in AML stem cells leading to IDHmut-like DNA hypermethylation. Nature 551(7680):384–388
Hoek KS, Goding CR (2010) Cancer stem cells versus phenotype-switching in melanoma. Pigment Cell Melanoma Res 23(6):746–759
Brabletz T (2012) To differentiate or not — routes towards metastasis. Nat Rev Cancer 12(6):425–436
Hoek KS, Eichhoff OM, Schlegel NC, Dobbeling U, Kobert N, Schaerer L et al (2008) In vivo switching of human melanoma cells between proliferative and invasive states. Can Res 68(3):650–656
Caramel J, Papadogeorgakis E, Hill L, Browne GJ, Richard G, Wierinckx A et al (2013) A switch in the expression of embryonic EMT-inducers drives the development of malignant melanoma. Cancer Cell 24(4):466–480
Shakhova O, Zingg D, Schaefer SM, Hari L, Civenni G, Blunschi J et al (2012) Sox10 promotes the formation and maintenance of giant congenital naevi and melanoma. Nat Cell Biol 14(8):882–890
Zhang G, Herlyn M (2014) Linking SOX10 to a slow-growth resistance phenotype. Cell Res 24(8):906–907
Han S, Ren Y, He W, Liu H, Zhi Z, Zhu X et al (2018) ERK-mediated phosphorylation regulates SOX10 sumoylation and targets expression in mutant BRAF melanoma. Nat Commun 9(1):28
Roesch A, Vultur A, Bogeski I, Wang H, Zimmermann KM, Speicher D et al (2013) Overcoming intrinsic multidrug resistance in melanoma by blocking the mitochondrial respiratory chain of slow-cycling JARID1B high cells. Cancer Cell 23(6):811–825
Held M, Bosenberg M (2010) A role for the JARID1B stem cell marker for continuous melanoma growth. Pigment Cell Melanoma Res 23(4):481–483
Ahn A, Chatterjee A, Eccles MR (2017) The slow cycling phenotype: a growing problem for treatment resistance in melanoma. Mol Cancer Ther 16(6):1002–1009
Perego M, Maurer M, Wang JX, Shaffer S, Müller AC, Parapatics K et al (2017) A slow-cycling subpopulation of melanoma cells with highly invasive properties. Oncogene 37(3):302–312
Haferkamp S, Borst A, Adam C, Becker TM, Motschenbacher S, Windhovel S et al (2013) Vemurafenib induces senescence features in melanoma cells. J Investig Dermatol 133(6):1601–1609
Milanovic M, Fan DNY, Belenki D, Dabritz JHM, Zhao Z, Yu Y et al (2018) Senescence-associated reprogramming promotes cancer stemness. Nature 553(7686):96–100
Dou Z, Berger SL (2018) Senescence elicits stemness: a surprising mechanism for cancer relapse. Cell Metab 27(4):710–711
Leikam C, Hufnagel AL, Otto C, Murphy DJ, Muhling B, Kneitz S et al (2015) In vitro evidence for senescent multinucleated melanocytes as a source for tumor-initiating cells. Cell Death Dis 6:e1711
Muoio DM (2007) TXNIP links redox circuitry to glucose control. Cell Metab 5(6):412–414
Knoll S, Furst K, Kowtharapu B, Schmitz U, Marquardt S, Wolkenhauer O et al (2014) E2F1 induces miR-224/452 expression to drive EMT through TXNIP downregulation. EMBO Rep 15(12):1315–1329
Goldberg SF, Miele ME, Hatta N, Takata M, Paquette-Straub C, Freedman LP et al (2003) Melanoma metastasis suppression by chromosome 6: evidence for a pathway regulated by CRSP3 and TXNIP. Cancer Res 63(2):432–440
Cheng GC, Schulze PC, Lee RT, Sylvan J, Zetter BR, Huang H (2004) Oxidative stress and thioredoxin-interacting protein promote intravasation of melanoma cells. Exp Cell Res 300(2):297–307
Sullivan WJ, Mullen PJ, Schmid EW, Flores A, Momcilovic M, Sharpley MS et al (2018) Extracellular matrix remodeling regulates glucose metabolism through TXNIP destabilization. Cell 175(1):117
Wilde BR, Ayer DE (2015) Interactions between Myc and MondoA transcription factors in metabolism and tumourigenesis. Br J Cancer 113(11):1529–1533
Author information
Authors and Affiliations
Contributions
KL and SJ conceived and designed the experiments; KL, MT, ST, CW and QS performed the experiments; KL, ML, XS and TW analyzed the data; KL and SJ wrote the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors confirm that there is no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Li, K., Tang, M., Tong, S. et al. BRAFi induced demethylation of miR-152-5p regulates phenotype switching by targeting TXNIP in cutaneous melanoma. Apoptosis 25, 179–191 (2020). https://doi.org/10.1007/s10495-019-01586-0
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10495-019-01586-0